-
Tomato (Solanum lycopersicum) is one of the most important vegetable crop species widely cultivated worldwide. Tomato fruit can be eaten fresh or processed into sauce, fruit juice, salads and other manufactured foods, and thus plays an important role in daily diets[1]; tomato fruit also contains an abundance of nutrients such as lycopene, carotene, lutein and antioxidant substances, which exhibit anticancer and antiaging activities[2]. In addition, tomato has the advantages of a relatively short life cycle, a small genome, and high genetic diversity, and acts as a classic model plant species for molecular studies on plant growth, development and defense responses.
Jasmonic acid and its derivatives, such as jasmonic acid-isoleucine (JA-Ile) and methyl jasmonate (MeJA), are collectively referred to as jasmonates (JAs)[3,4]. The biosynthesis and signaling pathways of JA are well characterized in Arabidopsis. JA biosynthesis subsequently occurs in plastids, peroxisomes, and the cytoplasm. The JA precursor linolenic acid (18:3) is produced through the action of fatty acid desaturase (FAD) and phospholipase A1 (PLA), and catalyzed to 12-oxo-phytodienoic acid (OPDA) by lipoxygenase (LOX), allene oxide synthase (AOS), and allene oxide cyclase (AOC), which are located in the chloroplast[4−7]. Subsequently, OPDA is successively transported into the cytosol and peroxisomes with the help of the chloroplast outer envelope-localized transporter JASSY and the peroxisomal ATP-binding transporter COMATOSE 1 (CTS1)[8,9]. OPDA proceeds to be converted to JA in the peroxisome with the action of the enzymes 12-oxo-phytodienoic acid reductase 3 (OPR3) and OPC-8:0 CoA Ligase (OPCL) followed by three rounds of β-oxidation[10−13]. In addition, in chloroplasts, hexadecatrienoic acid (16:3) is catalyzed to produce dinor-OPDA (dnOPDA), which is metabolized to JA through two rounds of β-oxidation[14]. Moreover, OPDA is also metabolized to dnOPDA and converted to 4,5-didehydro-JA (4,5-ddh-JA), which is subsequently catalyzed to form JA under the action of OPR2 in the cytosol[14,15]. Afterward, JA is released into the cytoplasm, undergoing further modification (e.g., conjugation with amino acids, methylation, esterification, hydroxylation, and O-glycosylation) to produce various derivatives [e.g., (+)-7-iso-JA-Ile, MeJA, JA-glucosyl ester, 12-OH-JA, and 12-O-Glc-JA][11,16−18]. With the assistance of ABCG-type JASMONATE TRANSPORTER1 (JAT1), the bioactive JA form JA-Ile is delivered to the nucleus and activates the JA signaling pathway[3, 19].
JASMONATE ZIM DOMAIN (JAZ) proteins recruit the corepressors NOVEL INTERACTOR OF JAZ (NINJA) and TOPLESS (TPL) to repress downstream transcription factors (TFs)[20−23]. The F-box protein COI1 together with SKP-LIKE proteins (ASK1/2), Cullin1 and Rbx1 are involved in the formation of the SCFCOI1 E3 ligase complex, which is essential and necessary for the perception and transduction of the JA signaling pathway[24, 25]. COI1 also associates with JAZs. They form coreceptors to perceive JA-Ile, which facilitates the degradation of JAZ through the 26S proteasome pathway[26, 27]. The reduction in JAZs allows the release of diverse JAZ-interacting TFs (such as MYC2) to regulate various JA-mediated responses[28−31].
Recently, several tomato orthologs of Arabidopsis JA biosynthesis and signaling genes have been characterized and investigated, for example, the JA biosynthesis genes SUPPRESSOR OF PROSYSTEMIN RESPONSE2 (SPR2, encoding a fatty acid desaturase), LoxD, AOC and OPR3, the JA signaling genes COI1/JAI1, JAZs and MYC2, and a number of genes involved in JA pathway have been identified[32−38]. These have greatly helped to determine the function of JA in tomato. In this review, we summarize the current progress on the roles and molecular mechanisms of JAs in controlling growth and development, secondary metabolism, and defense against stresses in tomato (Figs 1 & 2).
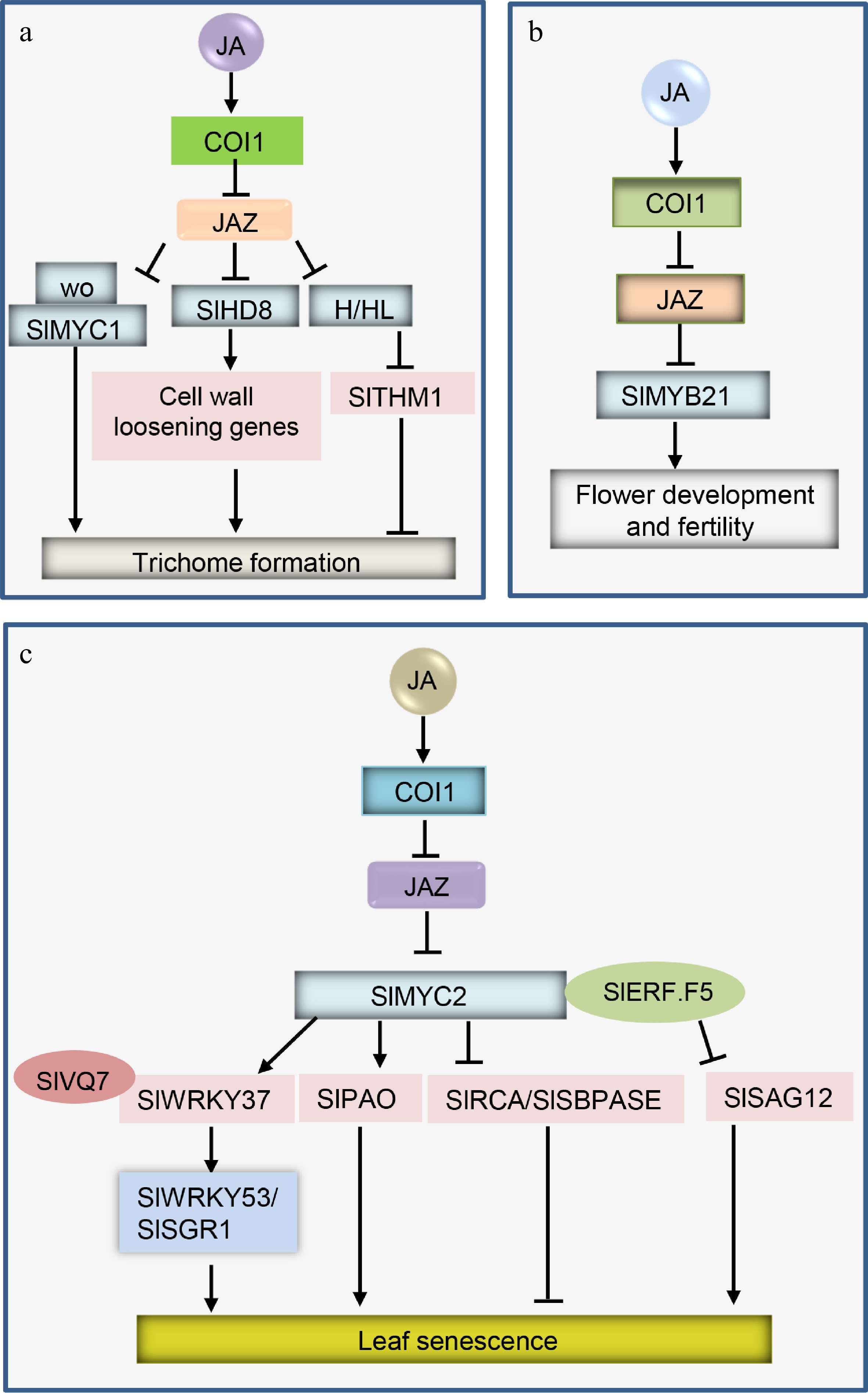
Figure 1.
Simplified models of JA signaling in tomato trichome formation, flower development and fertility, and leaf senescence. The bioactive JA is perceived by COI1-JAZ and triggers the degradation of JA repressors JAZs via the 26S proteasome pathway. JAZ degradation releases various downstream TFs to control JA-mediated responses, for example, (a) the release of wo/SlMYC1, SlHD8, and H/HL, to regulate trichome formation in tomato; (b) the release of SlMYB21 to control tomato flower development and fertility; (c) the release of SlMYC2 to promote tomato leaf senescence.
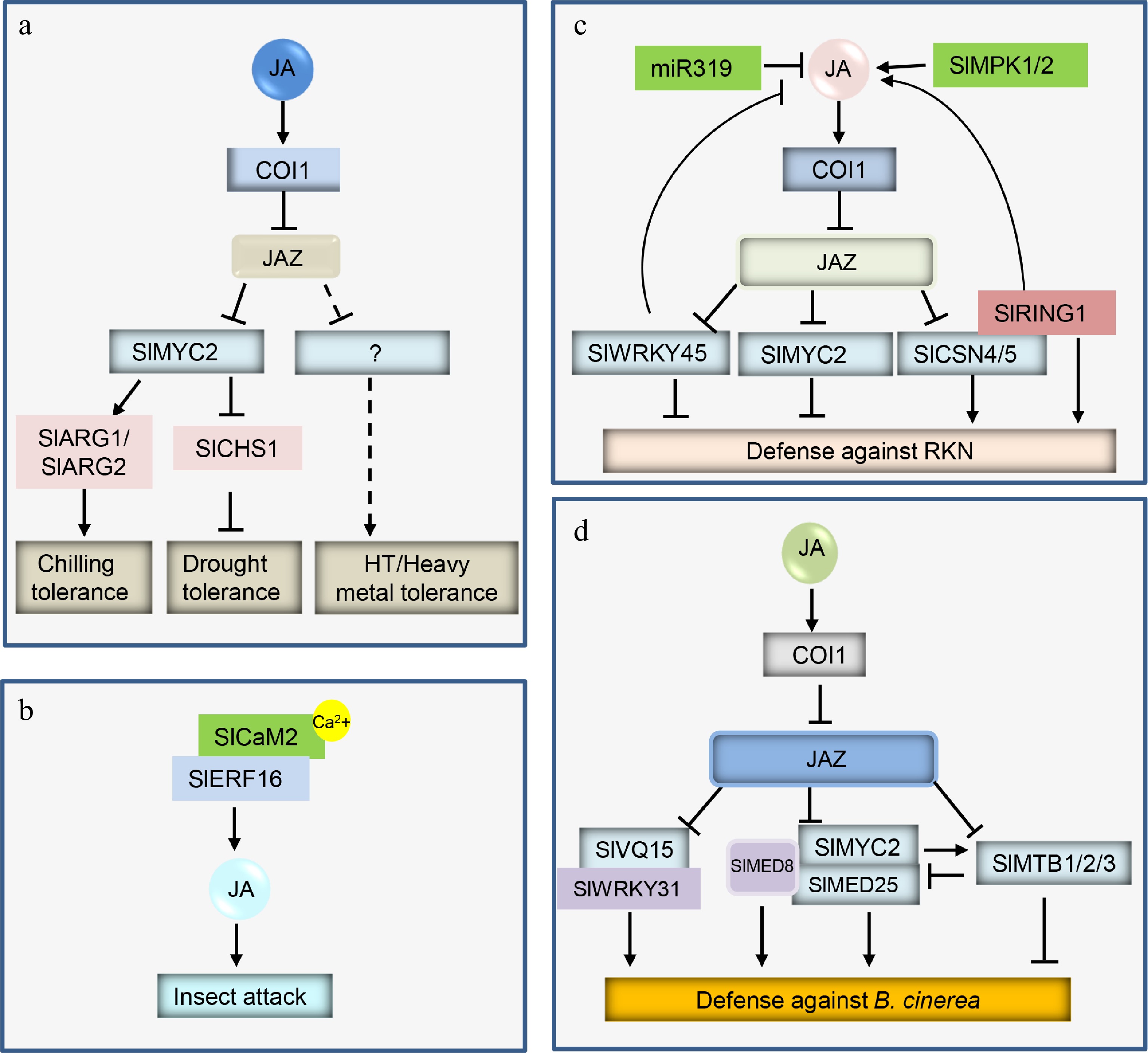
Figure 2.
Simplified models of JA signaling in tomato resistance to abiotic and biotic stresses. JA induces the degradation of JAZs through the 26S proteasome pathway, which releases different TFs to modulate diverse responses. (a) The released SlMYC2 controls tomato tolerance to cold and drought stresses. (b) Insect attack activates Ca2+ signals to promote the interaction of SlCaM2 and SlERF16, which promotes SlERF16 to activate JA biosynthesis for herbivore defense. (c) The released SlMYC2 and SlWRKY45 negatively control tomato resistance to RKN, while SlCSN4/5 positively controls this resistance; in addition, miR319 inhibits JA accumulation, whereas SlMPK1/2 induces JA biosynthesis. (d) The released SlVQ15-SlWRKY31 module, SlMED8-SlMYC2-SlMED25, and SlMTBs finely tune tomato defense against B. cinerea.
-
Leaf senescence is essential for plant growth and development, before which the nutrients and energy in aging leaves are transferred to developing tissues or fruit for redistribution and reutilization. The regulatory network of leaf senescence is finely tuned and complex.
Exogenous application of MeJA promotes leaf senescence together with decreased chlorophyll content and repressed photosynthesis in tomato[39]. Recent studies discovered the molecular mechanism of JA-mediated leaf senescence in tomato partially through SlMYC2, a key SlJAZ-interacting TF (interacting with SlJAZ1-SlJAZ11)[38−41]. SlMYC2 directly binds to the promoters of SlPAO (regulating chlorophyll degradation), SlRCA and SlSBPASE (these two genes participating in carbon assimilation), and promotes the expression of SlPAO while attenuating the transcript levels of SlRCA and SlSBPASE[39]. Silencing of SlMYC2 inhibits JA-induced leaf senescence by repressing chlorophyll degradation and inducing photosynthetic carbon fixation, suggesting that SlMYC2 plays a positive role in JA-induced leaf senescence[39].
SlWRKY37 participates in JA and dark-induced tomato leaf senescence. With JA and dark treatments, knockout of SlWRKY37 represses leaf senescence, while overexpression of SlWRKY37 accelerates leaf senescence[40]. JA induces the expression level of SlWRKY37. Furthermore, SlMYC2 promotes SlWRKY37 expression by directly binding to its promoter, whereas SlWRKY37 binds to and activates SlWRKY53 and SlSGR1 (a senescence-regulated gene and chlorophyll degradation-related gene, respectively) to promote leaf senescence. JA-induced SlVQ7 associates with SlWRKY37 to increase the transcriptional activity of SlWRKY37 in the regulation of SlWRKY53 and SlSGR1[40].
In addition, JA combines with other hormones, such as ethylene (ET), to control tomato leaf senescence. The ETHYLENE RESPONSE FACTOR F5 (SlERF.F5), a key gene of the ET signaling pathway, plays a negative role in leaf senescence[41]. SlERF.F5-RNAi plants present increased transcript levels of JA singling pathway genes (e.g., SlJAZ1/JAZ2/JAZ4/JAZ7/JAZ11/MYC2). SlERF.F5 interacts with SlMYC2 to integrate JA and ET signals and thus modulates leaf senescence in tomato[41].
Taken together, the findings from the abovementioned studies reveal that JA plays an indispensable role in leaf senescence.
Trichome formation
-
Trichomes are specialized from epidermal cells, composed of single or multiple cells, and can protect plants from biotic and abiotic stresses[42−44]. Tomato has seven types of trichomes, which are divided into nonglandular (II, III, and V) and glandular (I, IV, VI, and VII) types[45]. Glandular trichomes have a strong ability to synthesize and secrete a large number of special metabolites (e.g., acyl sugars, terpenoids, alkaloids, and flavonoids), most of which are related to defense responses[46−48]. JA plays a crucial role in trichome development in tomato.
Exogenous application of JA promotes the elongation and formation of trichomes in new tomato leaves and stems[49,50]. The suppressor of prosystemin-mediated responses 8 (spr8) mutant, with deficiency in the JA biosynthetic gene LoxD, exhibits defective development of type VI trichomes[32]. Tomato jasmonic acid insensitive 1-1 (jai1-1) mutant deficient in COI1 produces smooth and hairless fruit, and presents largely reduced type VI trichome density across its leaves and sepals[34]. jai1-1 also affects the ability of trichome glands to synthesize compounds. The accumulation of monoterpene is significantly decreased in sepals of jai1-1 compared with wild-type plants, and no monoterpene is present in jai1-1 fruit[34].
SlJAZ4, with high expression in trichomes, negatively regulates trichome length, as indicated by the observations that overexpression of SlJAZ4 results in shorter trichomes of types II, V, and VI compared with those of the wild type[49]. SlJAZ4 interacts with the homeodomain-leucine zipper (HD-ZIP) protein HOMEODOMAIN PROTEIN 8 (HD8), which is expressed in trichomes and induced by JA treatment. Clustered, regularly interspaced, short palindromic repeats (CRISPR)/CRISPR-associated 9 (Cas9)-mediated gene editing of SlHD8 significantly reduces the length of most types of trichomes and affects trichome morphology. In slhd8 mutants, the length of type II, V, and VI trichomes is reduced to one-fourth, three-quarters, and half of that of wild-type trichomes, respectively, and type II and V trichomes have more cells. Moreover, SlHD8 directly binds to and activates cell wall-loosening genes, whose expression is repressed by SlJAZ4[49].
Overexpression of SlJAZ2 decreases the transcript levels of the HD-ZIP gene SlWolly (SlWo) and the B-type cyclin gene SlCycB2, both of which are involved in the formation of type I trichomes in tomato[51], and reduces the formation of trichomes in stems[52]. SlJAZ2 physically associates with two C2H2 zinc finger proteins, Hair (H) and Hair-like (HL), via the C-terminal regions of H and HL[53]. H and HL synergistically and positively function in the regulation of trichome development in different tissues. HL mainly participates in trichome formation in hypocotyls and leaves, whereas H mainly participates in trichome formation in stems and sepals[53]. MeJA-induced trichome formation is obviously blocked in h/hl mutants. H and HL attenuate the expression of SlTHM1, a repressor in trichome development, through binding to the promoter of SlTHM1. However, this repressive function can be suppressed by SlJAZ2[53].
The number of type VI and VII trichomes is markedly reduced in tomato CRISPR/Cas9-generated wo mutants[54]. The application of MeJA barely increases the number of type VI trichomes in wo. Knockout of the bHLH TF SlMYC1 results in a reduced number of type VI trichomes in tomato leaves and stems. SlMYC1 interacts with Wo to form a regulatory module, and they coordinately regulate trichome development. SlJAZ2 associates with SlWo-SlMYC1 and disrupts their interaction. JA regulates trichome formation through induction of the degradation of SlJAZ2 to release SlWo-SlMYC1[54].
Flower development and fertility
-
Flowers are the reproductive organs of angiosperms. They play essential biological roles in the release of mature pollen grains to the stigma for fertilization. JA controls flower development and female fertility in tomato.
Silencing of the JA synthetic gene SlAOC results in early flower abortion, deficient seed development and female sterility[35]. Transgenic tomato lines with silencing of OPR3 (SiOPR3) produce fewer seeds that are less viable than the wild type, while exogenous treatment with MeJA restores the seed production of SiOPR3 lines[36]. Overexpression of SlJAZ2 has no effect on tomato fertility but causes an earlier floral-transition phenotype; flower buds appears at 4 weeks of age in SlJAZ2-overexpressing transgenic lines but appears at 39 days of age in wild type[52]. Compared with the wild type plants, slmyc2 mutants generated by CRISPR/Cas9 technology have more flowers, and a lower rate of fruit setting[55].
jai1-1 is female sterile, and male reproductive function (e.g., pollen viability, pollen germination, morphology and color of anther cones) is also impaired[34]. ET-biosynthesis and related genes present increased and earlier expression in stamens of jai1-1, which may result in the premature dehiscence of stamens and inefficient pollen development in jai1-1[56]. The concentrations of JA and JA-Ile in wild type flower buds increase during flower bud development stages, peaking in day-5 buds, while flower buds with delayed opening of jai1-1 contain an extremely low content of JA and JA-Ile[57]. Ovules in jai1-1 display abnormal morphology with increased amounts of callose and undergo programmed cell death in the nucellus[58]. Transcript profiling on different ovule development stages of wild type and jai1-1 shows that SlMYB21 is largely expressed in ovules of wild type but is almost undetectable in those of jai1-1. Further studies revealed that SlMYB21 interacts with SlJAZ9. SlMYB21 overexpression partially rescues the fertility of Arabidopsis myb21 mutants. Slmyb21 mutants exhibit altered flower development with incompletely open petals, which constitute abnormal ovule phenotypes similar to those of jai1-1, and are female sterile[58]. In addition, overexpression of AtMYB24-SRDX (a chimeric repressor of SlMYB21) in tomato delays flower opening, causes defects in the elongation of pollen tubes, and results in insufficient male and female fertility[57].
Fruit development and quality
-
JA modulates tomato fruit development, as indicated by the observation that slmyc2 mutants exhibit abnormal fruit shapes and reduced fruit firmness[55,59]. JA is also an essential regulator of tomato fruit quality. Application of JA improves tomato fruit quality with enhanced accumulation of nutrition- and flavor-related compositions (e.g., glucose, fructose, soluble sugar, soluble protein, starch, total phenol, lycopene, and flavonoids)[59]. The content of these compositions and the sugar-acid ratio are increased in SlMYC2-overexpressing tomato fruit[59]. Moreover, the concentration of lycopene is obviously reduced in fruits of the JA biosynthesis-deficient mutants def1 and spr2, and is increased in the constitutive JA biosynthesis transgenic line 35S::PS[60]. Consistently, the transcript levels of lycopene biosynthesis-related genes display a similar trend[60].
-
In addition to participating in the regulation of the abovementioned tomato growth and development processes, JA also controls seed germination, root growth, stem development, leaf initiation, internode formation and so forth. For instance, the application of exogenous JA has inhibitory effects on seed germination in tomato[61]; JA inhibits tomato root growth, which is blocked in jai1-1 mutants[62]; the application of JA and overexpression of SlMYC2 reduces the height, stem thickness, and photosynthesis of tomato seedlings[59]; and SlJAZ2-overexpressing tomato lines present increased leaf initiation but decrease plant height and internode length[52].
-
Secondary metabolites are essential for plant growth and development. They protect plants from insect attack and pathogen infection, and attract pollinators for pollination and seed transmission.
Flavonoids contribute to regulation of plant development and defense. SlMYB14 positively regulates flavonoid accumulation by directly binding to and controlling the expression of the flavonoid biosynthesis-related gene SlPAL[63]. JA promotes the expression of SlMYB14, which is dependent on SlMYC2[63].
The MYB-bHLH-WD40 (MBW) complex is involved in controlling anthocyanin biosynthesis. IIIf bHLH TFs SlJAF13 and SlAN1, members of the MBW complex, act as positive regulators to control anthocyanin synthesis through SlJAF13 binding to the promoter of SlAN1[64]. SlJAZ2 affects the formation of the MBW complex, and attenuates the transcriptional activity of SlJAF13 and SlAN1, whereas SlJAF13 associates with SlMYC2 to repress the SlMYC2-activated expression of SlJAZ2. Together, these actions form a negative feedback loop to precisely regulate the accumulation of anthocyanins[64].
Caffeoylputrescine (CP), which probably functions in plant reproduction, is induced by JA treatment in tomato leaves and flowers. In jai1-1, the accumulation of CP is blocked, whereas 35S::PS transgenic lines with constitutive biosynthesis of JA present increased concentrations of CP[65]. However, the molecular mechanism of JA-regulated accumulation of CP remains unclear.
The toxic compounds steroidal glycoalkaloids (SGAs) are mainly synthesized in solanaceous species and are involved in pathogen defense. Exogenous application of JA induces the expression level of SGA biosynthesis-related genes, which is affected in jai1-1[66]. The AP2/ERF TF GLYCOALKALOID METABOLISM 9 (GAME9) functions alone or together with SlMYC2, binds to the promoter of SGA biosynthesis-related genes and controls the accumulation of SGA[67].
-
Drought is recognized as one of the main stress factors, and severely affects agricultural production. JA plays essential roles in helping diverse plants resist drought stress[68]. In tomato, drought induces the transcript levels of SlMYC2[69]. Overexpression of SlMYC2 enhances tomato tolerance to drought stress, and promotes stomatal closure and the accumulation of abscisic acid (ABA) and JA[69]. SlMYC2 negatively affects the content of flavanol through binding to and inhibiting the flavanol biosynthesis-related gene SlCHS1, which subsequently negatively regulates tomato resistance to drought stress[69]. In addition, SlMYC2 directly represses SlPP2C1, a negative modulator in ABA signaling, and SlRR26, a type-B response regulator RR in the cytokinin (CK) pathway, which integrates JA, ABA and CK to fine-tune drought stress[70].
High-temperature stress
-
High temperature disrupts the structure of cell membranes, denatures proteins, reduces photosynthetic efficiency, and causes abnormal biochemical and physiological metabolism. A recent study discovered that high temperature promotes the exsertion of tomato stigma, resulting in failure to set fruit[71]. Under high temperature, the content of JA is obviously reduced in stamens because of the repressed transcription of SlFAD2 and SlFAD3. The application of JA restores high temperature-induced tomato stigma exsertion in a dose-dependent manner via the JA/COI1 signaling pathway[71]. However, further studies on the details of JA function in tomato resistance to high temperature are needed.
Cold stress
-
Cold stress constricts plant growth and development by triggering the massive production of reactive oxygen species (ROS), the inactivation of proteins and enzymes, and the modification of membrane structure. Chilling and freezing are two typical types of cold stresses. Foliar application of JA increases the contents of proline and lycopene, and the activities of antioxidant enzymes, which alleviates chilling injury in tomato fruit[72]. These effects of JA are attenuated in SlMYC2-silenced tomato fruit[72]. Min et al. further found that JA protects tomato fruit from chilling tolerance by inducing the biosynthesis of polyamines[73]. SlMYC2 binds to and activates the polyamine biosynthesis-related genes SlARG1, SlARG2, SlADC and SlODC. Silencing of SlMYC2 significantly reduces the content of polyamines, which results in a more sensitive phenotype to chilling stress[73]. In addition, JA cooperatively acts with melatonin (MT) to alleviate cold damage to tomato[74]. Cold stress increases JA and MT accumulation in tomato, and the addition of exogenous JA or MT enhances the tolerance of tomato to cold[74]. JA promotes increased transcript levels of MT biosynthetic genes, whereas knockdown of SlMYC2 suppresses JA-induced expression of MT biosynthetic genes, MT accumulation and tomato tolerance to cold. In addition, JA acts downstream of ABA to participate in phytochrome A (phyA) and phyB antagonistically mediated cold tolerance in tomato[75]. Additionally, JA and SlMYC2 induce the expression of ABA biosynthesis gene 9-CIS-EPOXYCAROTENOID DIOXYGENASE2 (NCED2), and ABA accumulation to elevate tomato tolerance to cold[76].
Heavy metal stress
-
The accumulation of the heavy metal lead (Pb) in plants alters the structure of chloroplasts, blocks the absorption of nutrients, reduces the efficiency of photosynthesis, and impairs plant growth and development. JA mitigates the toxicity of Pb by increasing the contents of secondary metabolites, photosynthetic pigments, and organic acids[77]. However, the molecular mechanism of JA-regulated tomato tolerance to Pb needs to be further investigated.
Biotic stresses
Defense against herbivorous insects
-
The application of JA enhances tomato defense against flea beetles (Epitrix hirtipennis), and potato aphids (Macrosiphum euphorbiae) by activating polyphenol oxidase and proteinase inhibitors (PIs)[78]. Deficiency in JA biosynthesis and signaling pathways results in reduced resistance to herbivorous insects. For instance, spr8 mutants exhibit increased susceptibility to cotton bollworm (Helicoverpa armigera)[32]. The resistance of spr2 mutants to tobacco hornworm (Manduca sexta) larvae is compromised[33]. acx1 (a JA biosynthesis-deficient mutant) presents increased susceptibility to tobacco hornworm (Manduca sexta) attack[79], and def1 mutants have reduced resistance to tomato fruitworm (Helicoverpa zea), although exogenous application of JA restored their resistance[80]. Compared with the wild type, jai1-1 is more susceptible to the two-spotted spider mite (Tetranychus urticae)[34]. In turn, herbivorous insects manipulate JA to protect themselves. For example, the whitefly (Bemisia tabaci) causes tomato to produce volatiles, activates salicylic acid (SA)-mediated defenses and attenuates JA-regulated defenses, which leads to the neighboring tomato plants more susceptible to whiteflies[81]. Moreover, other environments utilize JA to affect tomato responses to insect attack. For instance, ultraviolet (UV) radiation enhances tomato resistance to thrips (Frankliniella occidentalis), but this is compromised in def-1 mutants, indicating that UV modulates tomato defense against herbivory possibly through activation of JA signaling[82].
A recent study revealed that JA acts with ET to modulate tomato resistance to cotton bollworm (Helicoverpa armigera)[83]. ET promotes the transcript levels of ETHYLENE RESPONSE FACTOR 15 (ERF15) and ERF16, which directly bind to and activate the JA biosynthetic genes SlLOXD, SlAOC, and SlOPR3, leading to rapid accumulation of JA to defend against herbivore attack. JA also integrates Ca2+ signals and ERF16 to control tomato defense responses[84]. Insect attack promotes Ca2+ influx, electrical activity, and the expression of CALMODULIN2 (CaM2) and ERF16. SlCaM2 interacts with SlERF16, increases the transcriptional activity of SlERF16, and results in high accumulation of JA, which regulates defense against the cotton bollworm Helicoverpa armigera in tomato[84].
Defense against nematodes
-
The root knot nematode (RKN) Meloidogyne incognita is a plant parasite widely distributed worldwide. Application of JA increases tomato defense against the RKN M. incognita[85], and the JA-deficient mutant spr2 is extremely sensitive to M. incognita[86,87].
Several studies have reported that some factors control tomato resistance to M. incognita by manipulating the biosynthesis and/or signaling pathway of JA. For example, miR319-overexpressing transgenic tomato lines present decreased concentrations of JA and reduced resistance to M. incognita[88]. Both SlCSN4 and SlCSN5, two subunits of the COP9 signalosome, are strongly induced in response to M. incognita infection[89]. SlCSN4 and SlCSN5 physically interact with SlJAZ2, and positively regulate the transcript levels of SlJAZs. Silencing of SlCSN4 or SlCSN5 reduces the basal level and M. incognita-induced accumulation of JA, which leads to decreased resistance to M. incognita[89]. The E3 ubiquitin ligase RING1 associates with SlCSN4 and stabilizes it under M. incognita infection[90]. Overexpression of SlRING1 promotes tomato resistance to M. incognita through stimulation of the biosynthesis of JA[90]. SlWRKY45 is involved in both JA biosynthesis and signaling pathways. SlWRKY45 interacts with most SlJAZs (SlJAZ1/2/3/4/5/6/7/11), binds to and represses the JA biosynthesis-related gene SlAOC to control JA content. Overexpression of SlWRKY45 reduces the resistance to M. incognita, whereas CRISPR/Cas9-mediated gene editing of SlWRKY45 enhances defense against M. incognita, suggesting that SlWRKY45 acts as a negative regulator in tomato defense response to M. incognita. In addition, M. incognita infection triggers electrical and ROS signal delivery from roots to leaves and activates mitogen-activated protein kinases 1/2 (MPK1/2) to promote the accumulation of JA in leaves, which contributes to defend against M. incognita[91].
Other hormones, such as ABA, and strigolactones (SLs), are partially involved in the JA pathway to modulate tomato defense against M. incognita. M. incognita resistance in tomato is positively regulated by JA and negatively controlled by ABA, whereas in response to M. incognita, both JA and ABA are inhibited by SL. SL positively regulates resistance to M. incognita by repressing SlMYC2, which is a negative gene of M. incognita resistance and is controlled by ABA[92].
Defense against pathogens
-
JA controls tomato resistance to pathogens. For instance, the tomato mutants spr2, def1, opr3, and acx1, whose production of JA is impaired, exhibit decreased resistance to Botrytis cinerea[36,93]. jai1 is severely sensitive to the oomycete pathogen Pythium with 100% mortality[94], and is also more susceptible to B. cinerea and Fusarium species[38,93,95].
Recent studies have greatly helped to elucidate the molecular mechanism of JA-mediated tomato resistance to B. cinerea. SlMYC2 plays vital roles in JA-mediated tomato defense against B. cinerea. B. cinerea infection induces the expression of SlMYC2, which is blocked by the mutation of COI1. Silencing of SlMYC2 resultes in increased susceptibility to B. cinerea. Knockdown of both SlJA2L and SlERF.C3, two B. cinerea-induced genes, increases the susceptibility to B. cinerea. Molecular analysis showed that SlMYC2 directly binds to the promoters of SlJA2L and SlERF.C3, which targets the late wounding-responsive gene THREONINE DEAMINASE (TD) and the pathogen-responsive gene PR-STH2[38]. In addition, SlMYC2 plays a positive role in tomato fruit resistance to B. cinerea, as indicated by the findings that knockdown or knockout of SlMYC2 exhibits extremely sensitive to B. cinerea tomato fruit[55,96].
Multiple factors interact/regulate/bind to SlMYC2 and are involved in JA-controlled defense responses to B. cinerea. For example, SlMYC2 functions cooperatively with MEDIATOR SUBUNIT 25 (SlMED25), and promotes the expression of the JA-induced bHLH factor MYC2-TARGETED BHLH 1/2/3 (MTB1/2/3)[97]. Conversely, SlMTB1/2/3 disrupts the formation of SlMYC2-SlMED25, and interferes with the ability of SlMYC2 to bind to its target gene[97]. SlMED8 forms homodimers and associates with SlMYC2 and SlMED25 to participate in JA-regulated defense responses[98]. Overexpression of SlMED8 increases tomato resistance to B. cinerea, whereas downregulation of SlMED8 reduces defense against B. cinerea in tomato. In addition, Jaiswal et al. reported that JA-induced immunity to B. cinerea relies on the tomato receptor-like cytoplasmic kinase TRK1, which interacts with SlMYC2 and promotes its accumulation[99].
A recent study found that JA mediates tomato defense against B. cinerea through the SlVQ15-SlWRKY31 module[100]. SlVQ15 cooperates with SlWRKY31 to positively modulate tomato resistance to B. cinerea. SlJAZs (e.g., SlJAZ2/5/6/7/11) interact with SlVQ15, which interferes with the formation of SlVQ15-SlWRKY31 and disrupts SlVQ15 to promote the transcriptional activity of SlWRKY31[100].
In addition, JA acts as a defense signal to negatively control resistance to Pseudomonas syringae pv. tomato DC3000 (Pst DC3000). For instance, CRISPR/Cas9-generated SlJAZ2 lines with edits to the Jas domain (SlJAZ2ΔJas) present abolished JA/COI1-mediated degradation, and are more resistant to Pst DC3000[101]. Similarly, jai1 exhibits a significantly enhanced defense responses against Pst DC3000[102].
-
In recent decades, several studies have revealed that JA acts as a plant growth regulator as well as a defense signal to participate in diverse plant growth and developmental processes and defense responses. The molecular mechanisms of JA-mediated roles have been extensively studied in the model organism Arabidopsis thaliana; however, many aspects of JA action in tomato and other horticultural crop species remain to be investigated. (i) JA regulates tomato resistance to wounding and pathogens through the JA-COI1-MYC2 pathway, and SlMYC2 positively modulates wounding and pathogen-responsive genes[38]. This is distinctly different from what occurs in Arabidopsis, in which AtMYC2 positively controls wounding-responsive genes and negatively regulates pathogen-responsive genes. The specific mechanism of JA action in tomato is still largely unclear and needs to be extensively explored. (ii) A large amount of evidence demonstrates that in Arabidopsis, JA functions in combination with other hormones to synergistically or antagonistically control various physiological processes, whereas the molecular basis of the crosstalk between JA and other hormones in tomato is relatively uncharacterized and needs to be further investigated. Future research on the roles and precise molecular mechanism of JA in tomato will benefit agricultural production.
This work was supported by grants from the Project of Cultivation for young top-notch Talents of Beijing Municipal Institutions (Grant No. BPHR202203099), the National Natural Science Foundation of China (31902026), Beijing Natural Science Foundation (6194030), and the Beijing Natural Science Foundation project-Key Project of Science and Technology Plan of Beijing Education Commission (KZ202010020027). We apologize for being unable to discuss many additional excellent and relevant research papers due to space limitations.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Huang H, Qiao H, Ma X, Zhao W, Sun L, et al. 2023. Roles of jasmonates in tomato growth, development and defense. Vegetable Research 3:14 doi: 10.48130/VR-2023-0014
Roles of jasmonates in tomato growth, development and defense
- Received: 19 December 2022
- Accepted: 21 March 2023
- Published online: 04 May 2023
Abstract: Tomato (Solanum lycopersicum), one of the most important vegetable crop species, is popular worldwide due to its richness of nutrients and flavors. It also serves as a classic model plant species for investigating the mechanisms of plant growth, development, and defense processes. Jasmonates (JAs) are well characterized as defense phytohormones involved in the regulation of plant resistance to abiotic and biotic stresses, and also play indispensable key roles in controlling plant growth and development. Here, we summarize the recent progress in understanding JA function in governing tomato growth and development (e.g., leaf senescence, trichome formation, flower development and fertility, and fruit development and quality), secondary metabolism, and defense against stresses (e.g., drought, heat, cold, insects, nematodes, and pathogens).
-
Key words:
- Tomato /
- Jasmonates /
- Growth /
- Development /
- Stress












