-
Global demand for cash-crop products has been a major driver of land use/land cover change in the tropics, and the expansion of these kinds of cash crops has come largely at the expense of tropical forestland[1,2]. Since the mid-20th century, the tropical forests of Southeast Asia have witnessed a massive conversion to tree cash crops, such as oil palm (Elaeis guineensis) and rubber (Hevea brasiliensis) monocultures[3−5]. Xishuangbanna Prefecture is located in southwestern China, bordering Laos and Myanmar. Rubber trees were introduced to Xishuangbanna in the 1940s and rapidly increased their area from1980−2020[5−7]. In the last thirty years, 50% of natural vegetation has been lost, with more than 82% of the land being replaced by rubber plantations[5]. The area increasing of rubber plantations in Xishuangbanna has significant influences on local ecological, social and economic systems as well as food security, and cultural practices[8−11]. The land-use change from mixed native forest to plantation monoculture alters soil quality, including soil organic matter content, carbon, nitrogen pools, and biomass[12−16]. However, few studies have been conducted regarding the consequences of land-use change on soil microbial communities.
Investigating the impacts of land-use change from natural vegetation to plantation on soil microbial communities is vital to understanding the consequences of the recent large-scale land-use changes in tropical forests, forming the bedrock of future forest restoration. Soil microorganisms are the driving force behind nutrient supply in soils, and any kind of environmental anthropogenic perturbation can lead to distinct effects on the abundance, composition, and activity of microorganisms responsible for nutrient transformations in soils[17]. Microbes played as soil biomarkers to indicate the nutrient transformation and availability in the soil ecosystem[18−19]. Numerous studies have been carried out on soil microbial community properties of disturbed forests in temperate and tropical regions, including land-use change and chronic fertilizer additions[20−24].
Most former research on soil microbial diversity focused on a single microbial domain. However, fundamental differences in physiology, ecology, and function have been found not only between bacterial and fungal communities but also among different microbial sub-groups, suggesting that patterns of each group are controlled by separate biotic and abiotic factors[25]. Bacteria play an important role in soil carbon and nutrient cycles[26]. Several studies have documented that soil bacterial communities are sensitive to variations in soil pH, soil C-to-N ratio, moisture, and temperature[27−29]. In contrast to bacteria, soil fungi play a critical role in plant litter decomposition. Consequently, the diversity and composition of soil fungi are commonly affected by the aboveground plant community. As a predominate fungal group, basidiomycetes have primary roles in carbon decomposition due to their ability to produce a series of enzymes. Therefore, soil basidiomycetes might be more sensitive in their response to the conversion, potentially capable of serving as an indicator of soil health.
To date, few studies have been conducted to investigate changes in soil bacterial and fungal communities following the conversion of tropical forest to a native plantation forest. Therefore, in this study, we utilized denaturing gradient gel electrophoresis (DGGE) analyses to investigate soil bacterial and fungal communities in three native tropical rainforests and adjacent rubber plantation monoculture forests in Xishuangbanna, southwestern China. The main objective of this study was to assess the influence of land-use from natural forests to plantations on soil bacterial and fungal communities, including main decomposer communities.
-
The study sites were located in the Xishuangbanna Tropical Botanical Gardens in Yunnan Province of China. This region receives an average annual precipitation of 1,560 mm, 80% of which occurs between May and October (wet season). The average annual temperature is 21.4 °C. The study area is located at 21°56 N, 101°15 E with an elevation of 550 m asl. This region has experienced large-scale deforestation over the last several decades, and it is common practice to convert forestland to agricultural land or rubber plantation. The results of previous studies on soil characteristics are presented in Table 1. The seasonal tropical rainforest (SR, dominated by Pometia tomentose and Terminalia myriocarpa) and rubber plantation (RP, Hevea brasiliensis) were selected to conduct the field experiment. The distance between the two stands was about 500 m, and there was no significant difference in rainfall characteristics or geological properties between the stands. Horizon soil was sampled by removing litter and collecting two soil cores (25 mm diameter 100 mm depth) every 2 m along three 10 m transects run randomly through each of the stands (SR1-3 and RP1-3). Soil cores were immediately transported to the laboratory and stored at 4 °C for < 4 days before sieving. The two soil cores from each sample point were then mixed to form a single soil sample, which was then sieved (< 2.0 mm) to remove large pieces of organic matter and stored at −80 °C.
Table 1. The soil characteristics of seasonal rainforests (SR) and rubber plantations (RP) of Southwest China.
SR RP Organic C (g kg−1) 26.4 ± 1.2* 19.9 ± 1.5 Organic N (g kg−1) 2.4 ± 0.1 1.6 ± 0.1 Inorganic N (mg kg−1) 9.1 ± 0.6 7.1 ± 0.7 C/N 11.0 ± 0.3 12.4 ± 0.3* PH 5.8 ± 0.1* 4.7 ± 0.1 Soil aggregate
distribution> 2 mm 59.0 64.2* 0.25−2 mm 12.3* 6.8 0.25−0.053 mm 11.9* 7.0 < 0.053 mm 16.7 22.0* LOC (g kg−1) 1.4 ± 0.1* 0.9 ± 0.1 MBC (g kg−1) 0.9 ± 0.1* 0.6 ± 0.1 Litter dry weight (kg ha−1) 3,000−5,000 5,000−7,000 * indicated p < 0.05. DNA extraction and DGGE analysis
-
DNA extractions were performed on 0.5 g of soil samples using the Ultra CleanTM Mega Prep Soil DNA kit (Mo Bio Labs, Solana Beach, CA, USA) following manufacturer protocol. The purified DNA was detected by agarose gel electrophoresis. The purity and usefulness of the DNA samples were ultimately determined by successful PCR amplification.
Bacterial partial 16S rRNA genes were amplified using the primers 341f-GC and 907R[30]. The fungal partial 18S rRNA genes were amplified from community DNA by PCR using general fungal primers FR1-GC and FF390[31]. The Basidiomycota ITS rRNA genes were amplified from community DNA by PCR using the general fungal primers 4B and ITS3-GC[32,33]. PCR was conducted in 25 ml reactions using final concentrations of 0.1 mM for each primer, 0.2 mM for each dNTP, 0.1 mg ml−1 for BSA, 1.5 mM for MgCl2, 1 × PCR buffer, and 0.02 U ml−1 Expand High-Fidelity DNA polymerase (Roche Diagnostics, Indianapolis, IN); 5 ml of diluted genomic DNA was added to each PCR. Samples were diluted (1:50−1:200) to achieve optimal PCRs for each sample (strong PCR product but no background smear of DNA in an agarose gel stained with ethidium bromide). Reactions were performed using the following program: 5 min at 95 °C, followed by 35 cycles of 45 s at 95 °C, 45 s at 50 °C, and 2 min at 72 °C, followed by 10 min at 72 °C. The PCR processes for each primer set were modified according to the related paper. Optimal PCRs were run in triplicate and pooled.
The PCR was performed as the above process by changing conditions according to references listed in Table 1. DNA concentration was quantified in pooled PCRs using a microplate PicoGreen assay following manufacturer instructions (Invitrogen, Carlsbad, CA). 100 ng of PCR product was subjected to DGGE in a DCode Universal Mutation Detection System (Bio-Rad, Hercules, CA) according to Table 1. An aliquot of 100 bp ladder (100 bp PCR Molecular Ruler) (Bio-Rad) was also run in three lanes of each gel. Gels were stained with SybrGreen (Invitrogen) and digitally photographed with a UV Transilluminator Box (Bio-Rad). Denaturing gradient gel electrophoresis was performed twice using different sample combinations in the second set of gels to facilitate aligning lanes between gels. Gel images were analyzed in Quantity One (Bio-Rad). Duplicate DGGE profiles were used to verify band presence and alignments. A matrix of relative band intensity was exported from Quantity One, with samples as rows and band categories as columns.
DGGE profiles were analyzed using CANOCO 4.5. The diversity of microbial communities at respective habitats was estimated based on the number and relative intensity of DGGE bands. Phylotype (band) richness was evaluated from totally different DGGE bands. Multivariate analyses were done using canonical correspondence analysis (CCA) for treatments and community composition. To assess the effects of treatment and environmental factors, DGGE analysis profiles were analyzed by CCA of Hellinger distances, using 9,999 random permutations of sample identity. Statistical differences among DGGE profiles were assessed using multi-response permutation procedures[34] in PC-ORD software. NMDS analysis used PC-ORD to show differences across all sites. The multiple Response Permutation Procedure (MRPP) is a nonparametric procedure like MANOVA for testing the hypothesis of no difference between two or more pre-existing groups[35]. MRPP yields a P-value to evaluate how likely it is that an observed difference is due to chance, as well as the chance-corrected within-group agreement (A), which describes within-group homogeneity compared to random expectation[35]. An A value equal to 1 is found when all items within a group are identical; when heterogeneity within groups equals expectation by chance, A = 0. In instances where there were no significant differences among plots with roots (p > 0.1), these treatments were combined and compared with the rootless treatments in a two-way analysis.
-
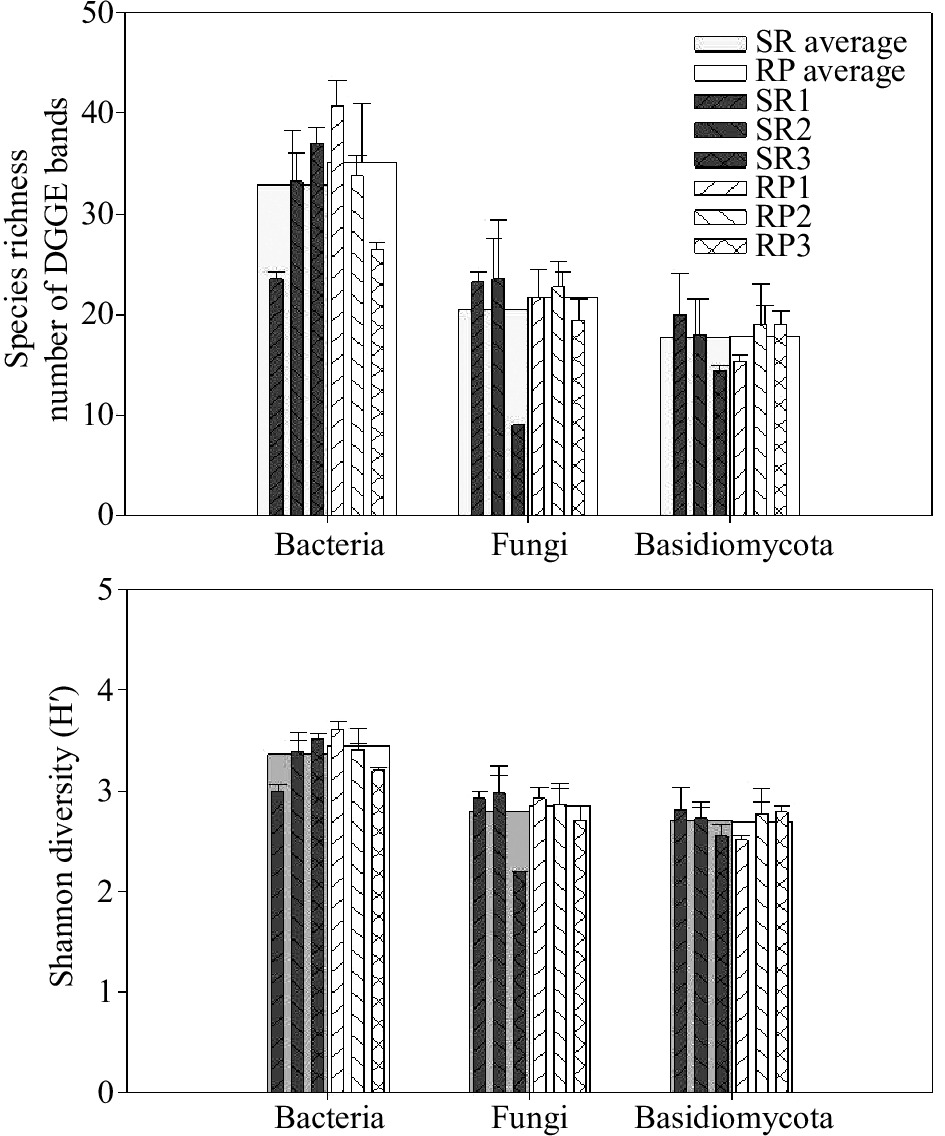
Denaturing gradient gel electrophoresis (DGGE) was used to examine differences in microbial community composition between rubber plantations and natural forests. Species richness, as calculated from the number of the bands, was slightly higher on average in rubber plantations than natural forests. However, differences between pairs of SR-to-RP were significant and contrasting. For example, SR1 was marked richer in bacterial species than RP 1, while the opposite trend was found between SR3 and RP3 (Fig. 1). When CCA analysis was used to test for differences between all forests, general bacterial community composition did differ significantly between rubber plantation and natural forest (p < 0.01) and significantly interacted with sites (p < 0.01) (Table 2). Patterns of the bacterial communities were separated into two clusters by the NMDS analysis: rubber plantations and primary forests (Fig. 2). The pairwise comparisons results using MRPP revealed that all the SR forests were significantly different from NR forests within sites. When comparing different sites, bacterial community composition was found to be significantly different in many comparisons, except for two pairs: SR1 vs. RP3 and SR3 vs. RP2 (Table. 3).
Table 2. Pairwise comparison of microbial communities between seasonal rain forest (SR) and rubber plantation (RP), or among the sample forest type.
Bacteria Fungi Basidiomycota SR1 vs. RP1 * ** NS. SR2 vs. RP2 ** ** NS. SR3 vs. RP3 * NS. ** SR1 vs. RP2 * ** ** SR1 vs. RP3 NS. * * SR2 vs. RP1 ** * NS. SR2 vs. RP3 * * * SR3 vs. RP1 * * NS. SR3 vs. RP2 NS. * NS. SR1 vs. SR2 * NS. NS. SR2 vs. SR3 * * * SR3 vs. SR1 * NS. * RP1 vs. RP2 * * NS. RP1 vs. RP3 * * ** RP2 vs. RP3 * * * * Indicated p < 0.1; ** indicated p < 0.5; *** indicated p < 0.01. 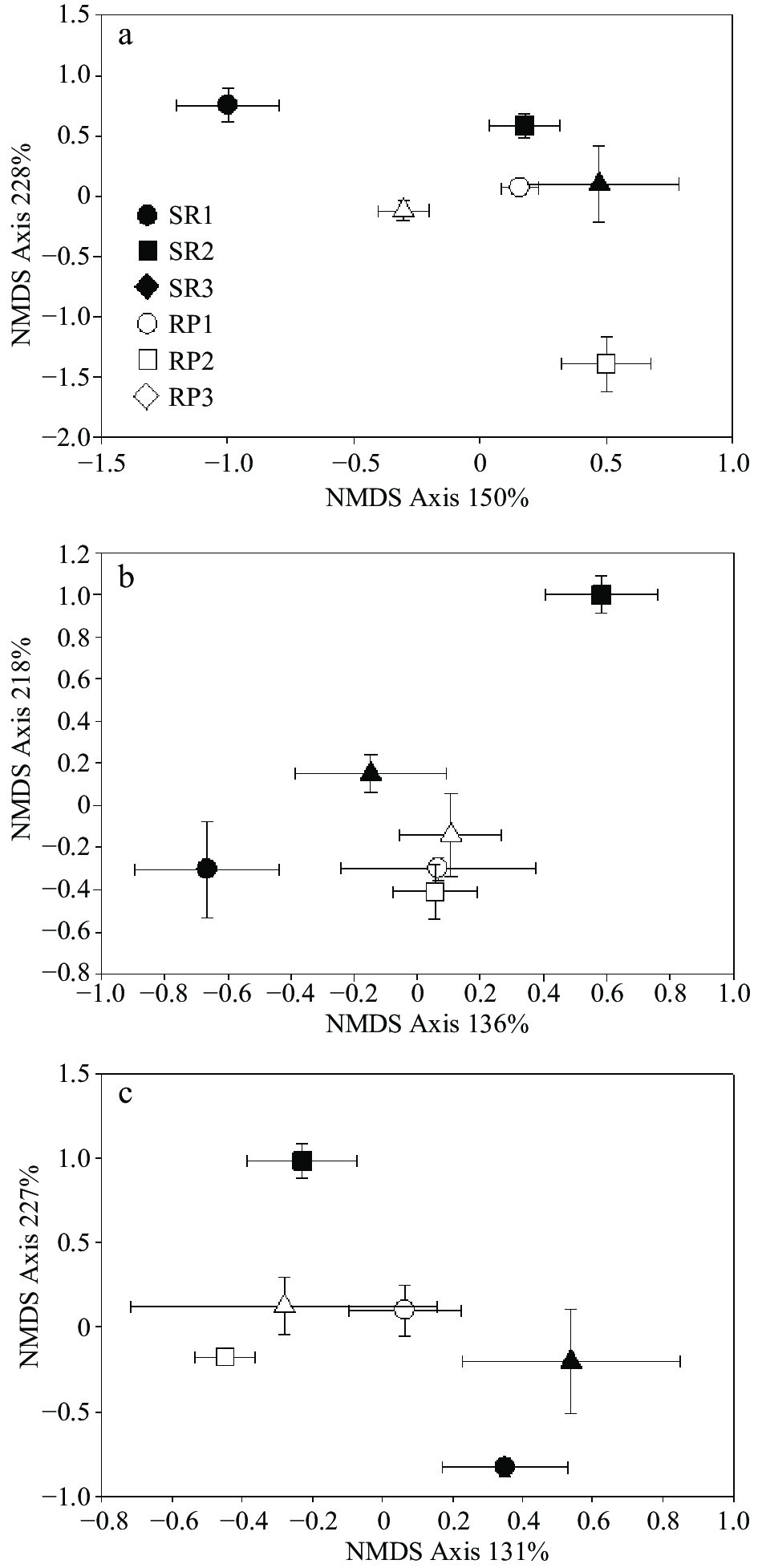
Figure 2.
NMDS analysis of (a) bacterial, (b) fungal and (c) Basidiomycota community. The empty and solid points indicate rubber plantation and rainforest samples. Circle, square and triangle indicate paired rubber plantation vs. rainforest (replicated locations 1, 2, 3).
Table 3. CCA analysis of microbial communities between seasonal rain forest (SR) and rubber plantation (RP), or among the sample forest type.
Bacteria Fungi Basidiomycota Effect of SR-to-RP 1.917** 1.882** 0.930 Effect of Site 2.918** 1.744** 1.425 Effect of SR-to-RP* site 2.640** 1.922** 1.111 * Indicated p < 0.1; ** indicated p < 0.5; *** indicated p < 0.01. Fungal community
-
Like bacterial richness, the species richness, as calculated from the number of the bands, showed slightly higher in rubber plantations than natural forest in average value. The fungal species richness was very low in the SR3 forest, but similar among the rest (Fig. 1). The CCA analysis revealed that general fungal community composition did differ significantly between rubber plantation and natural forest (p < 0.01), but significantly interacted with sites (p < 0.01), which was also like bacterial community (Table 2). As shown in the NMDS analysis, the patterns of the fungal community were separated according to forest conversion when comparing rubber plantations to a primary forest in each site (Fig. 2). In addition, the fungal community showed higher heterogeneity in rubber plantations than in primary forests (Fig. 2). The pairwise comparisons results using MRPP revealed that all the SR forests were significantly different from NR forest within sites, except for SR3 vs. RP3. When comparing different sites, bacterial community composition was found to be significantly different in many comparisons, except for two pairs: SR1 vs. SR3 and SR3 vs. SR2 (Table. 3).
Basidiomycota community
-
Species richness was between rubber plantation and natural forest in average value. However, differences between pairs of RS-to-SR were significant and contrasting. For example, SR1 had a higher amount of Basidiomycota species than RP 1, while the opposite trend was found between SR3 and RP3 (Fig. 1) The CCA analysis revealed that the Basidiomycota community composition was not different between rubber plantation and natural forest (p > 0.05), which was consist with bacterial and general fungal communities (Table 2). NMDS showed that the Basidiomycota community composition was similar between rubber plantations but different between primary forests, with higher heterogeneity within a single forest (Fig. 2). However, pairwise comparisons present that the Basidiomycota community was significantly different between SR3 and RP3. When comparing different sites, Basidiomycota community composition was found significantly different in 5 comparisons (SR2 vs. RP1, SR3 vs. RP1, SR3 vs. RP2 SR1 vs. SR2, and RP1 vs. RP2), while similar in the other seven pairs (Table. 3).
-
Conversion of native forest land to rubber plantations is known to have significant impacts on soil characteristics because of changes in litter input and root exudates[28]. Consequently, the soil microbial community should respond to these changes. Previous research has documented that forest conversion has caused significant differences in soil chemical (e.g., pH, C/N, Organic C, LOC) and physical (e.g., soil texture) characteristics. Our data have revealed a significantly different community structure between rubber plantations and primary forests, which is mainly due to variation in soil properties. This result is grounded in several biogeographical studies, which have suggested that soil bacterial communities are significantly affected by soil pH, soil C/N and soil texture[27,36]. Like bacteria, fungal communities were also significantly affected by soil characteristics, especially soil C/N and texture. As an important carbon decomposer, the fungal community has a significant correlation with soil C/N, which is an important indicator of the substrate's quantity. In addition, soil fungi play an important role in soil texture by producing mycelium to tide up the soil particles[37]. Compared to natural forest, rubber plantation has a higher number of large aggregators, which might imply that the fungal community harbors more mycelium fungi than the primary forest. Different from the above two, we found no significant difference of Basidiomycota communities between rubber plantation and primary forests. However, we also found significant heterogeneity within a single pair, which might overlook the differences between forest and rubber plantation. Therefore, we can tell from the results that different microbial groups differ in their response to forest conversion along with abiotic factors.
Effect of biotic factors on soil microbes under conversion
-
In addition, the observed community changes were likely to be driven largely by known differences in the mycorrhizal flora of native tree taxa and rubber plantations. Our data showed a clear cluster pattern in the rubber plantation of separated sites in the fungal and Basidiomycota community. We presume that the dominant fungal phyla may be ectomycorrhizal fungi correlated with rubber species. A good example was the study of the ECM fungal community when the native forest changed into a plantation. Following the introduction to facilitate the establishment of plantations in the Southern Hemisphere, many pine-specific ectomycorrhizal fungi are now well established in exotic locations such as Australia, and, for the most part, there is little evidence that they transfer from plantation pines to native forest and vice versa[38]. When native forest changed into a plantation, AM fungal communities would significantly change and consequently impact the bacterial and fungal communities around them. Such responses have been studied in both incubation and field experiments[39]. In addition, soil animals, especially earthworms, have been found in significantly higher amounts in rubber plantations[37,40]. Previous studies have suggested that earthworms could be a critical disturbance factor to fungi, especially for some Basidiomycota species with long mycelium[41]. Therefore, the fungal community in rubber plantations faces more environmental pressures than in primary forests.
Effect of land-use changes on microbial community
-
We found that the effects of site and interaction were more significant than the land-use type change. Other than differences in original forest data, humus activities could be an important factor, especially considering fertilizer and fungistatic agent[42]. To improve the growth of rubber plantations and prevent diseases, farmers must use fertilizer and fungistatic agents regularly. These reagents have huge impacts on the microbial community, including changing the structure and reducing the abundance[43]. Owned by different farmers with different use protocols, some arrangements result in significantly higher site effects. With the existence of more than 450,000 ha of rubber plantations in Xishuangbanna, it is becoming increasingly important to understand the impacts of land-use changes from natural forests to rubber plantations on soil biological properties. Our results indicate that conversion of native rainforest to monoculture plantation significantly alters soil bacterial and fungal communities. Further work will be required to understand the functional significance of these changes and their long-term impacts on soil quality and productivity.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2022 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visithttps://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Shi L, Zhai D, Chen H. 2022. Converting tropical rainforest to native rubber plantations alters soil bacterial and fungal communities. Circular Agricultural Systems 2:2 doi: 10.48130/CAS-2022-0002
Converting tropical rainforest to native rubber plantations alters soil bacterial and fungal communities
- Received: 31 August 2021
- Accepted: 21 February 2022
- Published online: 08 April 2022
Abstract: Driven by soil biochemistry and plant community composition, soil microbial communities reflect land management and environmental conditions. To evaluate the effects of land-use change on soil microbial diversity, we used denaturing gradient gel electrophoresis (DGGE) combined with sequencing to compare bacterial and fungal community profiles between rubber plantation (RP) and nearby seasonal rainforests (SR). Rainforest soil generally had higher soil total C and microbial biomass C concentration, smaller soil aggregate proportions, and a soil pH below rubber plantation soil. The bacterial and fungal richness and diversity were similar after converting primary forests to rubber plantations. However, the composition of bacterial and fungal communities has significantly changed in rubber plantations. Basidiomycota, the predominant group of fungi, was significantly different between primary forests and rubber plantations. However, Basidiomycota showed higher heterogenetic distribution in the rainforest under rubber plantations. In conclusion, land-use changes mainly affect soil microbial community composition and heterogeneity distribution patterns, especially for saprotrophic fungi, which consist of changes in litter inputs and soil C conditions.
-
Key words:
- Bacteria /
- Fungi /
- Basidiomycota /
- Rubber plantation /
- Forest conversation