-
Rosa is one of the most economically important genera in ornamental horticulture. The genus' natural distribution is widespread throughout the northern hemisphere[1]. While taxonomic classification within the genus is considered difficult with past efforts recognizing more than 300 species, Rosa is now generally considered to contain around 140 to 195 species[2]. The currently accepted taxonomic system was devised by Rehder[3] and revised by Wissemann[4]. The system contains four subgenera: Hesperhodos (two species), Platyrhodon (one species), Hulthemia (one species), and Eurosa (also named Rosa; 180 species). Subgenus Eurosa is further divided into 10 sections. Despite the large number and wide distribution of species, the origin of modern garden roses (R. hybrida) is believed to have originated primarily from 10 species in the subgenus Eurosa (Table 1)[5, 6]. Three more taxa believed to be involved in the origin of modern cultivars are likely hybrids, including R. damascena (R. gallica × R. moschata or R. phoenicea), R. alba (R. canina × R. gallica), and R. centifolia (R. ruba × R. moschata) × R. alba[6]. Because of the long history of hybridization and interbreeding cultivated varieties, the cultivated rose gene pool is relatively isolated from the wild gene pool[7].
Table 1. Previously published values for relative genome size and ploidy levels of Rosa species that contributed to modern cultivated R. hybrida.
Species Section Ploidy Relative genome size (pg) 1Cx genome size (pg) Reference R. canina Caninae 5 2.84−3.07 0.57−0.61 [1] R. chinensis Indicae 2 1.27 0.64 [1] R. foetida Pimpinellifoliae 4 1.95 0.49 [2] R. gallica Gallicae 4 2.20 0.55 [2] R. gigantea Indicae R. moschata Synstylae R. multiflora Synstylae 2 1.15 0.58 [1] R. phoenicea Synstylae R. rugosa Cinnamomeae 2 1.14 0.57 [1] R. wichuraina Synstylae 2 1.13 0.57 [2] Mean 0.57 ± 0.05 Genome size data from (1) Roberts et al.[13] and (2) Yokoya et al.[37] No published genome size data for species with blank categories. The base monoploid chromosome number in rose is seven[8]. Most R. hybrida-derived species are diploid (2n = 2x = 14) while R. gallica and R. foetida are tetraploid (2n = 4x = 28), and R. canina is pentaploid (2n = 5x = 35; Table 1). Although aneuploidy among garden roses may not be as rare as previously thought[9, 10], roses in the section Caninae provide the most instances of aneuploidy[11]. The nuclear DNA 1Cx-value in subfamily Rosoideae is approximately 0.66 pg[12], while the reported nuclear DNA 1Cx-values within Rosa range from 0.39 to 0.65 pg[13] and most species involved in the origin of modern roses contain 1Cx-values around 0.5 to 0.6 pg (Table 1).
Meiosis in rose varies based on the ploidy as well as homology between chromosomes[14]. Heteromorphic bivalents are often observed in cultivated roses, especially in tetraploid varieties[10]. Both interstitial and terminal deletions alter chromosome structures, which may play a substantial role in the genetic diversity among cultivated varieties. The presence of both heteromorphic bivalents and univalents significantly affect fertility in tetraploid cultivated roses, with a higher frequency of univalents or heteromorphic bivalents being positively correlated with pollen sterility[10].
While some modern rose cultivars are triploids or diploids, most elite cultivars are tetraploids. Ploidy differences between parents can affect both the cross-success rate and the resulting offspring's fertility. Unreduced gamete production has been observed at multiple ploidy levels in rose. Unreduced gametes in triploid genotypes led to the development of the first tetraploid Bourbon and Hybrid Tea roses[15]. Pentaploid and hexaploid progeny has resulted from crosses between triploids and tetraploids, indicating unreduced gamete production in the triploid and tetraploid parent, respectively[16]. Pollen size observations revealed likely unreduced gamete production in tetraploid cultivars 'Anna' and 'Sweet Promise'[17]. Pollen observation in a different study revealed possible 2n gamete production in 20% of diploids and 4% of tetraploids[18]. 2n pollen has also been observed in studies of dihaploid roses, potentially allowing breeders to more efficiently introgress wild, diploid germplasm into cultivated tetraploid gene pools[19, 20].
Species in the section Caninae are either tetraploid, pentaploid, or hexaploid and can exhibit an unusual form of meiosis as well as apomixis[21]. During their unique meiosis, the chromosomes form seven bivalents, with the remainder forming univalents. During microsporogenesis, the univalents are eliminated, causing the pollen to only carry seven chromosomes originating from the bivalents (n = 7). In megasporogenesis, a combination of bivalents and univalents form the egg cell (n = 2n − 7). This results in matrilineal inheritance among offspring[22]. Successful hybridization between Caninae species and species in the other sections depends on the direction of the cross as well as the number of chromosomes in each species[23].
Genome size can give insights into genome evolution and taxonomic relationships[24−28]. Also, genome size data can be used to estimate ploidy levels among closely related taxa when calibrated with cytogenetic work[29−33]. Flow cytometry provides an accurate and efficient method to determine relative genome size and ploidy level of plant tissues[34]. Previous reports on genome size in roses have focused primarily on species with little information for specific cultivars and breeding lines[13, 35−37].
Rosa hybrida is a diverse cultivated taxon with thousands of registered cultivars across dozens of horticultural classes[1]. However, limited cytogenetic information and lack of ploidy level knowledge can hinder breeding efforts. The objectives of this study were to determine genome sizes and estimated ploidy for a broad collection of rose cultivars and breeding lines and evaluate flow cytometry as a reliable method for determining ploidy in rose.
-
Flow cytometry was performed on 154 rose cultivars and 20 rose breeding lines to determine relative 2C holoploid genome size, 1Cx monoploid genome size, and ploidy estimates (Table 2). Of the accessions screened, we found that 11 were diploids (2n = 2x = 14), 24 triploids (2n = 3x = 21), and 139 tetraploids (2n = 4x = 28) (Table 2 and Fig. 1).
Table 2. Relative genome size and estimated ploidy determined using flower cytometry and chromosome counts for rose cultivars and breeding lines.
Taxa/cultivar Source/accession Relative 2C genome
size (pg ± SEM)Estimated ploidy
level (x)1Cx value
(pg)10037N017 TAMU 1.62 ± 0.05 3 0.54 10038N001 TAMU 0.98 ± 0.01 2 0.49 16015N005 TAMU 2.30 ±0.01 4 0.58 16502N012 TAMU 1.87 ± 0.06 4 0.47 17005N171 TAMU 1.52 ± 0.03 3 0.51 17010N049 TAMU 1.43 ± 0.05 3 0.48 17020N143 TAMU 1.41 ± 0.03 3 0.47 17035N035 TAMU 1.55 ± 0.02 3 0.52 17037N063 TAMU 1.38 ± 0.04 3 0.46 170400N082 TAMU 1.42 ± 0.05 3 0.47 17300HT9R5P093 TAMU 0.98 ± 0.02 2 0.49 18032HT9R2P82 TAMU 2.28 ± 0.03 4 0.57 19006HT9R5P018 TAMU 1.88 ± 0.05 4 0.47 19006HT9R5P019 TAMU 1.99 ± 0.06 4 0.50 19007HT9R2P080 TAMU 2.25 ± 0.09 4 0.56 19007HT9R5P050 TAMU 2.14 ± 0.07 4 0.54 19007HT9R5P112 TAMU 2.17 ± 0.09 4 0.54 19012HT9R5P137 TAMU 1.41 ± 0.03 3 0.47 19025HT9R8P166 TAMU 1.54 ± 0.03 3 0.51 1907HT9R5P069 TAMU 2.19 ± 0.04 4 0.55 About Face™ 'Wekosupalz' RRG 2.23 ± 0.01 4 0.56 Apricot Drift® 'Meimirrote' JCRA 2.20 ± 0.03 4 0.53 Artistry™ 'Jacirst' RRG 2.12 ± 0.05 4 0.53 At Last® 'Horcogjil' MHCREC 2.40 ± 0.10 4 0.60 'Belinda's Dream' Biltmore Estate 1.86 ± 0.02 3 0.62 Blushing Knock Out® 'Radyod' Biltmore Estate 2.28 ± 0.09 4 0.57 'Blushing Queen' Biltmore Estate 2.44 ± 0.03 4 0.61 Blush™ Veranda® 'Korfloci04' Biltmore Estate 2.45 ± 0.03 4 0.61 Brass Band™ 'Jaccofl' RRG 1.95 ± 0.004 4 0.49 Brigadoon™ 'Jacpal' RRG 2.16 ± 0.02 4 0.54 Carefree Beauty™ 'Bucbi' MHCREC 2.31 ± 0.04 4 0.58 Carefree Delight® 'Meipotal' MHCREC 1.72 ± 0.03 3 0.57 Carefree Spirit 'Maizmea' Biltmore Estate 2.42 ± 0.08 4 0.60 'Cecile Brunner' Biltmore Estate 1.16 ± 0.24 2 0.58 Cherry Parfait™ 'Meisponge' RRG 2.23 ± 0.05 4 0.56 'Chrysler Imperial' RRG 2.30 ± 0.07 4 0.58 Cinderella™ Fairy Tale 'Korfobalt' Biltmore Estate 2.42 ± 0.06 4 0.60 Claire Austin 'Ausprior' JCRA 1.98 ± 0.03 4 0.49 Crimson Bouquet™ 'Korbeteilich' RRG 2.18 ± 0.05 4 0.55 Crocus Rose 'Ausquest' JCRA 1.93 ± 0.03 4 0.48 Crown Princess Margareta® 'Auswinter' JCRA 1.90 ± 0.03 4 0.47 Darcey Bussell 'Ausdecorum' Biltmore Estate 2.38 ± 0.01 4 0.60 Day Breaker™ 'Frycentury' RRG 2.16 ± 0.05 4 0.54 Dick Clark 'Wekfunk' RRG 2.19 ± 0.05 4 0.55 Doris Day® 'Wekmajuchi' Biltmore Estate 2.35 ± 0.08 4 0.59 Double Delight® 'Andeli' RRG 2.03 ± 0.06 4 0.51 Double Knock Out® 'Radtko' MHCREC 2.40 ± 0.02 4 0.60 'Dr. Huey' MHCREC 1.86 ± 0.03 3* 0.62 Dream Come True™ 'Wekdocpot' RRG 2.13 ± 0.02 4 0.53 'Dublin' RRG 2.06 ± 0.03 4 0.52 Easy Does It® 'Harpageant' RRG 2.05 ± 0.03 4 0.51 Easy Elegance® All the Rage 'Bairage' JCRA 1.93 ± 0.01 4 0.48 Easy Elegance® Calypso 'Baiypso' JCRA 2.05 ± 0.03 4 0.51 Easy Elegance® Champagne Wishes 'Baicham' JCRA 1.96 ± 0.04 4 0.49 Easy Elegance® Chi™ 'Bailim' JCRA 1.61 ± 0.06 3 0.54 Easy Elegance® Como Park 'Baiark' JCRA 2.20 ± 0.08 4 0.55 Easy Elegance® Head over Heels 'Baieels' JCRA 1.53 ± 0.07 3 0.51 Easy Elegance® Kashmir 'Baimir' JCRA 2.03 ± 0.05 4 0.51 Easy Elegance® Music Box 'Baibox' JCRA 2.12 ± 0.03 4 0.53 Easy Elegance® My Girl 'Baigirl' JCRA 1.98 ± 0.02 4 0.50 Easy Elegance® Mystic Fairy® 'Baifairy' JCRA 1.59 ± 0.08 3 0.53 Easy Elegance® Paint the Town 'Baitown' JCRA 1.62 ± 0.01 3 0.54 Easy Elegance® Screaming Neon Red™ 'Baineon' JCRA 2.09 ± 0.08 4 0.52 Easy Elegance® Super Hero 'Baisuhe' JCRA 2.22 ± 0.04 4 0.56 Easy Elegance® Yellow Submarine 'Baiine' JCRA 2.09 ± 0.03 4 0.52 'Electron' RRG 2.15 ± 0.03 4 0.54 Elle® 'Meibderos' RRG 2.33 ± 0.08 4 0.58 Eureka™ 'Korsuflabe' RRG 2.22 ± 0.09 4 0.56 Fame!™ 'Jaczor' RRG 2.31 ± 0.05 4 0.58 Firefighter® 'Oradal' RRG 2.10 ± 0.03 4 0.53 First Editions® Campfire 'Ca 29' JCRA 2.11 ± 0.07 4 0.53 First Kiss 'Jacling' RRG 2.14 ± 0.003 4 0.54 Francis Meilland® 'Meitroni' RRG 2.26 ± 0.03 4 0.56 Funny Face™ 'Baiface' JCRA 2.16 ± 0.07 4 0.54 Gemini™ 'Jacnepal' RRG 1.99 ± 0.03 4 0.50 'Gene Boerner' RRG 2.17 ± 0.04 4 0.54 Gold Medal® 'Aroyqueli' Biltmore Estate 2.44 ± 0.01 4 0.61 Grand Champion™ 'Meimacota' JCRA 2.11 ± 0.07 4 0.53 'Hansa Rugosa' Biltmore Estate 1.10 ± 0.01 2 0.55 Home Run™ 'Wekcisbako' TAMU 2.21 ± 0.08 4* 0.55 Honey Dijon™ 'Weksproulses' RRG 2.22 ± 0.004 4 0.55 Honey Perfume™ 'Jacarque' RRG 2.10 ± 0.08 4 0.53 Hot Cocoa™ 'Wekpaltlez' RRG 2.20 ± 0.01 4 0.55 J06-20-14-03 TAMU 0.96 ± 0.02 2 0.48 Julia Child™ 'Wekvossutono' RRG 2.12 ± 0.02 4 0.53 Knock Out® 'Radrazz' MHCREC 2.23 ± 0.04 4* 0.56 'Kortersen' JCRA 2.22 ± 0.03 4 0.55 Lavener Veranda® 'Korfloci67' Biltmore Estate 2.47 ± 0.01 4 0.62 Lemon Fizz® 'Korfizzlem' TAMU 2.19 ± 0.05 4 0.55 Lichfield Angel 'Ausrelate' JCRA 2.09 ± 0.02 4 0.52 Limoncello™ 'Meijecycka' TAMU 1.74 ± 0.02 3 0.58 Lion's™ Fairy Tale 'Korvanaber' Biltmore Estate 2.31 ± 0.05 4 0.58 Look-A-Likes® Apple Dapple™ 'Meiplumty' JCRA 1.60 ± 0.03 3 0.53 'Louis Philippe' (China, Guérin 1834) JCRA 1.52 ± 0.02 3 0.51 Love and Peace™ 'Baipeace' RRG 2.07 ± 0.04 4 0.52 Lucetta 'Ausemi' Biltmore Estate 2.41 ± 0.04 4 0.60 Malvern Hills® 'Auscanary' JCRA 1.04 ± 0.04 2* 0.52 Marmalade Skies™ 'Meimonblan' RRG 2.29 ± 0.03 4 0.57 Mayor of Casterbridge 'Ausbrid' JCRA 2.02 ± 0.02 4 0.50 Melody Parfumee™ 'Dorient' JCRA 1.90 ± 0.05 4 0.48 Memorial Day™ 'Wekblunez' RRG 2.27 ± 0.07 4 0.57 Midas Touch™ 'Jactou' RRG 2.12 ± 0.04 4 0.53 Milestone 'Jacles' RRG 2.24 ± 0.07 4 0.56 Miracle on the Hudson® 'LGHR1' JCRA 1.98 ± 0.01 4* 0.49 'Mister Lincoln' RRG 2.14 ± 0.01 4 0.54 Moondance 'Meinivoz' RRG 2.09 ± 0.04 4 0.52 Mother of Pearl® 'Meiludere' Biltmore Estate 2.43 ± 0.02 4 0.61 Munstead Wood 'Ausbernard' JCRA 2.25 ± 0.06 4 0.56 'Mutabilis' MHCREC 1.27 ± 0.02 2 0.62 Neon Lights 'Jacout' RRG 2.31 ± 0.04 4 0.58 New Zealand 'Macgenev' RRG 2.38 ± 0.06 4* 0.60 Olympiad™ 'Macauck' RRG 2.23 ± 0.02 4 0.56 Opening Night™ 'Jacolber' RRG 2.28 ± 0.09 4 0.57 Oso Easy Double Red® 'Meipeporia' MHCREC 2.40 ±0.04 4 0.60 Oso Easy Hot Paprika® 'Farrowrsp' SMN 2.13 ± 0.05 4 0.53 Oso Easy Italian Ice® 'Chewnicebell' MHCREC 2019-021 2.32 ± 0.01 4 0.58 Oso Easy Lemon Zest® MHCREC 2.35 ± 0.02 4 0.59 Oso Easy Paprika® 'Chewmaytime' JCRA 2.19 ± 0.06 4 0.55 Oso Easy Peasy® 'Phyllis Sherman' MHCREC 1.28 ± 0.003 2 0.62 Oso Easy® Petit Pink 'Zlemarianneyoshida' MHCREC 2019-022 2.32 ± 0.07 4 0.58 Oso Easy® Double Pink 'Meiriftday' SMN 1.63 ± 0.06 3 0.54 Oso Easy® Mango Salsa 'Chewperadventure' MHCREC 2019-030 2.49 ± 0.08 4 0.62 Oso Easy® Urban Legend® 'Chewpatout' MHCREC 2019-020 2.31 ± 0.05 4 0.58 Oso Happy® Smoothie 'Zlecharlie' TAMU 0.96 ± 0.01 2 0.48 Pascali 'Lenip' RRG 2.15 ± 0.06 4 0.54 Peach Drift® 'Meijocos' JCRA 1.50 ± 0.04 3 0.50 Petite Knock Out® 'Meibenbino' JCRA 2.38 ± 0.06 4 0.59 Pink Double Knock Out® 'Radtkopink' MHCREC 2.28 ± 0.06 4 0.57 Pink Peace 'Meibil' RRG 2.01 ±0.09 4 0.50 'Pink Promise' RRG 2.23 ± 0.04 4 0.56 Pomponella™ Fairy Tale 'Korpompan' Biltmore Estate 2.41 ± 0.02 4 0.60 'Queen Elizabeth' RRG 2.15 ± 0.05 4 0.54 Queen of Hearts™ 'Korliolow' Biltmore Estate 2.42 ± 0.003 4 0.60 Queen of Sweden® 'Austiger' Biltmore Estate 2.50 ± 0.02 4 0.63 Rainbow Sorbet™ 'Baiprez' RRG 2.18 ± 0.01 4 0.55 Reminiscent™ Coral 'Bozfra221' SMN 2.29 ± 0.07 4 0.57 Reminiscent™ Crema 'Bozfra121' SMN 2.27 ± 0.02 4 0.57 Reminiscent™ Pink 'Bozfra021' SMN 2.23 ± 0.04 4 0.56 Ringo All-Star™ 'Cheweyesup' MHCREC 2.34 ±0.04 4 0.58 Ringo® Double Pink 'Chewdelight' SMN 2.17 ± 0.003 4 0.54 Rise Up Amberness™ 'Chewamberness' SMN 2.23 ± 0.04 4 0.56 Rise Up Lilac Days™ 'Chewlilacdays' SMN 1.53 ± 0.02 3 0.51 Rise Up™ Ringo® 'Chewgateway' SMN 2.12 ± 0.07 4 0.53 Rosa ×fortuniana MHCREC 1.26 ± 0.01 2 0.61 Royal Amethyst™ 'Devmorada' RRG 2.09 ± 0.02 4 0.53 Saint Patrick™ 'Wekamanda' RRG 2.33 ±0.02 4 0.58 Scentimental™ 'Wekplapep' RRG 2.07 ± 0.07 4 0.52 Scepter'd Isle 'Ausland JCRA 2.04 ± 0.04 4 0.51 Secret™ 'Hilaroma' RRG 2.07 ± 0.05 4 0.52 'Setina' JCRA 1.98 ± 0.04 4 0.50 Sexy Rexy® 'Macrexy' RRG 2.09 ± 0.06 4 0.52 Singin' in the Rain 'Macivy' RRG 2.26 ± 0.03 4 0.56 'Soaring to Glory' MHCREC 2.45 ± 0.003 4 0.61 'Spice' China Rose JCRA 1.10 ± 0.01 2 0.55 Strike it Rich™ 'Wekbepmey' RRG 2.26 ± 0.04 4 0.57 Sunny Knock Out® 'Radsunny' TAMU 1.76 ± 0.02 3 0.59 Sunorita® 'Chewgewest' SMN 2.21 ± 0.03 4 0.55 Sunrosa Red™ 'Zarsbjoh' JCRA 2.25 ± 0.02 4 0.56 Sunset Celebration™ 'Fryxotic' RRG 2.14 ± 0.04 4 0.54 Sunshine Daydream 'Meikanaro' RRG 1.91 ± 0.05 4* 0.48 Tahitian Sunset™ 'Jacgodde' RRG 2.24 ± 0.06 4 0.56 Tahitian Treasure™ 'Radtreasure' JCRA 2.09 ± 0.06 4 0.52 'Talisman' JCRA 1.94 ± 0.02 4 0.48 The Charlatan 'Meiguimov' JCRA 1.67 ± 0.04 3 0.56 'Tiffany' RRG 2.32 ± 0.11 4 0.58 Top Gun™ 'Wekmoridahor' TAMU 2.15 ± 0.02 4 0.54 Touch of Class™ 'Kricarlo' RRG 2.24 ± 0.07 4 0.56 Twilight Zone 'Wekebtidere' Biltmore Estate 2.41 ± 0.07 4 0.60 Voodoo 'Aromiclea' RRG 2.10 ± 0.02 4 0.52 Whisper™ 'Dicwisp' RRG 2.23 ± 0.04 4 0.56 White Drift® 'Meizorland' MHCREC 1.83 ± 0.01 3 0.61 White Knock Out® 'Radwhite' JCRA 1.98 ± 0.03 4 0.50 'White Queen' Biltmore Estate 2.26 ± 0.08 4 0.57 Wild Blue Yonder™ 'Wekisoblip' RRG 2.01 ± 0.04 4 0.50 Zaide™ 'Korparofe' Biltmore Estate 2.40 ±0.002 4 0.60 MHCREC: Mountain Horticulture Crops Research and Extension Center, Mills River, NC, USA. Biltmore Estate: Biltmore Estate Rose Garden, Asheville, NC, USA. JCRA: JC Raulston Arboretum, Raleigh, NC, USA. RRG: Raleigh Rose Garden, Raleigh, NC, USA. TAMU: Texas A&M University, College Station, TX, USA. SMN: Spring Meadow Nursery, Grand Haven, MI, USA. * Indicates ploidy level was confirmed with microscopy. 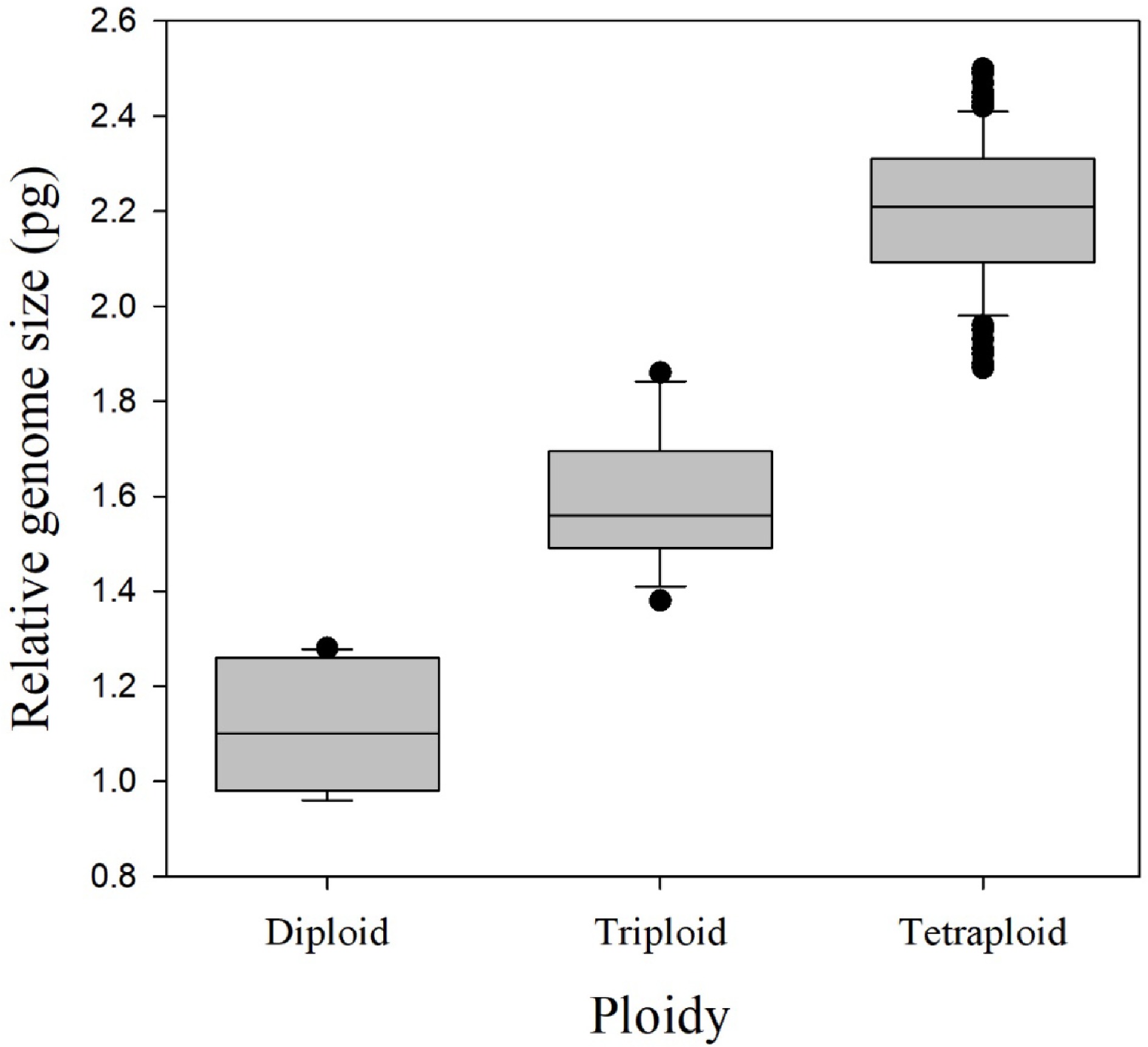
Figure 1.
Relative genome size for each of the three observed ploidy levels, for 174 rose species, cultivars, and breeding lines, determined by flow cytometry. Boxes display the lower and upper quartiles of the population with a horizontal line within the box showing the median. Vertical lines outside the boxes extend 1.5 times the interquartile range and data points outside of this range are considered potential outliers.
The mean 1Cx genome size for all cultivars was 0.55 ± 0.04 (SEM) pg with a range from 0.46 to 0.64 pg (Table 2). The 2C relative genome size range was 0.96–1.28 pg for diploids, 1.38–1.86 pg for triploids, and 1.87–2.50 pg for tetraploids, providing clear distinction between the genome size ranges of different euploid levels (Fig. 1). The mean relative genome sizes were 1.10 ± 0.13 pg, 1.59 ± 0.14 pg, and 2.20 ± 0.15 pg for diploids, triploids, and tetraploids, respectively.
Sixteen out of the 174 cultivars and breeding lines have had ploidy previously documented[38, 40]. Of these 16 accessions, our data confirm the previously reported ploidy of 10 accessions and dispute the ploidy of the other six accessions, i.e., Knock Out®, Home Run®, Miracle on the Hudson®, Blushing Knock Out®, Double Knock Out®, and Oso Easy Paprika®. Each of these six cultivars was previously reported as triploid (D. Zlesak, Per. Comm.)[38, 40]. However, the relative genome sizes determined by flow cytometry indicate that each cultivar is tetraploid (Table 2).
Chromosome counts were performed on seven of the accessions used for genome size determination including Knock Out®, Home Run®, Miracle on the Hudson®, Malvern Hills®, 'New Zealand', 'Dr. Huey', and Sunshine Daydream. Knock Out®[38, 40], Home Run®[38, 40], and Miracle on the Hudson® (D. Zlesak, Per. Communication) were chosen because of contradicting reports of ploidy between our flow cytometry estimates and previous reports. Malvern Hills® and New Zealand were chosen to calibrate relative genome size to ploidy for diploids and tetraploids, respectively (Table 2). 'Dr. Huey' and Sunshine Daydream were chosen to calibrate relative genome size to ploidy and help discern the transition point for relative genome size values for the largest triploid and the smallest tetraploid, respectively (Table 2). We found that Malvern Hills® was diploid, 'Dr. Huey' was triploid, and Knock Out®, Home Run®, Miracle on the Hudson®, 'New Zealand', and Sunshine Daydream were tetraploid (Fig. 2).
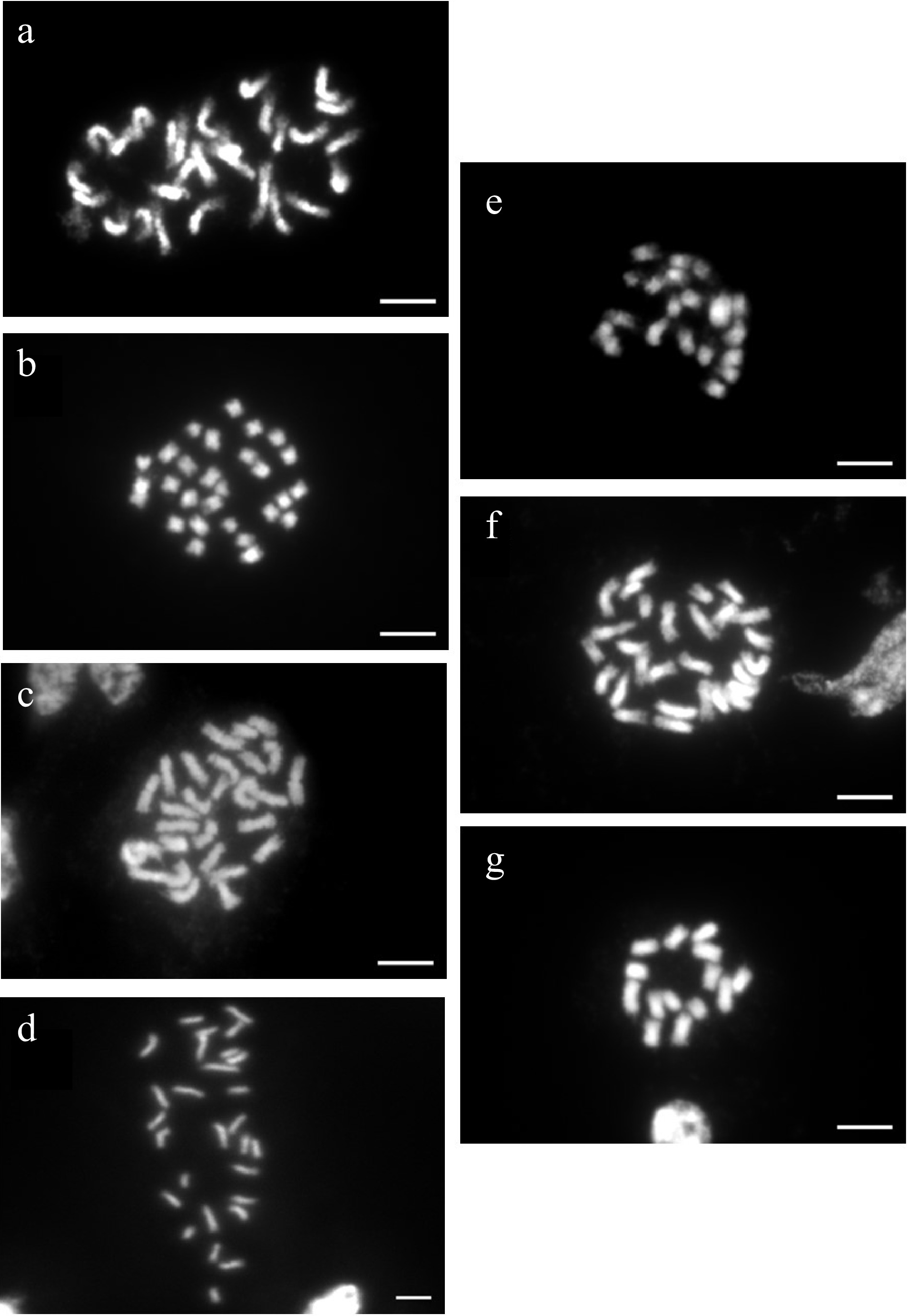
Figure 2.
Ploidy determination by counting chromosomes of metaphase cells of seven rose cultivars. Chromosome images of (a) tetraploid Miracle on the Hudson® (2n = 4x = 28), (b) tetraploid Home Run™ (2n = 4x = 28), (c) tetraploid New Zealand (2n = 4x = 28), (d) tetraploid Sunshine Daydream (2n = 4x = 28), (e) triploid 'Dr. Huey' (2n = 3x = 21), (f) tetraploid Knockout® (2n = 4x = 28), and (g) diploid Malvern Hills® (2n = 2x = 14). Bars = 10 µM.
-
Determination of ploidy using microscopy methods is tedious, challenging, and often difficult to obtain clear resolution of all individual chromosomes and accurate counts. Other indirect methods, such as pollen diameter and guard cell length[18, 40], can be correlated with ploidy but often lack the resolution to clearly differentiate between isoploids and anisoploids[40]. Furthermore, guard cell length only determines the ploidy level of the L-I histogenic layer, but gives no information about the L-II layer, from which pollen and egg cells are derived[41]. Although flow cytometry can be an effective method for estimating genome size and associated ploidy, this approach needs to be validated and calibrated among closely related species. Furthermore, variations in methodology including extraction buffers, fluorochrome stains, and internal standards can skew results from one study to another. For example, different fluorochrome stains may give slightly different estimates of genome size. Both propidium iodide (PI) and 4', 6-diamidino-2-phenylindole (DAPI) are effective and consistent for determining relative genome size and ploidy among closely related taxa[33] when methods are standardized.
This study found that the base genome sizes (1Cx) were relatively conserved in the roses tested and that 2C genome sizes could be used to distinguish among diploid, triploid, and tetraploid cytotypes. The mean 1Cx genome size for all cultivars included in this study was 0.55 ± 0.04 pg with a range from 0.46 to 0.64 pg (Table 2). The average 1Cx genome size for these cultivars aligned well with the mean 1Cx genome size for the 10 rose species that contributed to modern cultivars estimated to be 0.57 ± 0.05 pg (Table 1) even though both Roberts et al.[13] and Yokoya et al.[37] used Petroselinum crispum as a calibration standard and PI as a fluorochrome stain.
The gap in genomes sizes between triploids and tetraploids was relatively small with 1.38–1.86 pg for triploids and 1.87–2.50 pg for tetraploids. Because of this small gap between genome sizes for triploids and tetraploids, chromosome counts were completed to accurately identify ploidy for cultivars near this transition point. Using microscopy, we found 'Dr. Huey' (2C = 1.86 pg) to be triploid, and Sunshine Daydream (2C = 1.91 pg) to be tetraploid. For roses with genome sizes near this transition point, chromosome counts may be required to confirm ploidy.
In cases where our ploidy estimates differed from prior reports, our reassessment of chromosome numbers using microscopy (e.g., Knock Out®, Home Run®, and Miracle on the Hudson®) substantiated our estimates derived from flow cytometry. These discrepancies may be the result of mislabeling or incorrect identification of specific clones, cytochimeras, or the inherent difficulty in clearly visualizing and counting plant chromosomes.
Accurate ploidy-level assessment is essential to breeding species with a variety of ploidy levels. A cultivar's ploidy level can affect its fertility and potential usefulness in a breeding program. For example, Hibiscus syriacus cultivars 'Minerva', 'Diane', and 'Helene' were initially reported to be sterile, triploid cultivars (allo-hexaploid) when they were released by the US National Arboretum[42]. However, a recent flow cytometry analysis reported these cultivars found in nurseries to be fertile tetraploids[42]. The origin of the ploidy discrepancy, whether the result of initial misidentification, propagation of a mislabeled stock plant with various ploidy levels, or the reversion of a cytochimera, is unclear[42]. Flow cytometry is a useful, rapid tool for the determination of ploidy levels for the basis of breeding, ploidy manipulation, and uncovering past classification errors.
In addition to validating the application of flow cytometry for ploidy determination in roses, the results of this study provide an expanded database on the genome sizes and ploidy of 174 diverse rose cultivars and breeding lines. This research further contributes to the cytogenetic understanding of roses and a broader foundation for future breeding and improvement.
-
One hundred and fifty four rose cultivars and 20 rose breeding lines were included in this study (Table 2). Sixteen accessions with previously reported ploidy levels were included in this study to help calibrate genome size to ploidy level and to determine validity of previous reports[38−40]. Emerging flower bud tissue of these cultivars and breeding lines were collected from rose gardens, public breeding programs, and nurseries (Table 2). If emerging floral bud tissue was not available, emerging new leaf tissue was collected. Leaf tissue was collected from P. sativum 'Ctirad' grown in the greenhouse and used as the internal standard (2C DNA content = 8.76 pg[43]).
Flow cytometry
-
Relative genome size was determined using flow cytometry. Tissue samples were chopped using a double-sided razor blade in a 50 mm petri dish with 200 μL of extraction buffer (CyStain PI Absolute P; Sysmex Partec, Munster, Germany). P. sativum 'Ctirad' was chopped with Rosa tissue. The finely chopped tissues and buffer were poured through a 50 μm nylon-mesh filter into a test tube and stained with 800 μL 4',6-diamidino-2-phenylindole (DAPI) fluorochrome (CyStain ultraviolet Precise P Staining Buffer; Sysmex Partec). DAPI was used due to its speed, low toxicity, precision (low sample coefficient of variance), and repeatable results for determining ploidy[37]. Samples were tested in a completely randomized design using a flow cytometer (Quantum P; Quantum Analysis, Munster, Germany). 2C DNA content was calculated using the formula: DNA content of the internal standard × (mean fluorescence of sample / mean fluorescence of the internal standard). At least two replicates were used for each cultivar. The relationship between ploidy and genomic size was calibrated using roses in the same section with known ploidy as a reference[13].
Chromosome counts
-
Chromosome counts were conducted to confirm the relationship between genome size and ploidy[44]. Roots or young shoot tips of 0.5 cm in length were collected and placed in the pre-fixative solution containing 2 mM 8-hydroxyquinoline and 70 mg·L−1 cycloheximide in microcentrifuge vials. Vials were stored in the dark for 3 h at ambient room temperature and subsequently placed in a dark refrigerator at approximately 4 °C for an additional hour. Following the pre-fixative treatment, tissue was removed and rinsed thoroughly in deionized water. The tissue was then placed in Farmer's fixative solution containing 95% ethanol and glacial acetic acid (3:1 v/v) and left at 4 °C overnight or until tissues could be further processed.
Fixed tissues were removed from the fixative solution and rinsed with deionized water for 1 min per time in triplicate. After washing, the youngest, most tender tissues were placed on a microscope slide with 30 µL of a cell wall digestive enzyme mixture of 6% pectinase (Sigma Chemical; St. Louis, MO, USA) and 6% cellulase (Onozuka R-10; Yakult Honsha, Tokyo, Japan) in 75 mM KCl buffer (pH 4.0). Prepared slides with the cell wall digestive enzyme buffer were placed in an enclosed container with moistened paper towels and incubated at 37 °C between 3 h and overnight depending on the cultivar. The next day, the buffer was diluted with 1−3 drops deionized distilled water and any excess buffer was removed with a Kimwipe®. Then, two drops of a modified fixative buffer containing 95% methanol and glacial acetic acid (3:1, v/v) were added to the sample on each slide. Tissue was broken up using a small spatula, spreading the macerated tissue evenly on the slides in a circular pattern. Additional drops of the modified fixative buffer were added when needed.
Once the tissue was spread out, the extra fixative buffer was burned off and the slides were incubated at 37 °C overnight to be fully dried, then stored at room temperature. Dried slides were stained using Vectashield® Antifade Mounting Medium with (Vector Laboratories; Burlingame, CA, USA) with DAPI. Chromosomes of metaphase cells were counted using a compound microscope (Axio Imager.A2, Zeiss; Oberkochen, Germany).
The authors thank Raleigh Rose Garden, JC Raulston Arboretum, Spring Meadow Nursery, Biltmore Estate, and the Texas A&M Rose Breeding Program for sharing tissue for use in this study. The authors also thank Nathan Lynch for his technical assistance. This work was funded, in part, by the North Carolina Agriculture Research Service (NCARS), Raleigh, NC, the North Carolina Biotechnology Center, Research Triangle Park, NC, the Hatch project 02685 from the US Department of Agriculture National Institute of Food and Agriculture, and Spring Meadow Nursery, Grand Haven, MI, USA.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Harmon DD, Chen H, Byrne D, Liu W, Ranney TG. 2023. Cytogenetics, ploidy, and genome sizes of rose (Rosa spp.) cultivars and breeding lines. Ornamental Plant Research 3:10 doi: 10.48130/OPR-2023-0010
Cytogenetics, ploidy, and genome sizes of rose (Rosa spp.) cultivars and breeding lines
- Received: 27 December 2022
- Accepted: 06 March 2023
- Published online: 28 April 2023
Abstract: Roses (Rosa spp. L.) are valuable horticultural crops with global production, markets, and utilization. Cytogenetics of roses can be complicated with variation in ploidy among species and hybrids and occurrence of unreduced gametes, unbalanced (canina) meiosis, and aneuploidy. Most modern rose cultivars are complex, interspecific hybrids with unknown ploidy. Despite most breeding efforts being focused on crossing cultivated varieties, the genome size information is often only available at the generalized species level. The goal of this study was to survey cultivars and breeding lines to determine relative genome sizes and ploidy levels. Flow cytometry was used to determine relative genome size and ploidy levels of 174 accessions of shrub, hybrid tea, grandiflora, floribunda, polyantha, R. chinensis, and R. rugosa cultivars and breeding lines. Chromosome counts were performed to calibrate relative genome size to ploidy level and confirm previously published ploidy reports. 1Cx relative genome size ranged from 0.46 to 0.64 pg and the 2C relative genome size ranged from 0.96 pg to 1.28 pg for diploids, 1.38 to 1.86 pg for triploids, and 1.87 to 2.50 pg for tetraploids when using DAPI fluorochrome and Pisum sativum 'Ctirad' as the internal standard. Chromosome counts further substantiated these ranges and confirmed ploidy of cultivars that were in disagreement with earlier reports for three cultivars. These results provide an extensive database of genome sizes and ploidy for diverse cultivars and breeding lines of rose and establish/validate flow cytometry methods for future applications.
-
Key words:
- Rose /
- Ploidy /
- Rosa spp. /
- Cytology /
- Cytometry /
- Plant breeding /
- Cytogenetics












