-
Carbon catabolite repression (CCR) refers to the phenomenon in which microorganisms use up the more preferable carbon source before moving on to the secondary source, which is less preferable. The galactose utilization pathway (GAL) in yeast, for example, is triggered by the depletion of glucose in the culture to utilize other sources of energy, such as galactose[1,2]. The external galactose/glucose concentration ratio determines whether the GAL pathway is activated to control the energy source utilized by the microorganism in yeast cells[1,3]; this is known as ratio-sensing. Ratio-sensing is also responsible for how the cell protein resources are allocated[4]. A similar ratio-metric response can be seen in the bone morphogenetic protein (BMP) signaling pathway, whose function is the integration of competing signals in mammals[5]. A wide range of transporters play a role in yeast's ability to absorb sugar. HXT1 through HXT17 are the 17 members of the hexose transporter family (HXT) with different binding affinities for hexoses such as glucose[6]. One cell film galactose permease, Gal2p is equally partial in its restricting function for glucose and galactose[6,7], i.e., glucose and galactose could compete at the transporter level. The internal galactose sensor Gal3p is a potential additional mechanism. The transcription of GAL1, GAL2, GAL3, and GAL80, the GAL metabolic genes, are activated with the binding of Gal3p to galactose, releasing Gal4p from its sequestration by Gal80p[8−10]. The presence of glucose causes Mig1, its internal sensor, to repress Gal80p transcriptionally. The yeast culture-specific energy metabolic pathway indicates both the importance and viability of the utilization of galactose in the absence of glucose by the Gal pathway as a separate source of energy in yeast cells
In all kinds of cells, maintaining metabolic functions and cellular growth depends on gene expression and its regulation. S. cerevisiae's galactose utilization is controlled by cells. The GAL system controls both galactose induction and glucose repression. The genes in the GAL system produce the proteins that use galactose. The GAL system's induction and repression mechanisms have been extensively studied, and it is considered a model for eukaryotic transcriptional control. Because industrial media like lignocellulose and beet molasses contain galactose in the form of hemicellulose and raffinose, respectively, it is of commercial value to utilize the existing galactose by eliminating the glucose control on the galactose metabolism[11,12]. The overall process time will be reduced through reducing the lag phase between the use of glucose and galactose and the control of glucose on galactose metabolism.
-
Intracellular galactose uses an ATP-dependent mechanism to play the role of a signal molecule[13,14]. The interaction between Gal80, which downregulates the GAL system, and Gal3, a transducer protein, leads to the induction of the GAL system. The binding between the transcriptional activator Gal4 and Gal80 is permanent[15]. Gal4 initiates transcription in the GAL genes' promoter region by binding to an upstream activator sequence (UASG). The interaction between Gal3 and Gal80 frees the Gal4 activating domain from Gal80[16], leading to the expression of the structural GAL genes GAL1, GAL2, GAL7, and GAL10; As a result, galactokinase, UDP-glucose 4-epimerase, galactopermease, and galactose-1-phosphate uridylyltransferase (G1PUT) are synthesized. The transport of extracellular galactose across the cell membrane and the subsequent conversion of intracellular galactose to glucose-1-phosphate are both performed by these enzymes. Phosphoglucomutase encoded by GAL5 converts glucose-1-phosphate to glucose-6-phosphate. In the presence of galactose, the transcription of GAL1, GAL2, GAL7, and GAL10 increases by 1,000 times[17], whereas transcription of GAL5 only increases by three times[18].
S. cerevisiae is a single-celled fungus eukaryote. Similar to Escherichia coli, it is one of the most thoroughly studied eukaryotic model organisms in molecular and cellular biology. S. cerevisiae can survive in both unicellular and multicellular environments. This ability to thrive in different environments is ideal for population dispersal[19]. The preferred method of energy production in S. cerevisiae is fermentation. When glucose supply is insufficient, however, a shift to respiration is carried out using the ethanol produced from prior fermentation as a carbon source[20].
This ability results in an increased expression of genes for gluconeogenesis and a decrease in the expression of fermentation genes[21,22]. Several proteins, mainly the zinc cluster proteins, Cat8, Sip4, Rds2, as well as the Adr1 gene play a major role in this gene expression reprogramming. Studies have also shown Ert1 (ethanol-regulated transcription factor 1) to be a primary gene in gluconeogenesis regulation and fermentation[23].
-
Gal80 is a transcriptional repressor preventing Gal4p from activating the transcription that Gal1p or Gal3p have inhibited in the GAL genes in the absence of galactose[24]. It is a transcriptional repressor that regulates transcription in response to source of energy alteration[12]. Through a GAL upstream activation site (UAS-GAL) in their promoter, Gal3p, Gal4p, and Gal80p regulate the expression of the GAL metabolic genes GAL1, GAL10, GAL2, and GAL7. Even in media that do not induce gene expression, gal80-deficient cells continue to express GAL genes[8].
The Gal80 molecular function happens in the cytoplasm and nucleus. It enables kinase inhibitor activity and works on activating RNA polymerase II-specific DNA-binding sites. Additionally, it is involved in importing and metabolizing galactose by binding to their promoter regions, the transcriptional activator GAL4, which makes a set of enzymatic and regulatory genes active. At the point where galactose is the sole carbon source, the galactose-utilizing proteins are communicated at multiple times their level in the presence of glucose[17] (Fig.1).
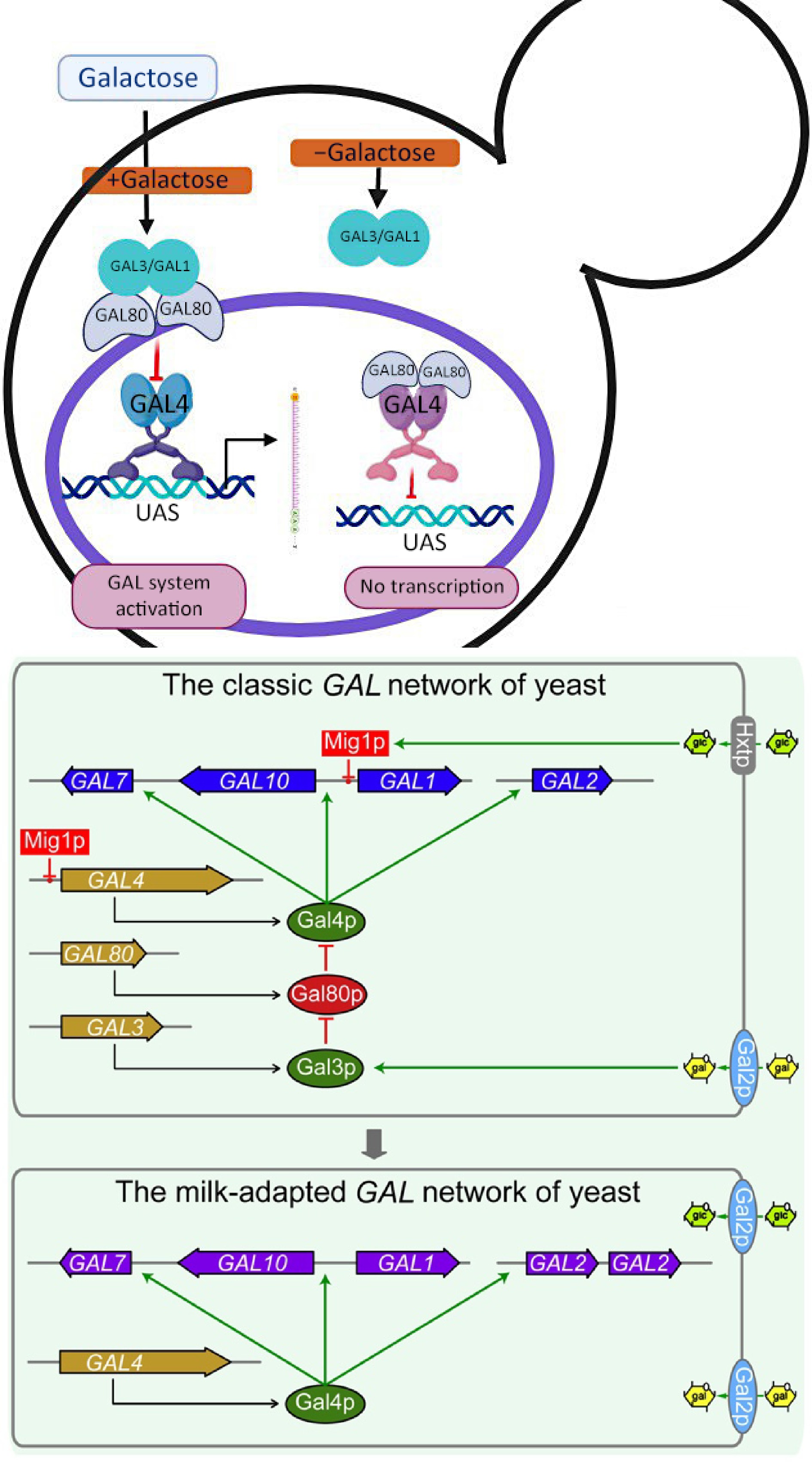
Figure 1.
Gal80 interaction with other genes in the GAL system. When galactose is present, it activates GAL3. GAL3 binds to GAL80 and detaches it from GAL4, and GAL4 initiates transcription. When galactose is not used as a source of energy, the GAL80–GAL3 interaction does not happen, and therefore, GAL80 inhibits GAL4 to start transcription[25].
-
This gene has 1,308 base pairs (bp) and is located between SUR7, a plasma membrane protein (upstream), and AIM32, a mitochondrial protein in redox quality control (downstream). There are two binding sites for the Gal80 protein in general yeast species: Gal4 and Gal3. In the absence of galactose, the GAL gene activator's (Gal4) transcriptional activation domain (AD) is inhibited by binding with Gal80, leading to the inhibition of GAL gene expression. Galactose relieves Gal80's inhibition of Gal4 by triggering an association between Gal3 and Gal80[15]. The Gal3p/Gal80p/Gal4p feedback mechanism also controls GAL80 transcription, as do the other GAL genes. However, GAL80's high basal transcription levels are due to having only one upstream activating site of the Gal gene (UAS-GAL) in its promoter. On the other hand, GAL structural gene expression is strictly regulated by multiple UAS-GALs in its promoters[26] (Fig. 2).
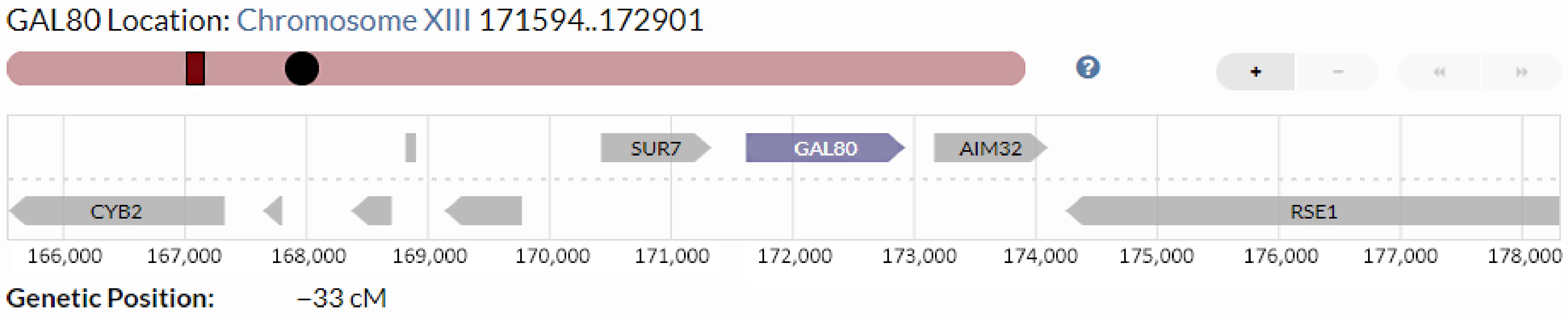
Figure 2.
Gal80 genetic position in the S. cerevisiae chromosome[27].
-
Certain GAL80 mutants cannot bind to Gal3 or Gal4. Although most Gal80 variants chosen for their impaired Gal3 binding show impaired self-association and binding to Gal4AD, mutants with impaired Gal4 binding maintain their Gal3 binding and self-association. Therefore, binding to Gal4 may require Gal80 self-association, and some Gal80 amino acids are determining factors for both Gal80 self-association and Gal80-Gal3 association. The competition between Gal3's binding with the Gal80 monomer and Gal80 self-association depletes Gal80 availability for Gal4 inhibition[26].
-
GAL4 activity is suppressed by forming a complex with the transcriptional repressor GAL80 (CPX-1044) when galactose is absent. The GAL1-GAL80 complex forms when galactose and ATP are present. This complex frees GAL80 from the GAL4-activation domain, allowing it to recruit the transcriptional machinery[28]. There is also the possibility of forming a tripartite complex (GAL4-GAL80-GAL1), which allows GAL4 to interact with promoters and counterbalance the influence of GAL80 on GAL4. GAL1 principally ties with ATP and galactose in the cytoplasm, and then moves into the nucleus to communicate with GAL80. GAL3-GAL80 (CPX-1042)[17], a paralogous complex transcriptional inducer of GAL genes with extensively higher activity, can also be formed. Although other evidence suggests that the complex is necessary for the continued expression of the GAL genes, the significance of GAL1-GAL80 is vague because GAL1 is not sufficiently expressed in the absence of galactose to function as an inducer[29].
-
In a study conducted by Cantone et al., IRMA, a synthetic network of five genes regulating one another 'for assessing reverse-engineering and modeling approaches', was built in S. cerevisiae. The cells were cultured in either a galactose or glucose medium. The network is active in the presence of galactose but deactivated by the Gal80-Gal4 interaction when grown in a glucose medium. It was found that the over-expression of Gal80 resulted in its binding to and repression of Gal4, even in the presence of glucose, and the downregulation of all the other genes (CBF1, SWI5, ASH1)[30].
-
The availability of galactose and glucose influences the transcriptional regulation of the yeast GAL and GAL genes, which are necessary for galactose metabolism. The transcription of these genes cannot be detected in yeast cells grown without galactose, but when galactose is present, it increases more than four-fold[31,32]. The activity of numerous negative and positive control components determines this uncommon acceptance proportion. These components are situated in the UASG, i.e., the 365-bp upstream activation sequence[33−35]. GAL4, an important transcription activator protein that binds to four related sites in the UASG control the rate of transcription when galactose is available[33−37]. Although in the absence of galactose GAL4 stays attached to UASG sites, GAL80 prevents it from initiating transcription[38]. Additionally, GAE1 and GAE2, the two GAL4/galactose-independent activating elements are inhibited by the negative control elements in the UASG, i.e., the GAL operators GALO1 to GALO6[37,39].
It is unknown how the GAL operators inhibit transcriptional activation. Although they activate transcription independently of GAL4 and without the requirement for galactose, similar to the GAL operators, GAE1 and GAE2 are located close to or overlapping the GAL4 binding sites. GAE1 and GAE2 reduce the basal-level transcription from the GAL promoter 20-fold compared to when the GAL operators located between the GAL4 sites and the GAL TATA element are removed and transcription is fully induced[32].
Even though we do not fully understand the physiological functions of GAE1 and GAE2, their inhibition by the GAL operators and the maintenance of the inducibility of GAL and GAL10 transcription are clearly necessary. In the fully induced levels when galactose is used alone in the yeast cell medium, GAL genes are repressed several hundred times more compared to when both galactose and glucose are available in the medium[40−44]. At least three distinct pathways control GAL gene repression by glucose. Reduced transport of galactose into the cell occurs through one pathway known as inducer exclusion[43,44]. There is a second pathway that reduces the apparent attraction of GAL4 to its UASG binding sites[42]. The UASG cis-acting sequence or sequences are necessary for a third, poorly understood pathway[41]. For instance, the deletion of the sequences between the GAL TATA element and GAL4-binding sites causes the GAL promoter to experience experiences 70 times less glucose repression[31].
The mechanism that GAL4 uses to start transcription is not well repressed by GAL operators. GAL4 is, in fact, susceptible to GAL operator repression, but to a smaller degree compared to other activators. Moreover, it has been demonstrated that GAL operator repression cannot be overcome without GAL4's high synergistic activating potential for the four binding sites in the UASG. GAL4 activity is reduced in the presence of glucose, and GAL10 and GAL1 transcription induction may be further repressed by GAL operators. As a result, there is likely to be at least one mechanism that acts in the cis for glucose repression. The negative and positive control element functions compete directly for transcriptional control of the GAL1 and GAL10 genes, as well as possibly other eukaryotic genes[45].
As mentioned before, Gal4 is a positive controlling transcription factor of gene expression of galactose-prompted genes[46]. This protein addresses a huge group of genes specific for yeast-positive transcription factors, the Gal4 family, which holds more than 50 factors including Oaf1, Pip2, Pdr1, Pdr3, and Leu3[47]. The activation of the GAL80 gene pathway directly inhibits the action of the GAL4 protein, and in the induction process, galactose plays the role of counteracting GAL8O; Galactose is not required for induction in Gal80 strains, and these mutants cannot induce GAL expression[48,49]. Regardless of the GAL80 allele, glucose significantly inhibits GAL4-directed transcription.
-
GAL70 and GAL7 gene transcription is activated when the GAL4 protein binds to four related 17-bp symmetric dyad sequences within the UASG. GAL4-protein-subordinate security of guanines in the UASG that yeast's GAL4 protein binds to upstream activation sites. The pattern observed in the GAL4-protein-dependent methylation protection supports the theory that the four connected dyad-symmetric 17 bp UASG sequences, which are the GAL4 protein's action locations, are recognized by the GAL4[47,50].
The manner of change of UASG sensitivity to methylation by DMS in yeast cells that are grown on glucose as their carbon source does not show dependence on GAU. This suggests that inhibiting the binding of the GAL4 protein to the UASG is one of the ways glucose is suppressed[36]. GAL4 is down-regulated by GAL80. A dimer of GAL80 binds to a GAL4 dimer causing its inhibition regardless of GAL4 being bound to the UAS, it can no longer activate transcription in the presence of GAL80. Gal4 coordinately regulates the transcription of GAL1, GAL2, GAL7, and GAL10 (galactose structural genes) in response to galactose. The Gal4p transcriptional activator binds as a dimer to the GAL gene promoter's upstream activation sites, whatever the source of carbon[51].
-
The yeast two-hybrid system is among the simple in-vivo genetic methods for detecting protein interactions. The development of these methods came as the result of awareness of activation (AD) and DNA-binding (DBD) domains in eukaryotic transcriptional activators that act on fusion with heterologous proteins[37]. A functional trans-activator complex is formed when DBD (bait) fusion and AD (prey) fusion proteins interact. Reporter genes with upstream cis-elements associated with the DBD are activated by this complex. The limitation on the use of transcriptional activators as baits is one of these strategies drawbacks. TUP1, the yeast general repressor protein, is recruited by gene-specific DNA-binding proteins to special promoters in a complex including the SSN6 co-repressor. In several studies, TUP1 was fused to Gal80 to see the effect of Gal4 expression, which, as expected, down-regulated the expression of the Gal4 reporter gene. This system is generally used in various organisms such as fruit flies, Aenorhabditis elegans, and Drosophila, among others.
Application of GAL80 in molecular research and biotechnology
-
Researchers utilize Gal80 mutants to dissect the regulatory networks that govern the GAL system and investigate how changes in the Gal80 function impact cellular function of galactose and glucose stimuli. Since Gal80 acts as a transcriptional repressor that controls the activity of the Gal4 transcription factor in response to glucose and galactose availability. Overexpression constructs of GAL80 would be useful to manipulate the activity of the GAL system and study its effects on target gene expression. By modulating GAL80 expression levels, researchers can dissect the regulatory mechanisms governing the GAL pathway and elucidate the signaling cascades and transcriptional network involved in cellular responses to changing environmental conditions. Also, Gal80 is a valuable model for studying protein-protein interactions (can be detected by yeasy-to-hybrid) as mentioned before and the dynamic interplay between transcriptional repressors and activators.
Biofuel production and the manufacturing of pharmaceuticals are just two out of several application of GAL80 in industry which often rely on engineered microbial strains capable of efficiently utilizing renewable carbon sources like galactose. GAL80-based genetic switches can be integrated into synthetic gene circuits for applications in synthetic biology and metabolic engineering. These engineered genetic circuits enable the precise control of gene expression in synthetic biological systems, offering new opportunities for designing novel biosensors, metabolic pathways, and gene therapies. By leveraging the Gal system's regulatory mechanisms, researchers can engineer microbial hosts with enhanced galactose utilization capabilities, thus improving the efficiency and sustainability of bioproduction processes. Furthermore, the Gal system's responsiveness to environmental cues makes it a promising target for metabolic engineering strategies aimed at optimizing bioproduction pathways and maximizing product yields.
Evolutionary conservation and homologs in other species
-
The Gal80 protein, known for its pivotal role in regulating the GAL system in S. cerevisiae, exhibits remarkable evolutionary conservations and homologies across diverse species. Extensive comparative genomic and phylogenetic analyses have revealed intriguing insights into the evolutionary history and functional significance of Gal80 orthologs in other organisms. Studies across fungal species have unveiled the widespread conservation of Gal80 orthologs, indicating its fundamental role in cellular physiology. Orthologs of Gal80 have been identified in various fungal taxa, including closely related yeasts such as Candida species and distantly related filamentous fungi like Neurospora crassaN. This conservation suggests a conserved regulatory mechanism for galactose metabolism across fungal lineages. Beyond fungi, homologs of Gal80 have been identified in higher eukaryotic organisms, such as plants and animals. In plants, orthologs of Gal80-like proteins have been characterized in model organisms such as Arabidopsis thaliana, where they play roles in sugar sensing and carbohydrate metabolism. Similarly, in animals, Gal80 homologs have been identified in diverse species ranging from insects to mammals, suggesting an evolutionarily conserved function in glucose and galactose homeostasis.
Comparative analyses of Gal80 orthologs provide valuable insights into the functional conservation and divergence of this regulatory protein. Additionally, evolutionary studies shed light on the co-evolution of Gal80 with other components of the GAL system and its integration into broader regulatory networks governing metabolism and stress responses. Leveraging insights from evolutionary studies, researchers can design novel strategies for metabolic engineering, gene expression control, and therapeutic interventions.
-
The GAL system is particularly useful in the two-hybrid technique due to its molecular methodology enabling immediate access to the gene encoding the interacting protein of interest. The tests' being in vivo allows the detection of interacting proteins in their native states. In yeast, GAL4 and GAL80 are responsible for galactose metabolism. GAL4 binds to at least 20 galactose genes (including GAL1, GAL10, and GAL3) in the presence of galactose. This binding is repressed via GAL80 in the absence of galactose by binding to GAL4 and inhibiting its transcription ability. Transcription of glucose renders all transcriptions of GAL genes inactive. The modular properties of GAL genes have prompted their use in the 2-hybrid method.
Limitations
-
One notable research gap lies in the comparative genomics of Gal80 orthologs across fungal species and other eukaryotes. While studies have identified homologs of Gal80 in various organisms, there is limited information regarding the evolutionary trajectories, structural variations, and functional adaptations of these orthologs. Additionally, the functional diversity of Gal80 proteins in different species remain poorly understood, with potential implications for cellular metabolism, stress responses, and developmental processes.
Future directions
-
Future studies should aim to address these research gaps by employing integrative approaches that combine comparative genomics, functional genomics, and biochemical analyses as well as protein interaction assays. Comparative genomic studies can elucidate the evolutionary relationships and conservation patterns of Gal80 orthologs across phylogenetically diverse taxa, providing insights into the ancestral functions and evolutionary innovations of this regulatory protein. Furthermore, functional genomics approaches, such as gene expression profiling and functional assays can unravel the diverse roles of Gal80 in cellular processes and regulatory networks.
-
The authors confirm contribution to the paper as follows: study conception and design: Tabaraei H, Salajegheh Tazerji S, Shahbinejad F; data collection: Tabaraei H, Salajegheh Tazerji S, Shahbinejad F, Mojtahedzadeh SM; analysis and interpretation of results: Tabaraei H, Salajegheh Tazerji S, Shahbinejad F, Sabbaghi SS, Diansaei M, Ghataei K; draft manuscript preparation: Tabaraei H, Salajegheh Tazerji S, Hajipour P, Shafiei M, Khalili MH, Karimaghaei M, Shahabinejad F, Nikaein D, Vazir B, Kabir F, Amraee G. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analyzed during this study are included in this published article.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Tabarraei H, Mojtahedzadeh SM, sabbaghi SS, Hajipour P, Shafiei M, et al. 2024. Molecular characteristic of yeast Gal80 as a transcriptional factor: a mini-review. Studies in Fungi 9: e008 doi: 10.48130/sif-0024-0008
Molecular characteristic of yeast Gal80 as a transcriptional factor: a mini-review
- Received: 04 August 2023
- Revised: 09 May 2024
- Accepted: 21 May 2024
- Published online: 16 July 2024
Abstract: The Gal80 gene is a transcriptional regulator of the Saccharomyces cerevisiae Gal4 transcriptional activator. In response to galactose, it binds to and renders Gal4 inactive, preventing it from activating transcription. In this paper the regulation of the Gal system and its induction and repression mechanisms are discussed, emphasizing the Gal4 and Gal80 genes and their interaction with one another. Gal80 is discussed in more detail, elaborating on its regulation through the feedback mechanism via the Gal3/Gal80/Gal4 genes, its structure and mechanism of action, functions in the cytoplasm and nucleus, post-translational modifications, role in DNA and protein stability, and its evolutionary conservations and homologies in other species. The variations and complexes of Gal80 are also discussed. The yeast-two-hybrid system is explained and its role is elaborated on in identifying a large number of protein interactions in vivo and the role of Gal genes in giving rise to this technique and being a major system in the 2-hybrid technique.
-
Key words:
- Gal80 /
- Yeast /
- Galactose utilization /
- GAL system /
- Saccharomyces cerevisiae












