-
It is well recognized that various reporters can be used to monitor gene expression and protein subcellular localization. For example, green fluorescent protein (GFP), red fluorescent protein (RFP), yellow fluorescent protein (YFP) and β-glucuronidase (GUS) have been widely used as reporters for gene expression or fusion proteins[1−3]. Although traditional GFP and GUS reporters are commonly used in plants for monitoring gene expression, there are limitations to their use. The fluorescent proteins GFP, RFP and YFP require a microscope for detection, and it is not suitable to observe the development state of the plants. The GUS reporter gene is invasive, since it requires killing the plants, and is not practical for use in the field. Consequently, it is necessary to develop new reporter systems that can be investigated noninvasively for cellular activities. The RFP identified in Discosoma sp. coral (DsRed2) was successfully used as a visual marker for cotton genetic engineering[4].
Betaine is a kind of natural plant product extracted from the amino acid tyrosine. Betalain biosynthesis has been well studied, and only three enzymatic reactions are required to convert tyrosine to betalain. The benzene ring of tyrosine is hydroxylated to produce L-3,4-dihydroxyphenylalanine (L-DOPA) by the catalysis of the P450 oxygenase CYP76AD1 (Fig. 1a), and L-DOPA is oxidized by CYP76AD1 to further generate cyclo-DOPA (Fig. 1a). In addition, L-DOPA-4,5-dioxygenase (DODA) catalyzes the formation of β-carbamic acid from L-DOPA. Subsequently, β-carbamic acid is condensed with cyclo-DOPA to produce betanidin, the process of does not require an enzyme (Fig. 1a)[5]. Furthermore, betalain has a very bright red color and could serve as a reporter for tracking gene expression or visualizing transgenic events. We assume that betalain, which requires no special facility and is visible to the naked eye, could be a more convenient reporter than previous reporters are. The possibility of improving tomato fruit color involves changing the regulation of carotenoids[6], flavonoids[7], anthocyanins[8] and the introduction of other pigment synthesis-related genes via genetic engineering. Components needed for the biosynthesis of the betalain-related genes Cyp76ad1 Bvdoda1 and Cdopa5gt were transferred into tomato, which ultimately resulted in the production of mature tomato fruits that were purplish red[9]. DR5 is an artificially synthesized and highly active auxin-response element[10]. Fortunately, Zhao et al. synthesized the betalain-related genes Cyp76ad1, Bvdoda1 and Cdopa5gt, which were subsequently transferred into Arabidopsis and driven by the DR5 promoter[11]. The results showed that the artificial open reading frame named Ruby is a very effective marker for noninvasively selecting transformation events in both rice and Arabidopsis. Moreover, Ruby can be used to visualize gene expression without any chemical treatments, and it is more useful and effective for analyzing plants in naturally growing fields.
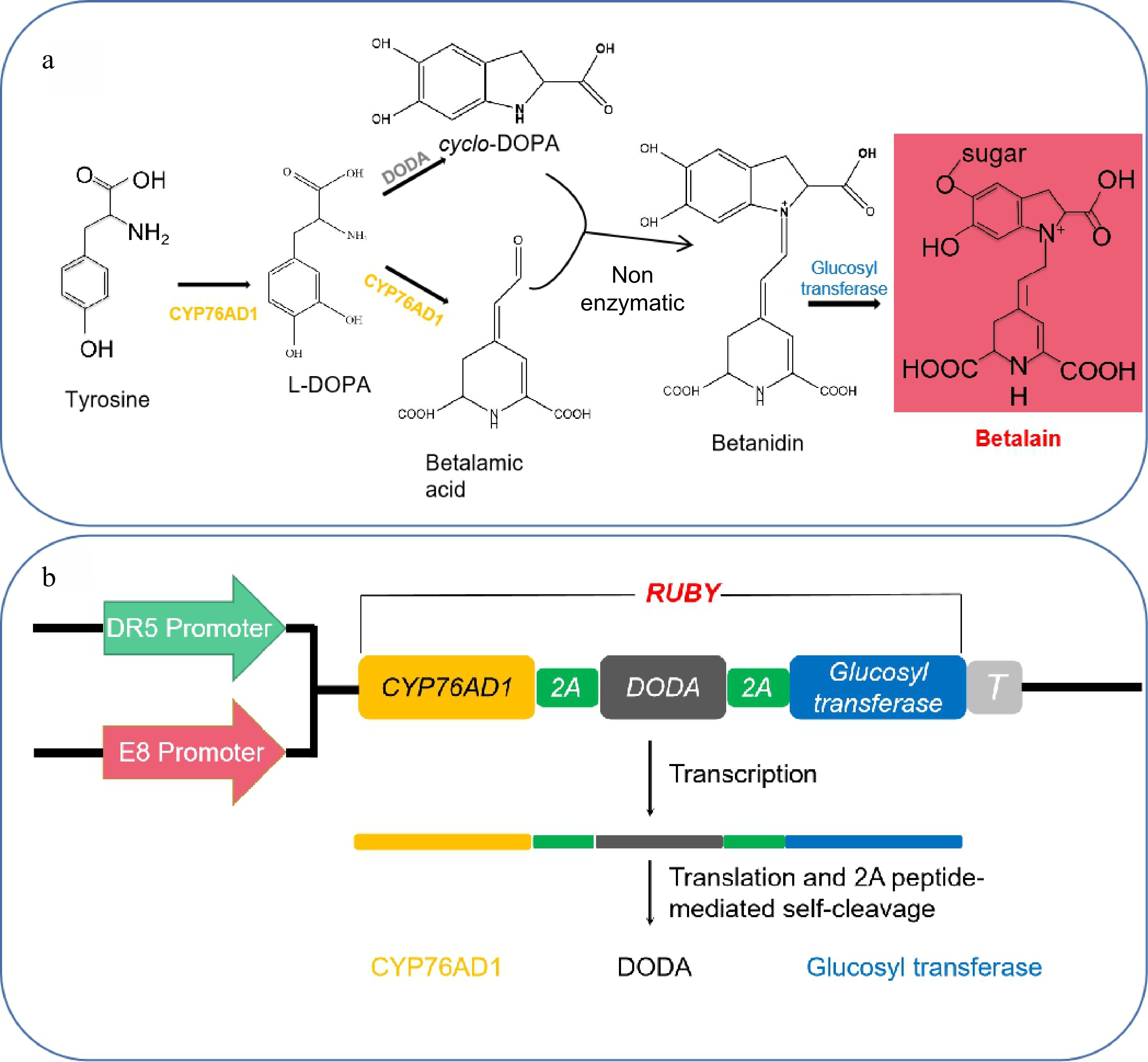
Figure 1.
Components required for betalain biosynthesis. (a) Chemical reactions for converting tyrosine into betalain, according to Zhao et al.[11]. (b) Two promoters, DR5 and E8, were used to drive the expression of the Ruby gene.
Deikman et al. confirmed that whether the E8 gene is specifically expressed during fruit ripening mainly depends on a 409−263 region of the E8 promoter[12]. The −1088 to −863 region of the E8 promoter is the binding region of DNA-binding protein. Knocking out this region significantly reduces the expression of the E8 gene in mature fruits. The results showed that the effective binding of −1088 to −863 to a DNA-binding protein is an essential factor for high expression of the E8 gene during tomato fruit ripening. Furthermore, the −2181 to −1088 region contains an ethylene-responsive factor gene-binding site.
The availability of fruit-specific promoters greatly promotes the ability to improve fruit quality by genetic transformation[13]. As one of the most widely used fruit-specific promoters, the E8 promoter has been applied to transgenic tomato fruits[14−16]. In this research, we clarified whether the E8 promoter can be used to regulate the expression of the Ruby gene in tomato. The original DR5 promoter was replaced by the E8 promoter to construct the E8::Ruby vector. In addition, to identify the auxin content and responses in different tissues of various developmental stages, a DR5 reporter system with the visual marker Ruby was constructed.
-
The plant materials used in this study had red, white or yellow colored fruit and came from the following four cultivars: the 'Moneymaker' cultivar, which is characterized by round red fruit of medium size, and which served as the control cultivar for the experiment; the white fruit bearing cultivar '06883', leaves yellow and fruit pre whitening, and which was breeding material obtained from our laboratory; the yellow fruit bearing cultivar '19458', which is characterized as a cherry tomato with round, yellow fruit, and which was also breeding material from our laboratory; and the cultivar 'Micro-Tom', which is a limited growth type with a production cycle of 4−5 weeks. All the tomato cultivars were grown under a 16 h/8 h light/dark photoperiod at 25−27 °C in the greenhouse. The positive transgenic plants, including E8::Ruby and DR 5::Ruby, were further used for monitoring tomato fruit color and the auxin response at different developmental stages.
Vector construction and plant transformation
-
The 2200 bp sequences of the E8 promoter was amplified by PCR, in which the E8 promoter replaced the DR5 promoter by double enzyme digestion with Sma I and Pst I restriction sites (Takara Bio, USA) to drive the expression of the Ruby gene. Both the DR5::Ruby and E8::Ruby plasmids were transformed into cotyledons of different tomato varieties (red, white and yellow fruit bearing) by Agrobacterium-mediated plant transformation according to a previous protocol. The transformed plants were subjected to PCR analysis. All the primers used are listed in Supplemental Table S1. The DR5::Ruby plasmids were donated by Dr. Zhao (Supplemental Fig. S1).
Identification of transgenic plants
-
Since betalain has a very bright red color, the Ruby gene can serve as a reporter to track gene expression or to visualize transgenic events. The tomato calli, primary transformants (T0) and transgenic seeds of E8::Ruby and DR5::Ruby were easily identified by their red color. In addition, the 2200 bp 5' flanking sequence of the E8 promoter and 2000 bp partial encode sequence of the Ruby gene were used to test the positive transgenic plants. Transgenic plants for DR5::Ruby and E8::Ruby were selected on Murashige and Skoog (MS) media containing kanamycin.
qRT‒PCR and IAA content quantification
-
Total RNA was extracted from the roots, stems, leaves, flowers and fruits of transgenic plants according to the instructions of TRIzol reagent, and then the first-strand cDNA was synthesized according to a cDNA synthesis kit. qRT-PCR was performed to analyze the expression levels of the Ruby gene at different developmental stages of tomato. Three biological replicates were included for each sample. The relative expression levels were analyzed using the 2−ΔΔCᴛ method, with the tomato Actin gene serving as an internal reference. All the primers used for qRT‒PCR analysis are listed in Supplemental Table S1. Endogenous indoleacetic acid (IAA) was extracted from the roots, stems, leaves, flowers and fruits of the transgenic plants. The IAA content was measured via high-performance liquid chromatography (HPLC).
-
Three enzymatic reactions are essential to convert tyrosine into betalain in the biosynthesis of betalain (Fig. 1a). The 2A-linked unit of CYP76AD1, DODA, and glucosyltransferase was named Ruby. It has a bright red color when the reporter system comprises a promoter and Ruby gene, which could serve as a marker for noninvasively selecting transformation events. Two different reporter systems with the fruit-specific promoter E8 and the auxin-responsive synthetic promoter DR5 were used to drive the expression of the Ruby gene (Fig. 1b). The DR5::Ruby plasmids were donated by Dr. Zhao[11]. The 2200 bp 5' band of the E8 promoter was cloned by PCR, in which the E8 promoter replaced the DR5 promoter to drive the expression of the Ruby gene.
RUBY is a very effective marker for noninvasively selecting transformation events in tomato
-
To identify the expression of auxin at different developmental stages, DR5::Ruby was transformed into Micro Tom tomato plants. In addition, to obtain new colored tomato fruits, E8::Ruby was transformed into different tomato varieties (red, white and yellow fruit bearing). As shown in Fig. 2a, the specific primer Hpt was used to screen the transformed plants. The PCR results were consistent with those of the Ruby gene, and the positively transformed calli were red (Fig. 2b). Furthermore, both DR5::Ruby and E8::Ruby clearly identified the transformed tomato calli (Fig. 2b).
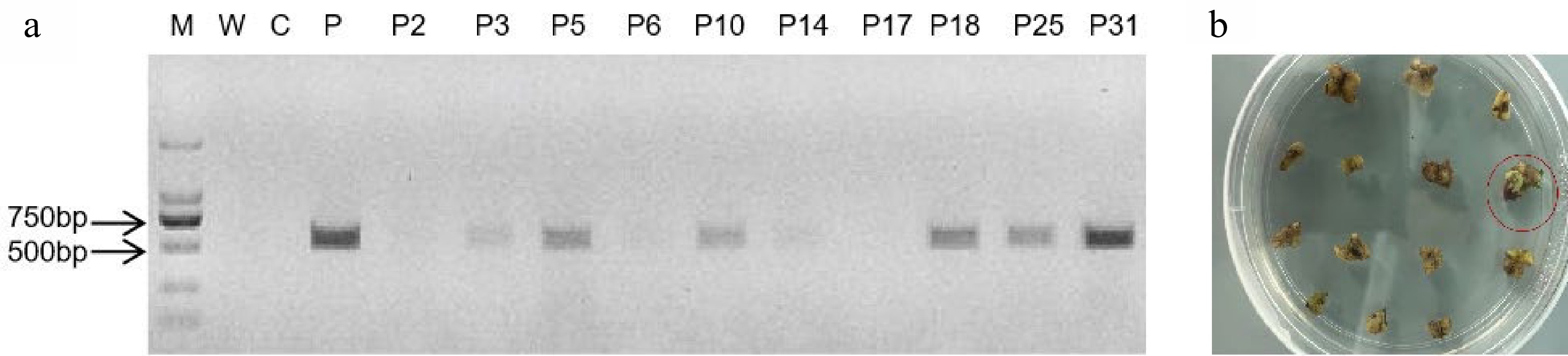
Figure 2.
Ruby was effective for noninvasively selecting transformation events in tomato. (a) The Hpt assay from seven regenerated plantlets; P3, P5, P10, P18, P25 and P31 were positive; P2, P6, P14 and P17 were negative. M, maker; W, water; C, negative nontransgenic control; P, positive plasmid control; (b) The Ruby gene was expressed in tomato calli. The red color was distinguished in positively transformed calli (red) and nontransgenic calli (green).
Tissue-specific activity of DR5 in the different tomato organs
-
DR5::Ruby reporter gene integration was identified by Ruby assay and PCR amplification. The Ruby assay was performed to assess the local auxin-response characteristics of the DR5 system in different tomato organs. The untransformed control seeds (Fig. 3a), stems (Fig. 3b), leaves (Fig. 2b), axillary buds (Fig. 3c) and fruits (Fig. 3d) were no different in their color. However, a pink color could be clearly observed at the apex of the seeds, hooks (Fig. 2a), axillary bud (Fig. 3c) stems, leaves (Fig. 2b) and fruits (Fig. 3d) of the transgenic plants. The above results showed the tissue-specific local accumulation of endogenous auxin in tomato. Hence, the nonuniformity of Ruby activity was consistent with the natural distribution characteristics of native auxin, and the results were also consistent with the qRT‒PCR results (Fig. 3f). In addition, to verify whether the auxin content could be visualized in the DR5::Ruby transformed plants, IAA was quantified in some tissues, and the results were in accord with the depth of color associated with Ruby in different tissues (Fig. 3g). Clearly, the accumulation of endogenous auxin at the immature green stage (IMG) was higher than that at the other stages (Fig. 3d & e). Furthermore, the results also showed that it is practical to use the DR5 system for auxin distribution and response observations in tomato.
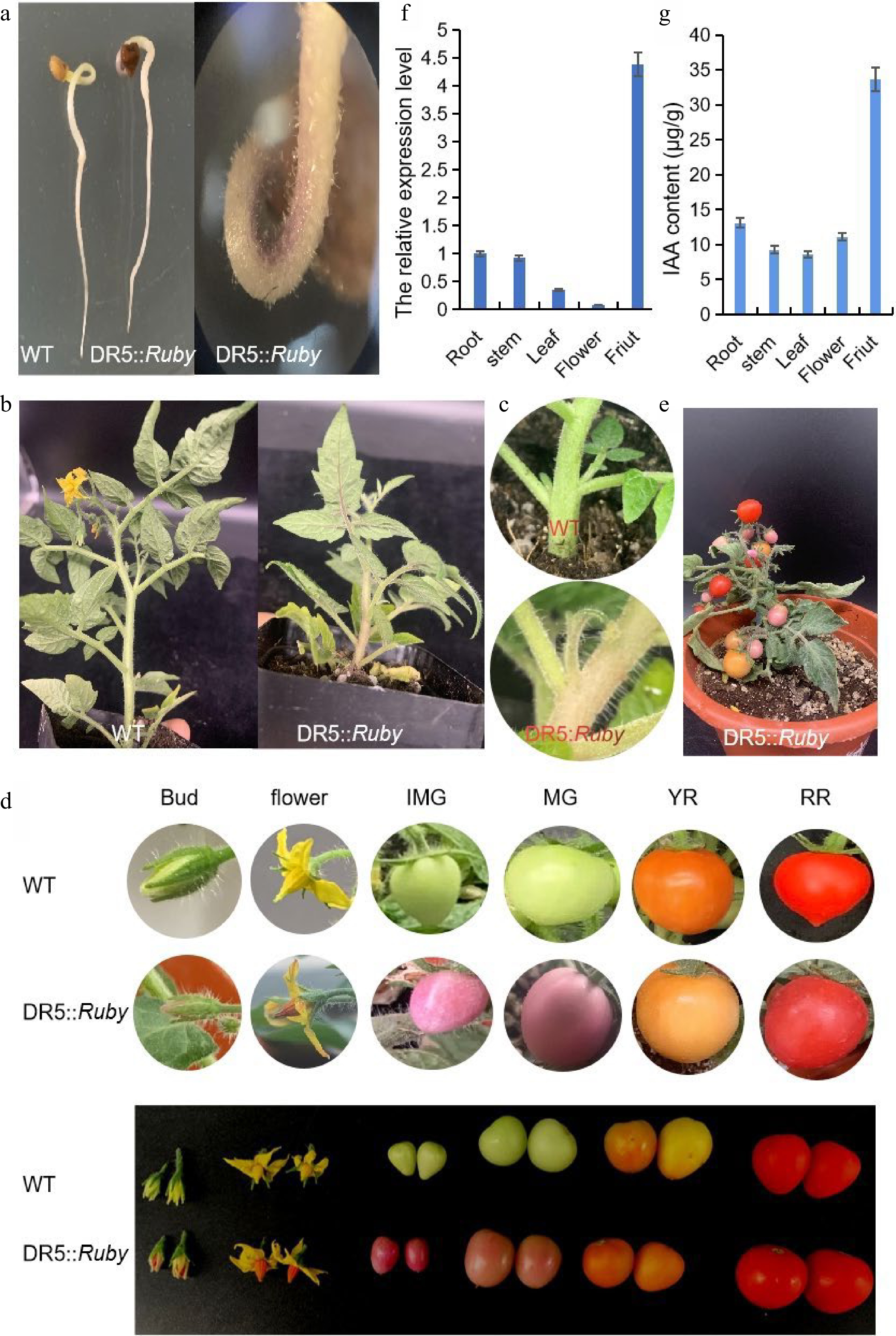
Figure 3.
Characteristics of DR5::Ruby transgenic tomato plants in different tissues. (a) Seeds; (b) Stems and leaves; (c) Axillary buds; (d) Flowers and fruits; (e) Plants; (f) qRT-PCR analysis of DR5 expression in different tomato organs. (g) The IAA content of different tissues. IMG, immature green stage; MG, mature green stage; BR, breaker stage; YR, yellow ripe stage; RR, red ripe stage.
E8::Ruby transformation into different tomato varieties (red, white and yellow)
-
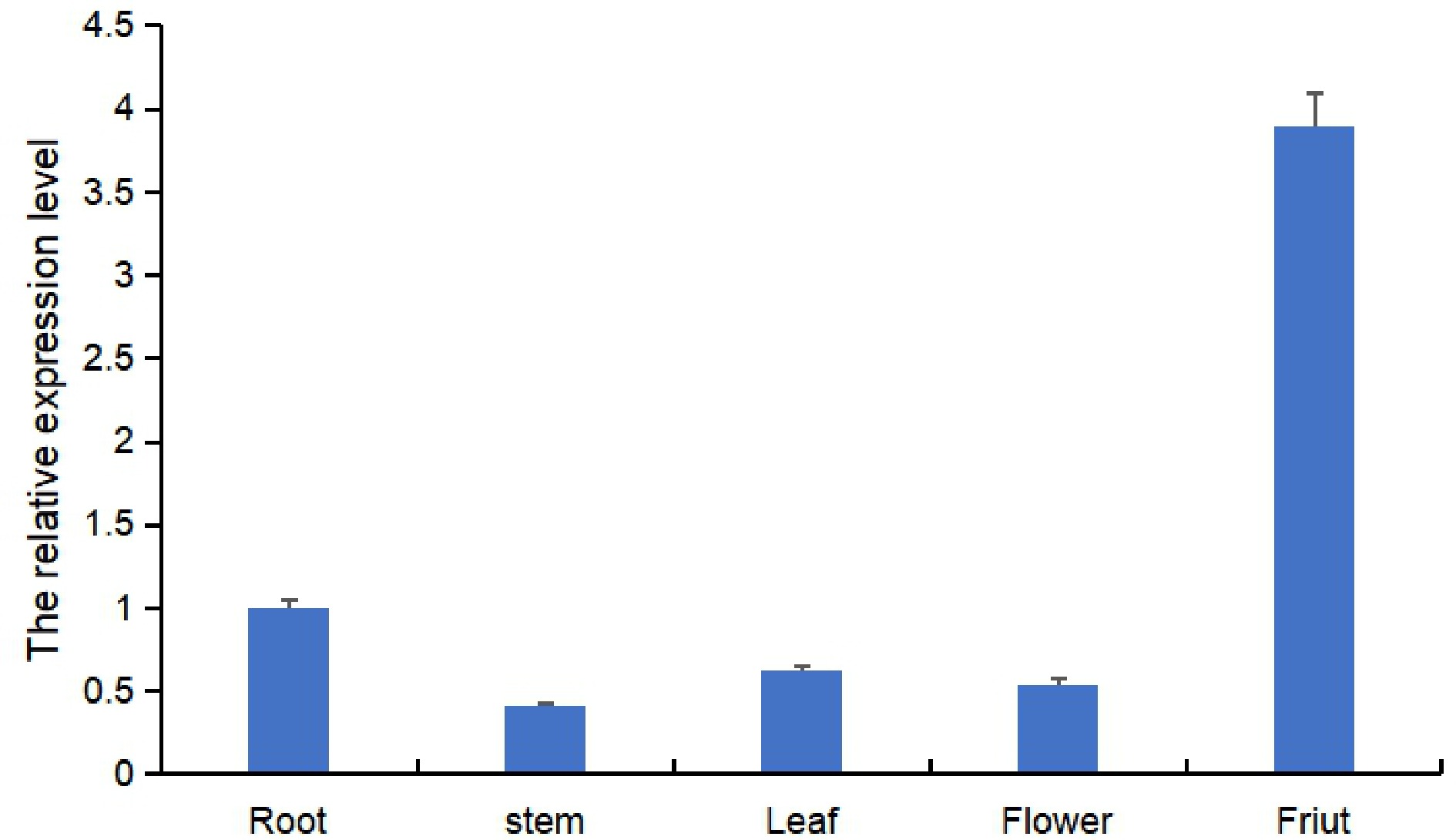
qRT-PCR was performed to identify the expression pattern of the Ruby gene in tomato. The expression pattern of Ruby in the roots, stems, leaves, flowers, seeds, and fruit was detected. As shown in Fig. 4, Ruby was expressed in all these organs, and the expression levels were higher in the fruits than in the other tissues.
Consistently, the expression pattern of Ruby was verified by the observation of bright red color in different tissues (Fig. 5a & c). The results showed that the bright red color was identified by the Ruby reporter assay in different organs of cultivars whose fruits are red, white and yellow, and the red color was brighter in the fruits than in the other tissues. Furthermore, the results were consistent with the qRT-PCR results. Nevertheless, the bright red color at the IMG stage of the white, red and yellow fruits was higher than that in other mature stages, and it was difficult to distinguish the bright red plants from control plants because they also showed red ripe (RR) fruits at the RR stage (Fig. 5b). Interestingly, the bright red color at the yellow ripe (YR) and RR stages was still higher than that of the WT, and it appears to be the purplish red variety compared with the WT. Thus, the results in the yellow cultivars were inconsistent with the results of red and white fruit bearing cultivars.
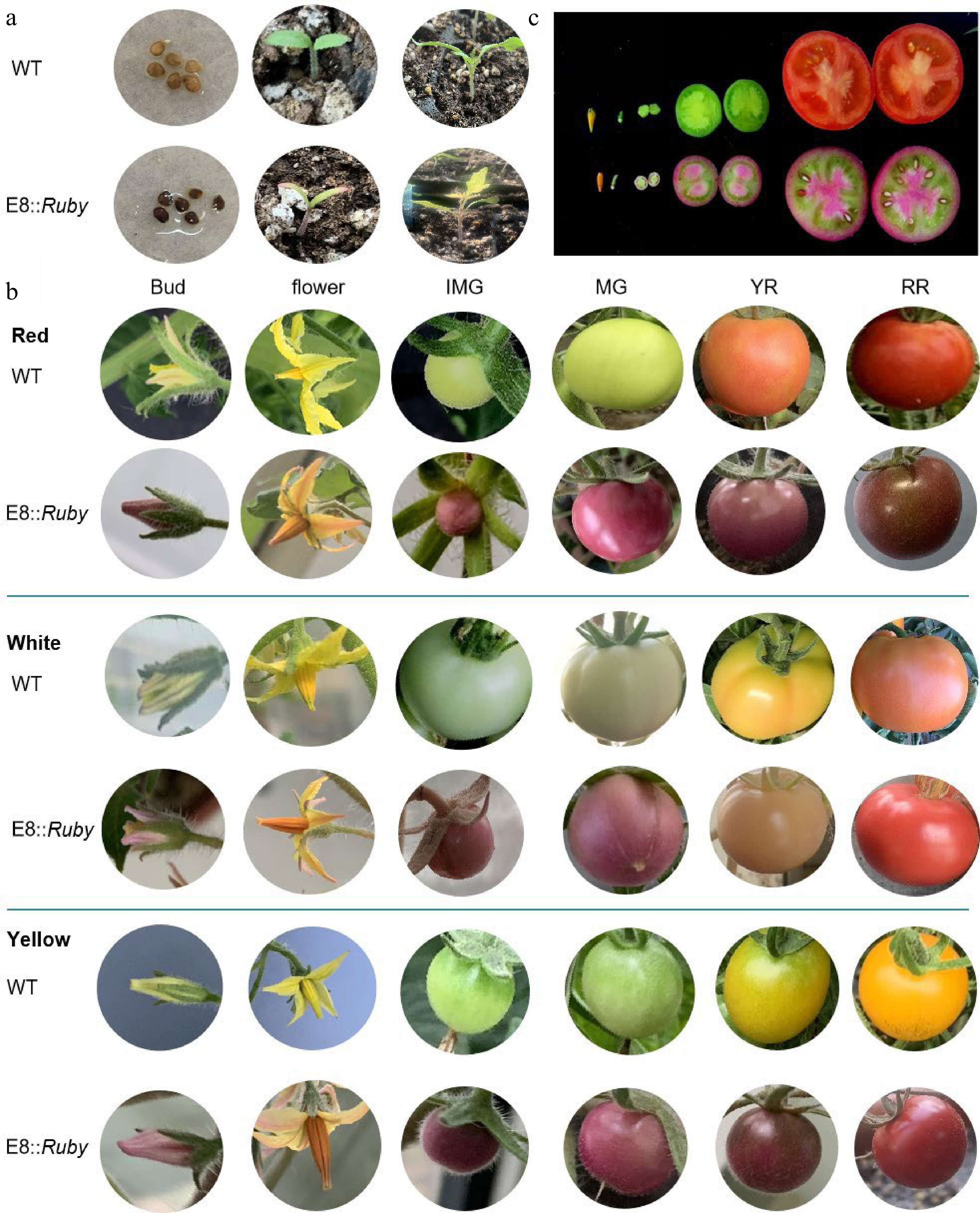
Figure 5.
Characteristics of E8::Ruby transgenic tomato plants in different tissues and different tomato varieties (red, white and yellow). (a) Seeds and seedlings; (b) Different tomato varieties red fruit bearing 'Moneymaker', white fruit bearing '06883' and yellow fruit bearing '19458'; (c) The section observation of the tomato fruits. IMG, immature green stage; MG, mature green stage; BR, breaker stage; YR, yellow ripe stage; RR, red ripe stage.
-
Auxin is essential for plant growth and development[17]. DR5, an auxin-specific promoter, has been reported to be an effective tool for monitoring the auxin expression and distribution of different organs in Arabidopsis, tomato, rice and Populus[18−20]. In this study, to noninvasively monitor the auxin response at different developmental stages in tomato, a DR5 reporter system with the visual marker Ruby was constructed. Consistently, the DR5::Ruby-transformed plants showed a pink color with an asymmetrical distribution of endogenous auxin in different tomato tissues. These results showed that the DR5 reporter system is a promising tool for monitoring the tissue-specific local accumulation of endogenous auxin in tomato.
The availability of fruit-specific promoters greatly benefits fruit quality by genetic transformation. The E8 promoter, as one of the most widely used fruit-specific promoters, is widely used for specific expression in mature tomato fruits. For example, Luo et al. transferred E8:AtMYB12 into Money Maker tomato to increase the flavonol content in the fruit and obtain orange fruit[21]. In addition, Scarano et al. introduced the grape stilbene synthase gene vvstsy into indigo tomato plants and developed bronze tomato plants[22]. Furthermore, Sun et al. overexpressed the Slan2-like gene comprising the E8 promoter in AC cultivated varieties, and purple tomato fruits were obtained with a purple peel and flesh[23]. The −1088 to −863 region of the E8 promoter is the binding region of DNA-binding protein, and knocking out this region significantly reduced the expression of the E8 gene in mature fruits. Inconsistently, all the −1088 region of the E8 promoter was cloned to construct the E8::Ruby vector in this study. The results showed that the E8::Ruby-transformed plants exhibited asymmetrical pink coloration in different tomato tissues of different tomato cultivars (red, white and yellow). In particular, it is brighter red at the RR stage in the yellow fruit bearing cultivars than in the red and white fruit bearing cultivars. Finally, all the results showed that DR5 and E8 are suitable systems for studying the application of the Ruby reporter gene in tomato.
-
In this study, DR5::Ruby transgenic tomato plants were used to further clarify the auxin content at different developmental stages in tomato. The results showed that the nonuniformity of Ruby activity was consistent with the natural distribution characteristics of native auxin. In addition, E8::Ruby was transformed into different tomato varieties (red, white and yellow) to obtain tomato plants from different germplasm backgrounds bearing new colors of fruit. The results not only showed that DR5 and E8 are suitable systems for studying the application of the Ruby reporter gene in tomato, but also will provide insight for the improvement of tomato fruit color via biotechnology.
We acknowledge financial support from the National Natural Science Foundation of China (32002059), the China Agriculture Research System (CARS-23-A11) and the Fellowship of China Postdoctoral Science Foundation (2020 M681068).We also acknowledge the donation of the DR5::Ruby plasmids by Dr. Zhao.
-
The authors declare that they have no conflict of interest.
-
# These authors contributed equally: Huanhuan Yang, Yiyao Zhang
- Supplemental Fig. S1 DR5: RUBY
- Supplemental Table S1 Primers used in the text.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Yang H, Zhang Y, Fu Q, Jia X, Zhao T, et al. 2023. The DR5 and E8 reporters are suitable systems for studying the application of the Ruby reporter gene in tomato. Vegetable Research 3:12 doi: 10.48130/VR-2023-0012
The DR5 and E8 reporters are suitable systems for studying the application of the Ruby reporter gene in tomato
- Received: 08 September 2022
- Accepted: 08 February 2023
- Published online: 03 April 2023
Abstract:
-
Key words:
- Tomato /
- DR5 /
- E8 /
- Ruby /
- Fruit color