-
Wine fermentation is a process carried out by wine microbial consortium (WMC), including yeast, lactic acid bacteria (LAB), and acetic acid bacteria (AAB), which consists of two stage alcoholic fermentation (AF) and malolactic fermentation (MLF). AF is a conversion of monosaccharides like glucose and fructose into ethanol and carbon dioxide (CO2)[1] by yeast, and out of them, S. cerevisiae shows the highest fermenting ability and ethanol tolerance in wine medium during AF[2]. Effective alcoholic fermentation depends on the presence of desirable microbial consortium (yeasts), suitable nutrition for microbial (yeast) growth, temperature, and prevention of the development of undesirable microorganisms[3, 4]. MLF takes place after AF by LAB, which reduces the acidity of wine by converting malic acid into lactic acid[5]. Furthermore, Oenococcus oeni is a notable LAB species responsible for this phase of fermentation. Different primary and secondary metabolites are formed during the fermentation of grape juice to wine due to several microbial activities and interactions. Some of these metabolites lead to cell-to-cell communication among wine microbial consortiums by providing chemical signals called quorum sensing[6]. Moreover, microbial cells coordinate their metabolic processes in response to the produced molecules called quorum sensing molecules (QSMs), in their environment. These QSMs are produced by wine microbial consortium, significantly yeasts, bacteria, and other filamentous fungus. This system was first discovered in bacteria, and recent evidence shows that QS coordinates processes like biofilm formation, pathogenesis, and secondary metabolite production in eukaryotes also significantly in fungi[7]. Quorum sensing mechanisms in wine production coordinate the production process of secondary metabolites[8], and these metabolites are responsible for the organoleptic features that determine wine's final quality and palatability[9]. Furthermore, microbes control their population density using the QS mechanism[10]. Every cell can sense the concentration of quorum sensing molecules (QSMs) in their environment and regulate their gene expression when QSMs reach their specific threshold levels, whereas QSMs are called 'autoinducers' as they promote their metabolism[7]. In spontaneous fermentation, several microorganisms naturally present in grape berries carry out the fermentation, resulting in random amounts of aromatic alcohols and metabolite synthesis, affecting the quorum sensing mechanism in different ways. Eventually, this spontaneous fermentation facilitates novel organoleptic profiles in wine. Furthermore, most of the regional wine varieties are produced based on spontaneous fermentation, leading to the development of specific organoleptic qualities in the final product[11]. Saccharomyces cerevisiae initiates the synthesis of quorum sensing molecules like tyrosol, 2-phenyl ethanol, and tryptophol. Furthermore, tryptophol auto stimulated the production of these aromatic alcohols. QS plays a significant role in wine fermentation. In the wine microbial consortium, yeast species are responsible for the increment of acidity, softer mouthfeel, and fruity aromas, while lactic acid bacteria synthesize favorable esters that improve the organoleptic characteristics of wine[5,12]. The diversity of microbial consortium decreases with fermentation due to the increase in acidic wine environment[3,13,14]. Previous studies have revealed that QSMs regulate the transcription mechanism of genes ARO8, ARO9, and ARO10. These genes are specific yet essential for the formation of aromatic alcohols. This review focuses on quorum sensing of wine microbial consortium in the fermentation process of spontaneous wine fermentation and how these signaling processes genetically manipulate and affect the organoleptic quality development of the regional wine products.
-
Grape skin is a preferable niche for different microorganisms, and some bacteria and parasitic fungi only live in the grapes, not in wine. Aspergillus, Botrytis, and Penicillium are the major fungi that affect the wine quality due to the production of mycotoxins[15]. Yeast, bacteria, and filamentous fungi are the main complex microbial consortia in grapes, and in the case of wine, yeast species, lactic acid bacteria, and acetic acid bacteria play significant roles[16]. Furthermore, the bacterial genera like Bacillus spp., Enterobacter spp., Enterococcus spp., Staphylococcus spp., Burkholderia spp., and Serratia spp., are present in grapes but they are unable to survive in wine due to the generated alcohol in the medium through the fermentation which creates toxicity to these bacterial species and inhibit the growth. The microbial strains with low poor alcoholic tolerance can't survive in wine. The fruit's maturity, variety, vineyard age, application of antifungal agents, climatic conditions, pH, and temperature during fermentation are the key factors that decide the types of microorganisms present in must or wine[17,18]. The grape microbiome consists of some spoilage agents like filamentous fungi[16,19,20]. The inhabitance and diversity of this grape microbial consortium are determined by harvest quality, harvesting period, and many biotic and abiotic factors[21]. Therefore, several aspects should be considered in beverage fermenting consortia since various strains carry different complex functions. A sufficient dynamic connection between the microbial groups determines the stability and efficiency of the microbial consortium involved in wine fermentation. Interactions between these communities significantly impact the quality and palatability of wine due to the metabolic activities of various microorganisms[16,22]. The involvement of several microorganisms, yeast, bacteria, and fungi enhances the complexity of various mechanisms like interactions, dynamism, transcriptomics, and metabolism that finally determine the organoleptic characteristics of wine[23].
Wine must, and grapes are the main niches of the complex microbiome responsible for the fermentation, flavor, and quality development of wine. The previous studies revealed that the fermentation process predominantly affects fungal communities compared to bacterial communities during spontaneous fermentation[24]. The significant types of wine yeast involved in AF are Saccharomyces and non-Saccahromyces. Generally, Hanseniaspora, Metschnikowia, Lachancea (Kluyveromyces), Candida, Pichia, and Saccharomyces are major wine microbial consortium that presents during the beginning of spontaneous fermentation[25, 26]. Along with the process of fermentation, S. cerevisiae controls the AF with the increase of ethanol content. Currently, the extent of expression of ARO genes in S. cerevisiae determines the amount of synthesis of QSMs responsible for the final organoleptic properties of wine. Later deacidification process begins with LAB Oenococcus, Leuconostoc Pediococcus, and Lactobacillus, which significantly impact the aging and quality of the wine[24].
Many research findings revealed that yeast is the most prevalent and valuable microorganism inhabiting grape bunches. For instance, the findings of Barata et al.[19], Kassemeyer & Berkelmann-Löhnertz[27] and Barata et al.[16], revealed the availability of 93 yeast species from 30 genera, isolated in 22 countries from 49 grape varieties. Further, 47 yeast species were identified from 22 various genera; Aureobasidium, Auriculibuller, Brettanomyces, Bulleromyces, Candida, Cryptococcus, Issatchenkia, Debaryomyces, Hanseniaspora, Kluyveromyces, Lipomyces, Metschnikowia, Rhodotorula, Pichia, Rhodosporidium, Saccharomyces, Torulaspora, Sporidiobolus, Sporobolomyces, Yarrowia, Zygoascus, and Zygosaccharomyces using Polymerase Chain Reaction-Denaturing Gradient Gel Electrophoresis (PCR-DGGE)[28]. Another study on grape berries, Aureobasidium pullulans, Candida magnoliae, Candida parapsilosis, Candida saitoana, Cryptococcus diffluens, Issatchenkia orientalis, Cryptococcus magnus, Rhodotorula glutinis, Hanseniaspora uvarum, Kluveromyces marxianus, Rhodotorula mucilaginosa, and Yarrowia lipolytica were identified as yeast microbial consortium by using Matrix-Assisted Laser Desorption/Ionization Time-of-Flight mass spectrometry (MALDI-TOF)[29]. Furthermore, Metschnikowia pulcherrima, Hanseniaspora vineae, Hanseniaspora guillermondii, Torulaspora delbrueckii, Lanchacea thermotolerans, Issatchenkia orientalis, Pichia kluyveri, Hanseniaspora uvarum, Cryptococcus flavescens, Aureobasidium pullulans, Rhodotorula glutinis, Cryptococcus magnus, Starmerella bacillaris, and Pichia membranifaciens were found to be the non-Saccharomyces species on grape berries. Population densities of yeast species on grape skin depend on their maturity levels[30]. Furthermore, vineyard factors like fruit color, grape variety, the health profile of berries, surface area of fruit, and nutritional availability during maturation also affect the biodiversity of yeast in grapes[3,31−34]. Moreover, the microbial consortium of a berry can be changed by the penetration of Botrytis cinerea like pathogens[19,35].
The research findings revealed that the second most prevalent microorganism inhabiting the skin of the grape is bacteria. More than 50 bacterial species have been isolated from grape berries, and most of the species belonged to two major phyla, Firmicutes and Proteobacteria. Firmicutes consist of gram-positive bacteria like Lactobacillaceae (Lactobacillus and Pediococcus), Bacillaceae (Bacillus), Enterococcaceae (Enterococcus Faecium), and Leuconostocaceae (Leuconostoc, Weiseilla, and Oenococcus). These species are called a technological group of lactic acid bacteria except Bacillus and Enterococcus spp.[16,36]. The important genera of wine fermenting LAB species are Oenococcus, Lactobacillus, Pediococcus, and Leuconostoc, and AABs are considered wine spoilage microorganisms. LAB reduces the acidity of wine through the decarboxylation of malic acid through both homofermentative metabolism and heterofermentative metabolism. Oenococcus onei can carry out homofermentative metabolism by producing lactic acid and carbon dioxide during MLF. Other species like Pediococcus pentosaceus and Pediococcus damnosus carry out heterofermentative metabolism and produce ethanol, acetic acid, and CO2. In the meantime, Lactobacillus casei and Lactobacillus plantarum follow a facultative heterofermentative metabolism pathway. These two species can survive in both aerobic and anaerobic conditions[37]. In the case of bacterial microbiota, the variations in the size of the population vary with the ripening process. When grapes become matured, gram-negative bacterial colonies decrease, and gram-positive increase[38]. Different bacterial groups in grapes depend on the harvest's health and hygienic state[39].
There are other microbial groups present in grapes considered as spoilage agents. The organoleptic features of the wine can be affected by filamentous fungi such as Aspergillus and Penicillium due to the production of mycotoxins[20, 40−42]. Other microorganisms like Plasmopara viticola, Erysiphe necator, and Botrytis cinerea cause diseases like grey mold, downy mildew, and powdery mildew, respectively[27]. Furthermore, Botrytis cinerea, Aspergillus, and Penicillium cause off-flavors in winemaking. Moreover, the previous study revealed that the combination of B. cinerea with some fungi like Penicillium and Rhizopus spp. cause grey rots in grapes. These grey rots badly affect the organoleptic properties of wine due to the production of 2-methylisoborneol, (−)-geosmin, 2-octen-1-ol, 1-octen-3-one, 1-octen-3-ol and 2-heptanol like chemical compounds[16]. Powdery mildew is a significant disease in grape berries caused by the fungus Uncinula necator which gives a strong mushroom odor due to the production of flavonoids and total phenolic compounds. Further, it revealed that 1-octen-3-one is responsible for such an effect. Furthermore, sour rot is another fungal disease that has been poorly understood due to its nature of simultaneous occurrence with other fungal diseases[16]. All these microorganisms coordinate with each other in their specific way to carry out quorum sensing mechanisms during winemaking.
-
Wine is a reservoir of several diverse groups of microorganisms that are favored by interactions among the microbial consortia. These significant microbial interactions are yeast–yeast interactions, bacteria-bacteria interactions, bacteria-yeast interactions, and fungi-yeast interactions. Apart from that, direct and indirect interactions also play a role in the fermentation process. Some direct interactions include quorum sensing, predation, physical contact, inhibition, parasitism, and symbiosis. Indirect interactions include neutralism, commensalism, amensalism, mutualism, and competition due to extracellular metabolites[43]. Except these filamentous fungi in the consortia also interact with themselves and with others, their growth rate decreases with wine fermenting progress, so their effect is not significant[44,45]. Among these interactions, quorum sensing takes special place due to manipulating the organoleptic qualities of the final wine product.
-
Since 1960 it has been known that microbial consortiums do not survive alone in their environment, and they are communicating among themselves, allowing the microbes to regulate their behaviors[6]. This advanced cell-to-cell communicating mechanism is known as QS. This is coordinated through small molecules called QSMs which diffuse into the medium and are gathered during the development of microbial colonies[7]. According to the concentration of QSMs in the external environment, every cell can regulate its phenotypes[46]. The effect on transcription and translation of genes responsible for aromatic alcohol production occurs due to the binding of these quorum sensing molecules (QSMs) with their target sensory proteins. When the secreted QSMs and growing populations reach a specific density, the binding threshold is achieved, as stated by the phrase 'quorum'. Several QS systems are identified as relevant to different QSMs in the environment[46, 47]. Every single cell can sense the concentration of QSMs in their environment and regulate their gene expression when QSMs reach their specific threshold level, as shown in Fig. 1[7]. Microbes control their population density by using the QS mechanism[10]. QSMs are called 'autoinducers' as they promote their synthesis and carry out the QS process through their different types of microbial interactions[7].QS grants an evolutionary benefit by letting the microbial colonies adapt to the evolving environment. Some authors also suggest the neo-Darwinian mechanism of evolution[10]. There are several metabolites present in the supernatant of conditioned culture media possessing the ability to perform the activity of QSMs in the QS system. Some special characteristics differentiate QSMs from other metabolites. The QSMs are formed and gathered in the extracellular environment, and their production takes place at a particular stage of growth and under specific environmental conditions. It has a unique mechanism to identify some conditions and respond based on the extracellular threshold values of QSMs. QSMs are nontoxic at their threshold level, which is required to bring out the specific response. The effects of QSMs in the extracellular environment can be easily differentiated from the primary metabolites of QSMs[9, 48−51].
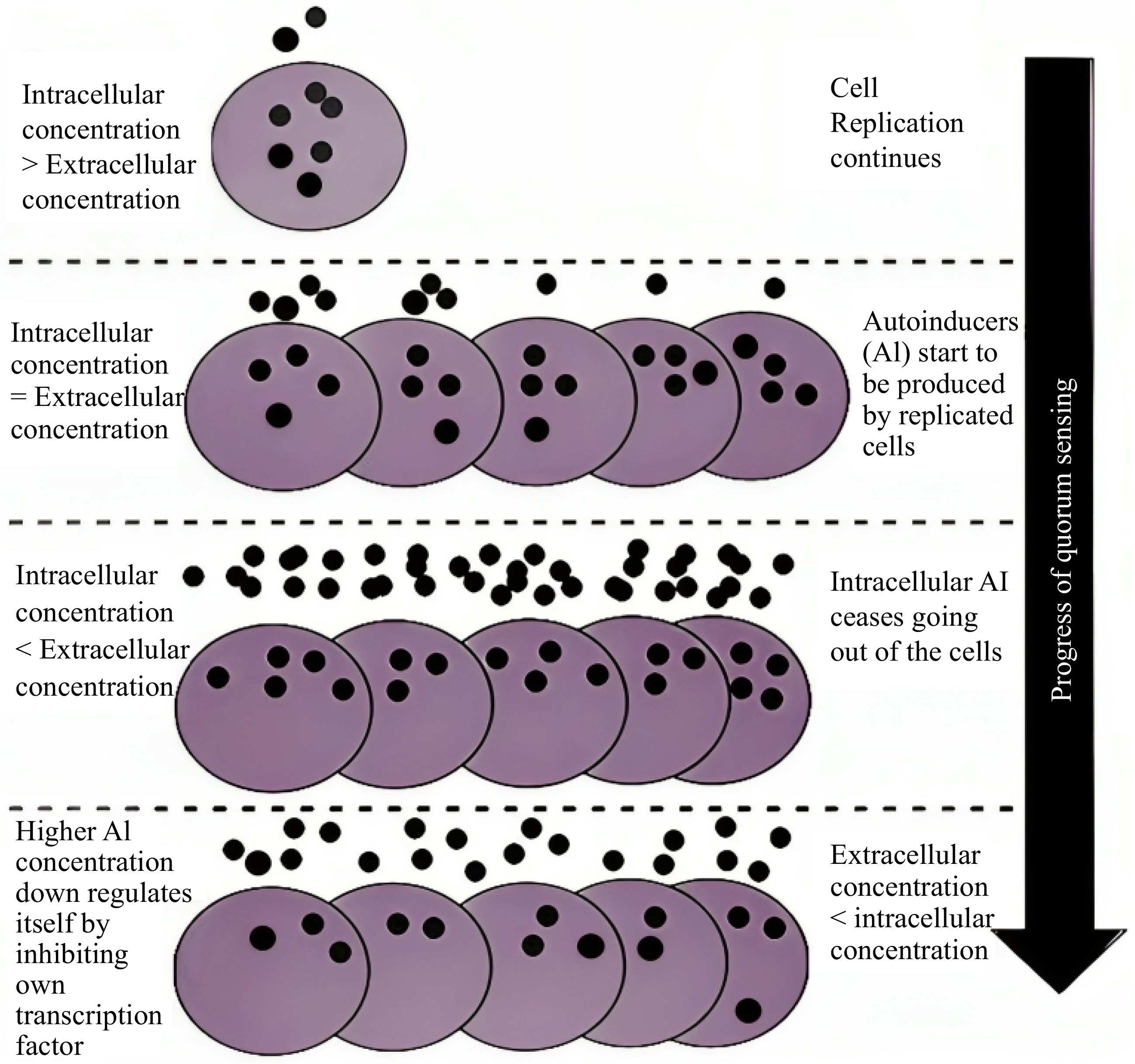
Figure 1.
Diagrammatic view of QS regulation[82].
-
QS in wine fermentation is a cell to cell communication between wine microbial consortiums, which is a mechanism of creation, synthesis, and detection of QSMs, and the concentration depends on the presence of related microorganisms in the wine[52]. This mechanism mediates the beginning of competence and the synthesis of secondary metabolites like ammonia, bicarbonate, farnesol, acetaldehyde, tryptophol, and phenylethanol which are the significant yeast QSMs[53]. In the yeast QS process, alcohols, small peptides, volatile compounds, lipids, and small molecules act as messengers in intraspecies communication, and aromatic alcohols are the only acting bodies of QSMs[54]. QS systems were first observed in a dimorphic fungus called C. albicans, and it regulated the transition of filamentous growth from yeast cells. Farnesol is one of the QSMs secreted during the development of C. albicans cultures. When the culture reaches a specific density, accumulated farnesol prevents the development of germ tubes[55]. Tyrosol is another QSM in C. albicans that induces cell growth and germ tube formations even in low density[56]. The QS system of C. albicans is different from S. cerevisiae. Anyhow, according to previous studies, S. cerevisiae plays a major role in the secretion of QSMs, affecting the aromatic profile of the wine. Especially, it converts phenylalanine and tryptophan into aromatic alcohols like 2-phenyl-ethanol and tryptophol, respectively, through the Ehrlich pathway[57]. These molecules could act as QSMs; hence they induce the phase shifting into the stationary phase. At the same time, they promote pseudohypha generation and rapid growth of cells. Like this QS controls wine's microbial population and organoleptic properties during winemaking concerning regions and available microbial consortium. There is no evidence available for the function of QSMs like tyrosol, tryptophol, or 2-phenylethanol by S. cerevisiae during AF. QSMs are synthesized when the phase change from exponential to stationary in the alcoholic fermentation process[58]. For instance, the mini fermentation results of Zupan et al.[58], further revealed that tyrosol, tryptophol, or 2-phenylethanol production of S. cerevisiae is high in the exponential phase of the fermentation and rapidly declined just before the stationary phase of alcoholic fermentation. Furthermore, the whole-genome microarray analysis results of Chen & Fink[59] showed tryptophol and 2-phenylethanol regulate the transcription of a set of genes that manipulate the entrance to the stationary phase. Furthermore, tryptophol stimulates the production of aromatic alcohol. Yeast like S. cerevisiae exhibits higher fermentative capacity and higher ethanol tolerance under the availability of quorum sensing molecule tyrosol. Furthermore, aromatic alcohols derived from amino acids mediate QS-type yeast regulations[60]. All these results showed that QS molecules control the correct entrance of S. cerevisiae cells to the stationary phase. Other than, S. cerevisiae some non-Saccharomyces species, H. uvarum, T. pretoriensis, and Z. bailii were able to produce phenylethanol, tryptophol, and tyrosol like quorum sensing molecules[58]. It has been postulated by a recent study that S. cerevisiae, Candida zemplinina, Hanseniaspora uvarum, Torulaspora pretoriensis, Zygosaccharomyces bailii, and Dekkera bruxellensis expressed species-specific mechanisms for the synthesis of tyrosol, 2-phenylethanol, and tryptophol (Fig. 2). At the same time, no QSMs were observed after one day of wine fermentation of C. zemplinina and D. bruxellensis[58]. Further, this research revealed that Zygosaccharomyces bailii were able to produce tryptophol at a similar rate to S. cerevisiae, which could be because both belong to the same family Saccharomycetaceae. Further, the results revealed, secondly, the highest phenylethanol and tyrosol production capability after S. cerevisiae was shown by this yeast species. The aromatic alcohol 2-phenylethanol has a significant impact on the aroma profile of a wine. On the other hand, Hanseniaspora uvarum has different QS kinetics, showing the early tyrosol synthesis initiation and the late start of tryptophol synthesis compared to H. uvarum. Furthermore, it showed a meager rate of production of phenylethanol without any peaks in the kinetics curve. All in all, these analyses revealed the heterogeneity of QSMs and their kinetics in all tested species. Therefore, further studies on the kinetics of QSMs and mechanisms may open new avenues in wine fermentation, facilitating the better quality of the wine[58]. Therefore, the combination of wine microbial consortium through various indigenous microorganisms for wine fermentation leads to the development of specific organoleptic qualities of the final wine product. In Gram-positive bacteria, QS-mediated signaling is coordinated by small peptides due to the intracellular mechanism detected by a sensory protein called histidine-kinase[61]. Lactones, terpenes, alcohols, and peptides are the prominent families of QSMs[62]. No scientific proofs are available on how S. cerevisae communicates signals, and it was stated that S. cerevisiae synthesized and released QSMs at a specific microbial density like C. albicans[55]. Tryptophols and 2-phenylethanol are present in C. albicans, but they do not express any density-dependent responses. Debaryomyces hansenii also secreted QSMs like tyrosols, 2-phenylethanol, and tryptophols to communicate with the other microorganisms in the environment.
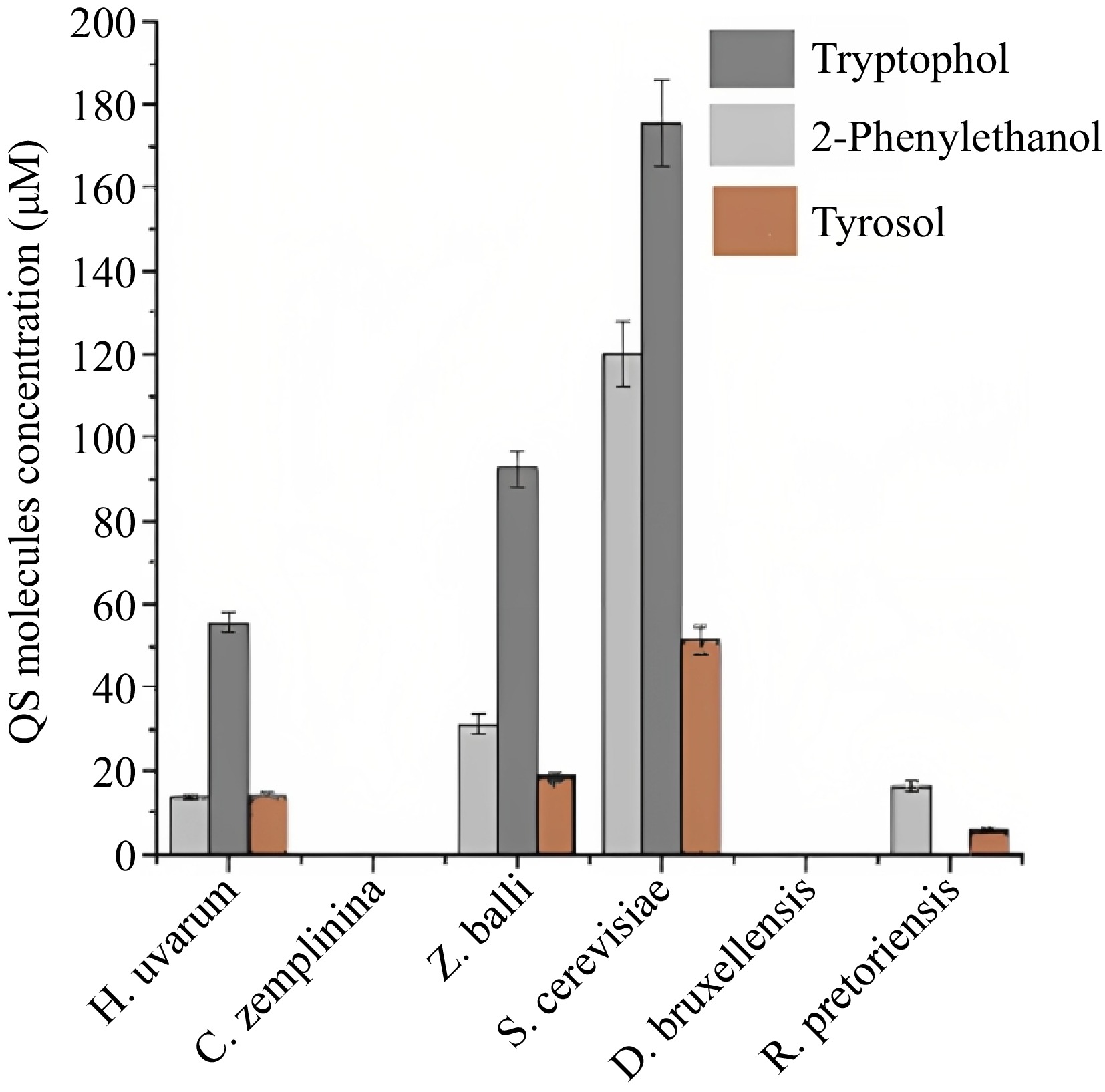
Figure 2.
Synthesis of QSMs of different wine yeast species in MS300 medium at 22 °C for 29 h in 2 mL microcentrifuge tubes (mini fermentation)[58].
Further, the study of Nissen et al.[43] revealed that, when S. cerevisiae and non-Saccharomyces are in a co-culture, yeast-yeast interactions are mediated by QSMs to reduce the early growth of non-Saccharomyces. Furthermore, this research revealed that it might be due to the cell to cell contact process of yeast in mixed cultures, and it demonstrated that the cell to cell contact process is not a single mechanism responsible for the early growth arrest non-Saccharomyces. Several other factors and mechanisms also involve the inhibition of growth of non-Saccharomyces in early fermentation. Acetaldehyde is a major QSM that mediates the cell to cell signaling process and regulates biomass synthesis, fermentation kinetics, and by-product formation[63]. Flocculation is a significant cell-to-cell associative mechanism in which colloidal cells come out of the medium to accumulate in a flake form. The effective yeast flocculation regulates the formation of compacted sediments and initiates the clarification mechanism in post AF[64]. An individual cell does not carry out the flocculation process[65]. A flocculent of K. apiculata communicates with a non-flocculent S. cerevisiae strain in mixed fermentation and facilitates co-flocculation in both strains. It has been revealed that S. cerevisiae, Dekkera spp., and K. apiculata are involved in co-flocculation with various bacterial strains[66]. A lectin-carbohydrate binding system regulates these types of co-flocculation mechanisms[65,66]. Due to this mechanism, the indigenous non-Saccharomyces species involved in early AF of spontaneous fermentation are controlled by S. cerevisiae, and as a result, regional wine varieties obtained different organoleptic qualities[65,66]. However, there have been no records for the QS mechanism and cell to cell contact of bacterial cells in the wine medium, and many kinds of research need to be carried out to clear this area. Hence different wine microbes have different QS systems, and this opens an avenue to conduct much research to identify some yeast genera specific QS systems. Further different region-specific grape microbiome also shows diversity in their QS systems. As a result of QS variations, they generate different metabolites in different stages of alcohol fermentation. This may lead to the region-specific organoleptic property development of the final wine product. Therefore, this is a highly blind area to be investigated through many kinds of research.
The quorum sensing capability of wine microbial consortium varies from region to region. Geographical features and climatic conditions of vineyards and wineries determine the grape microbial consortium, quorum sensing, and, eventually, the quality of the wine. Specifically, climatic features, training systems, soil types, and cultural behaviors interact, which determines the terroir and growth of different grape varieties[67]. Five regions of the world are considered the top five wine regions; hence more than 80% of wine exports took place from these regions. France, Italy, the United States of America, Australia, and Chile are the top five wine regions globally. Several well-known wine varieties from specific regions of these countries use selective grape varieties (Table 1)[11]. Other than these countries, Spain, Germany, Argentina, Portugal, South Africa, and New Zealand are also significant wine producers globally. Likewise, according to the information from the Fruit Crops Research and Development Centre, Department of Agriculture (Horana, Sri Lanka); Israel blue, Cardinal, Black muscat, Muscat MI, and French MI are the major grape varieties cultivated in Sri Lanka. Among these Muscat MI (red wine) and French MI (white wine) are used for winemaking, while others are used as table varieties. In Sri Lanka, better grape harvest can be obtained from well drained, deep soils in the dry zone under irrigation. In Sri Lanka, farmers use the pandol system, Geneva double curtain system (GDC), and other fence systems as training systems. Jaffna and Dambulla are some of the significant grape-cultivating regions in Sri Lanka. Similarly, worldwide, the climatic conditions, fertility of soil, geology, pH, and training systems determine the growth of specific types of grape varieties and the microbial consortium of grapes[67]. Not only grapes, other fruits like apples, pears, berries, peaches, kiwis, bananas, pineapples, lemons, limes, passion fruits, and oranges are also used to make wine where we can analyse QS with both spontaneous and inoculated fermentation[68]. Eventually, this microbial consortium affects wine fermentation, secretion of quorum sensing molecules, and quality of the wine. This is how the quality of wine varies from region to region.
Table 1. Significant regional grape varieties and their respective wine types all over the world[11].
Region Well known wine type Grape variety France Haut-Médoc Château Cantemerle, AC Cabernet Sauvignon blended with
Cabernet Franc and MerlotChÍteau Lafite – Rothschild, Latour Mouton-Rothschild Upper Loire Sancerre Rouge, AC Pinot Noir Bordeaux fringe country Bergerac, AC Merlot, Cabernet Càte de Beaune Aloxe-Corton, AC Pinot Noir Italy Tuscany Brunello di Montalcino, DOCG Sangiovese Vernaccia Di San Gimignano Vernaccia Veneto Soave, DOC Garganega Breganze, DOC Rondinella and Molinara United States of America California Mondavi Chardonnay(white), Cabernet Sauvignon Merlot Berringer SonomaCutrer Cabernet Sauvignon/Chardonnay Oregon Adelsheim Pinot Noir Australia Victoria Lindemans Chardonnay ChÍteau Tahbilk Cabernet, Shiraz South Australia Adams Chardonnay (white) Henschke Sauvignon (red) Wynns Cabernet Chile Central valley Santa Rita Cabernet Sauvignon Concha y Toro Merlot (red) Terra Noble Carmenieres Several research teams have showcased the finding of artificial communication within and between species by employing synthetic circuits that incorporate elements of quorum sensing systems. Engineered quorum sensing (QS)-based circuits find diverse applications, including the synthesis of biochemicals, tissue culture engineering, and the facilitation of co-culture fermentations. Moreover, they play a crucial role in the development of microbial biosensors for discerning and monitoring microbial species within the environment and microorganisms[52]. The biofilm formation, acid stress tolerance, bacteriocin production, competence, adhesion, morphological switches, and oriented growth are some of the traits directed by QS in foodborne microorganisms. Moreover, QS has been identified in fermented food producing microorganisms. So, it can be applied to small-scale processing of fermented foods like dairy products, sourdough, fermented vegetables, and wine[69].
-
Nitrogen source is the primary factor that affects the synthesis of quorum sensing molecules, and the amount of nitrogen present in the medium affects the synthesis of QSMs. For instance, high ammonium concentration reduces the synthesis of aromatic alcohols by suppressing the expression of decarboxylases, dehydrogenases, and transaminases. The production of aromatic alcohols increases when ammonium sulfate concentration is < 50 μM and > 500 μM. The low concentration of nitrogen determines the synthesis of tryptophol compared to tyrosol and 2-phenylethanol. Furthermore, nitrogen-activated protein kinase (MAPK) and protein kinase A (PKA) are the two major signaling pathways that transmit nitrogen deficiency signals, and it causes variations in cell growth, cell adhesion, cell elongation, and bipolar growth[55]. Ethanol is another major factor that affects the synthesis of QSMs. Growth of microbes and rate of synthesis of QSMs such as 2-phenylethanol, tryptophol, and tyrosol are reduced at 2%−8% ethanol concentration. Furthermore, ethanol slows down the initiation of aromatic alcohols synthesis. It was stated, that at the ethanol concentration of 8%, there was no detection of tryptophol, as the concentration of microbial cells was not up to the specific threshold value for the onset of synthesis of tryptophol[70]. Aerobic and anaerobic conditions also have a significant impact on QSMs synthesis. The results of Ghosh et al.[71], revealed that C. albicans synthesized twice the amount of 2-phenylethanol, tryptophol, and tyrosol in anaerobic conditions than that of aerobic conditions at 30 °C. Concordantly another study on S. cerevisiae demonstrated similar results during wine fermentation under anaerobic conditions[55].
Another factor is cell density which regulates the synthesis of 2-phenylethanol and tryptophol. Cell density depends on the expression of ARO9 and ARO10 genes which are also the regulators of the synthesis of aromatic alcohols[55]. Low yeast cell densities synthesized a low amount of 2-phenylethanol and tryptophol compared to yeast cells with high population density[55], whereas a similar pattern has been observed in S. cerevisiae as well[37]. In another analysis with a low and high initial concentration of yeast cell, two parallel fermentation processes were carried out, and it has been found that the very first QSMs 2-phenylethanol, tryptophol, and tyrosol were detected when the microbial population reached the specific level. Further, these results revealed the quorum sensing kinetics of the microbial cells in the medium[55]. For research purposes, QS can be applied on a small scale with suitable microbial consortium and medium. Continuous observation should be carried out while adjusting the conditions with sampling and analysis[55]. Furthermore, with the development of biotechnology, researchers are focusing on genetic analysis, which affects the wine's quorum sensing mechanism and quality.
-
A study revealed that ARO genes are the responsible factors that regulate the synthesis of quorum sensing molecules like tyrosol, 2-phenylethanol, and tryptophol. Furthermore, ARO8, ARO9, and ARO10 present in S. cerevisiae are the significant genes that regulate the production of the above QSMs and other aromatic alcohols, which determines the sensory qualities of wine[55]. The FLO11 present in flor yeast is another major responsible gene that regulates quorum sensing and determines the organoleptic properties of wine. Further, these yeast plays a significant role in a process called 'biological aging' of wine. Avebelj et al.[70] researched to evaluate the correlation of ARO gene expression in S. cerevisiae with the production rate of QSM during the fermentation period (Fig. 3).
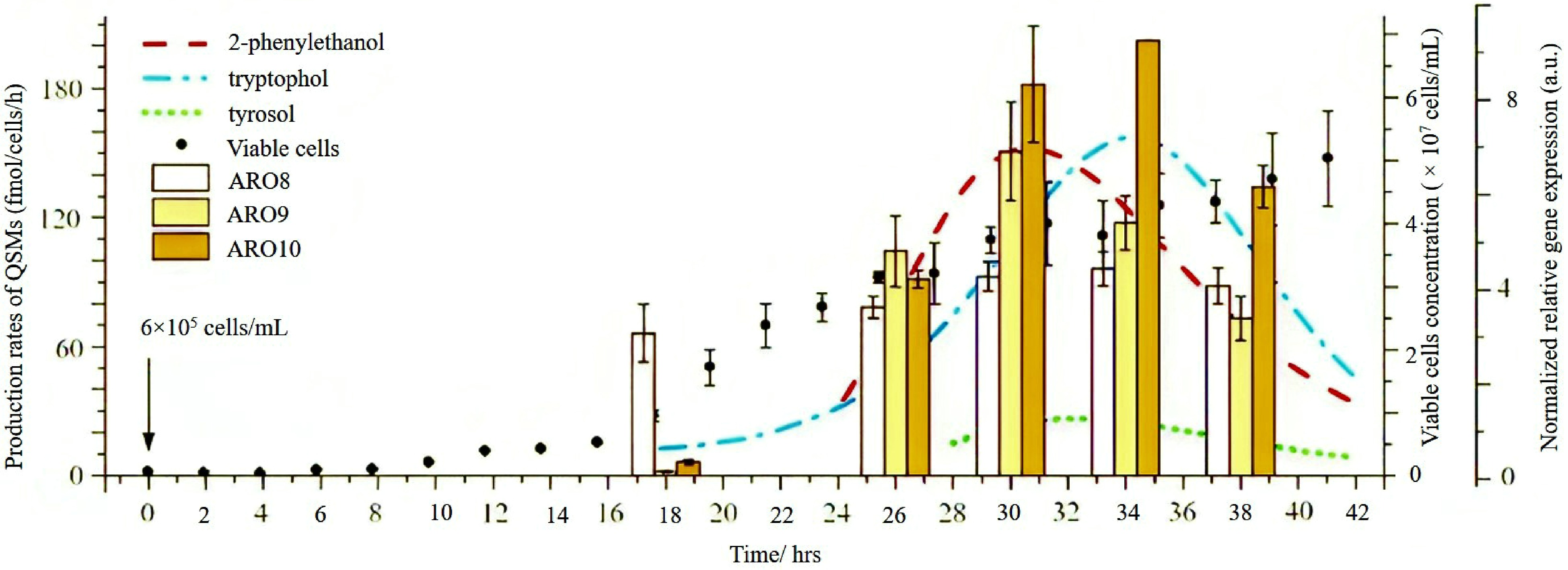
Figure 3.
The correlation between the production of QSM's and expression of ARO genes during mini fermentation by S. cerevisiae for 42 h with the initial concentration of 6 × 105 cells/mL[70].
In addition to the above correlation, they worked to show the correlation between the quorum sensing kinetics and cell concentration.
The viability of the microbial cells and synthesis of QSMs were observed for 42 h with specific time intervals. Furthermore, the gene expression level of ARO genes was compared to the kinetics of QSMs synthesized S. cerevisiae. Here analysis was carried out using both high and low initial cell concentrations. At first, the production of tryptophol started, followed by 2-phenylethanol during the exponential phase of cell growth at both concentrations. Among these QSMs, at first, 2-phenylethanol reached the peak synthesis, followed by tyrosol and eventually tryptophol. All these peaks' syntheses occurred in the declining growth phase of microbial cells. The rate of synthesis of QSMs is reduced when microbial cells reach the stationary phase. According to the available findings, this may be due to some autoregulatory mechanisms or some nutritional deficiencies[55]. Similarly, in expressing ARO genes with peak QSMs synthesis, both high and low initial cell concentrations were used, and observations were taken within a specific time. In both concentrations, the ARO8 gene expressed first and initiated the early production of QSMs. This occurred during the exponential phase by the aminotransferase enzyme, which was encoded by the ARO8 gene. Then the expression of ARO9 and ARO10 orderly initiated. According to the observation, ethanol and microbial cell concentration impact the beginning and effectiveness of QSMs synthesis. To support these observations expression of ARO genes was also monitored with ethanol addition during mini fermentation. In the beginning, the expression of ARO genes was only detected, confirming its impact on the early synthesis of QSMs. An enhanced rate of tryptophol synthesis was observed during the addition of 6% ethanol which can be correlated to the enhanced production of tryptophan in ethanol stress conditions[72]. At the same time, the ARO10 gene, which encodes phenylpyruvate decarboxylase was downregulated. The authors predict it may be because of ethanol on decarboxylase regulation. Moreover, at 8% ethanol, there was no tryptophol detection as the concentration of microbial cells did not attain the threshold level. Several new facts were revealed regarding gene expression and factors affecting QSMs synthesis during mini fermentation with S. cerevisiae. Therefore, these findings open a new avenue for research that focuses on studying the quorum sensing systems belonging to different yeast species involved in spontaneous fermentation and how the gene expression occurred during the fermentation.
Another essential gene that affects the quality of wine is FLO11. Among S. cerevisiae flor yeasts are something exceptional. Usually, this yeast grows on the surface of the wine, especially sherry and sherry-like wines[73]. Generally, flor yeasts carry out a 'biological aging' process during the storage period or after the alcoholic fermentation process. This is also called biofilm formation. Generally nutritional content of the wine determines the rate of biofilm formation, specifically during the breakdown of nitrogen and sugar at the end of alcoholic fermentation. It takes place when the cell metabolism shifts from fermentative to oxidative, called the 'diauxic shift'. This shift increased the expression of FLO11 genes, which resulted in enhanced cell surface hydrophobicity. This facilitates the generation of multicellular aggregates, which capture the CO2 released from fermentation and give aggregates floatability, thus enhancing biofilm formation[74]. This biofilm restricts the contact of wine with the external environment, thus enhancing wine's organoleptic properties.
-
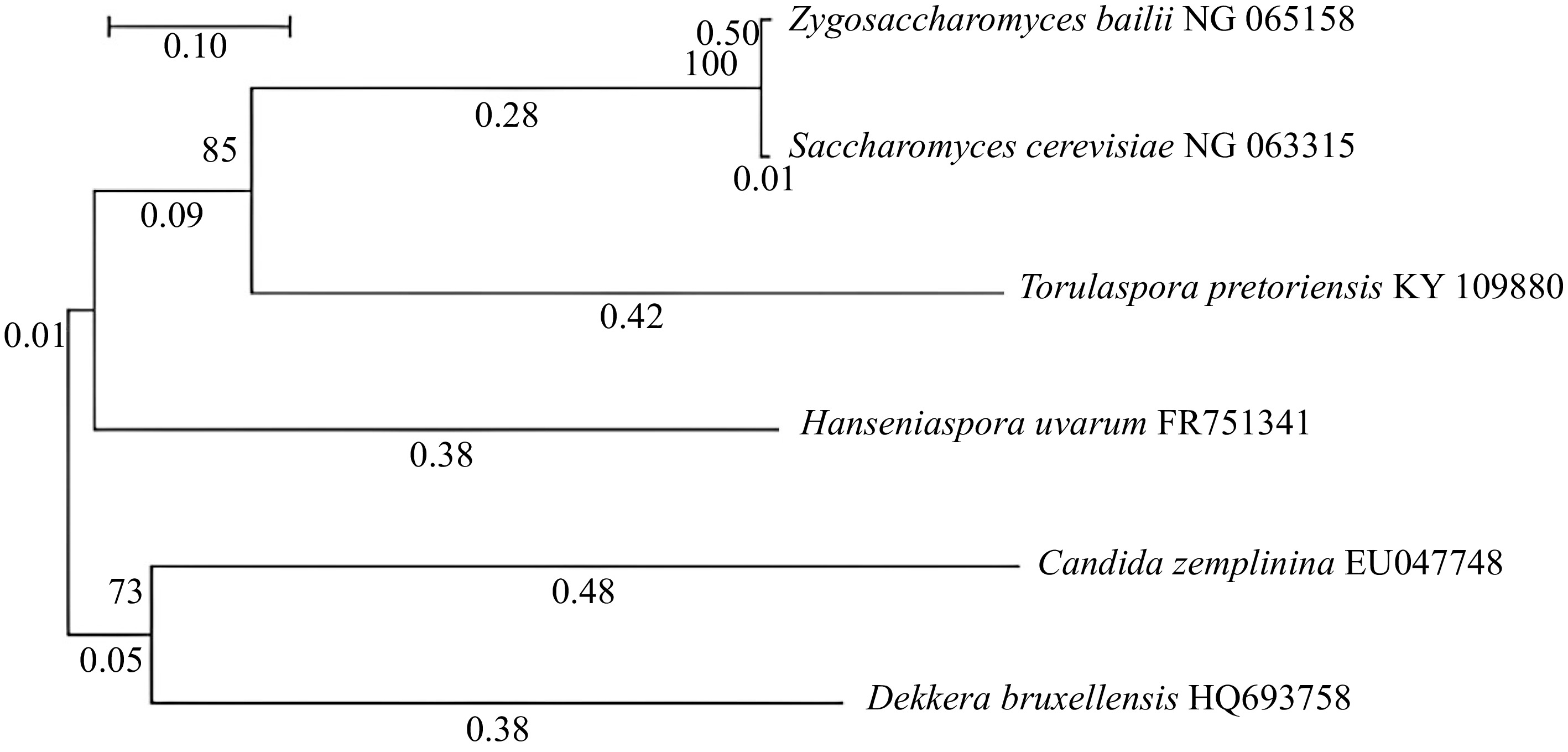
Although these ARO genes are from the same genus Saccharomyces, some showed distinct evolutionary characteristics, and some showed relative characteristics. Figure 4 expresses the evolutionary relationships among the essential genes of S. cerevisiae from different regions which are responsible for quorum sensing regulation. Here every strain in a respective group is closely related to each other. In the ARO10 cluster, both strains have almost similar characters, are equally closely related, and have a common ancestor. In the ARO9 cluster, ARO9 NM 001179267 and ARO9 DQ332024 are in a close relationship, and at the same time, YCK1 ARO9 ARO9 SPS100 Y13625 strain and the other two ARO9 strains have the same ancestor.
Furthermore, in the ARO8 cluster, all strains have close relative characters and common ancestors. Eventually, all these three clusters obtain a common ancestor. Here ARO9 NM 001179267, ARO10 NM 001180688, and ARO8 NM 001181067 genes are isolated from regions like Europe, North America, and Japan[75]. These results further revealed that any type of gene could be present in several locations. The existence of genes does not depend on the location. The capacity of synthesis of QSMs and the quality of wine depends on the expression of genes which is affected by the type of microorganisms and extracellular factors. Figures 2 & 4 revealed the correlation between the types of microorganisms, expression of ARO genes, and synthesis of QSMs[58, 70]. According to the above tree, the specific group of genes showed almost similar evolutionary relationships. Hence, it can be correlated that they will show the same genetic expression in a specific microorganism at specific extracellular conditions. In summary, to achieve novel organoleptic properties, fermentation should be carried out using different types of grapes with various combinations of microorganisms at suitable conditions. Like these, we can compare the evolutionary relationship among these essential genes and find out the brief connection among them. It opens up a way to compare the future significant genes responsible for effective quorum sensing for novel organoleptic properties development in wine and find out the best microbial combination.
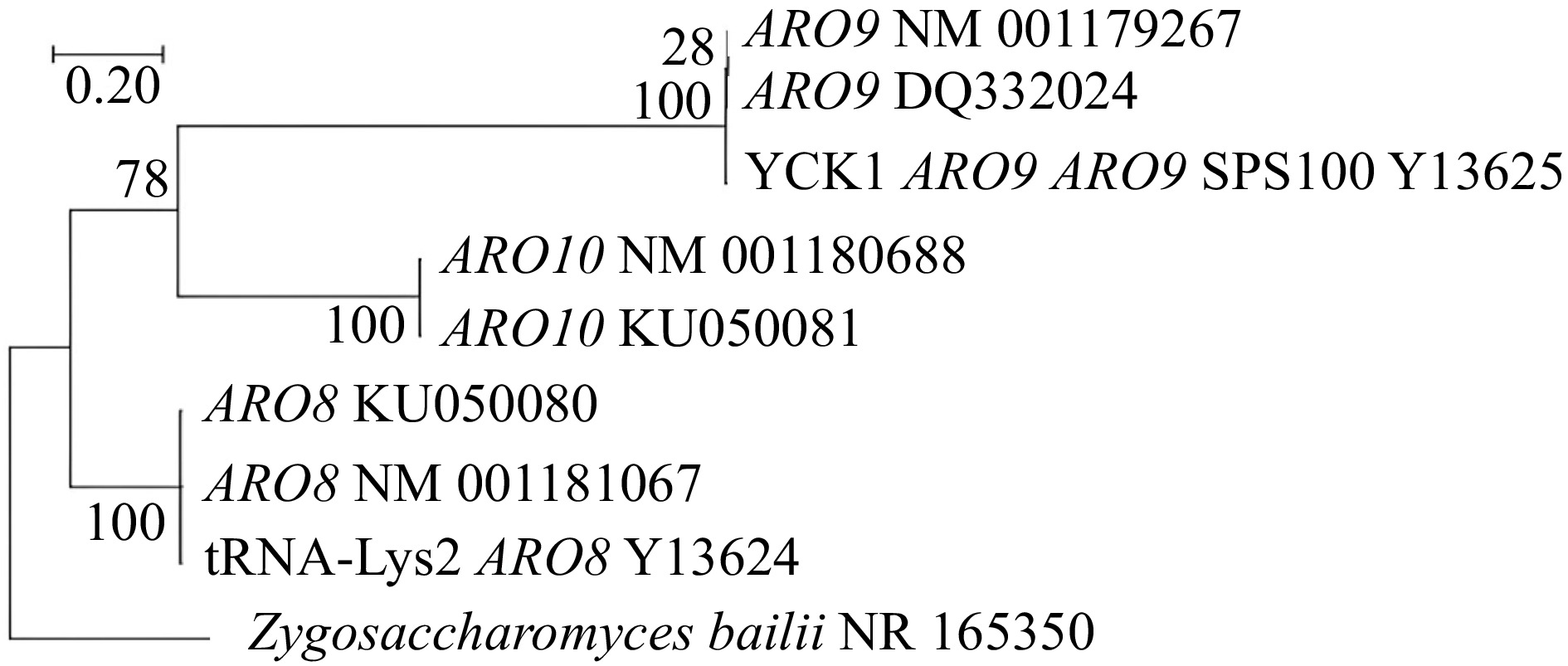
Figure 4.
Neighbour-joining tree of ARO8, ARO9, and ARO10 genes of S. cerevisiae from different regional strains which regulate QS.
A recent study analyzed the production of QSMs and quorum sensing kinetics of various wine yeast species during fermentation. Figure 2 shows the comparison of the synthesis of QSMs by various wine yeast varieties. It can be correlated to the following phylogenetic tree (Fig. 5). According to Fig. 2, S. cerevisiae and Z. bailli showed the highest QSMs synthesis capability[58]. Then H. uvarum and T. pretoriensis synthesized a considerable amount of QSMs, and finally, C. zemlinina, D. bruxellensis did not synthesize any QSMs. Z. bailli expressed the highest rate of tryptophol synthesis and showed similar QS kinetics for synthesized QSMs like S. cerevisiae. The above tree can prove this similarity as they are closely related to each other. Both belong to the same family Saccharomycetaceae, and hence express similar phylogenetic characteristics. Similarly, non-synthesizing of QSMs by C. zemplinina and D. bruxellensis is also correlated by the tree as they are closely related to each other and have long distance relationship with S. cerevisiae and Z. bailli.
At the same time, H. uvarum showed some dissimilarity in QS kinetics compared to S. cerevisiae and Z. bailli. It expressed early tyrosol production and late tryptophol production[58]. Similarly, the above tree also showed that H. uvarum expressed some distinct evolutionary characters from S. cerevisiae and Z. bailli. That means H. uvarum was not in a close relationship with them. Hence it can be considered that the microbial strains that show a similar range of QSMs synthesis and similar QS kinetics have almost similar genetic characteristics and evolutionary relationships. Every wine yeast species has different QSMs, and QS kinetics still need to be revealed. The observation and analysis rate of synthesis of QSMs and their concentration creates a new avenue in wine fermentation. Genetic studies and phylogenetic analysis can facilitate this to filter the species with similar QSMs production ability. This platform facilitates mixed wine fermentation with selective, effective, and suitably combined strains, which would enhance the organoleptic properties of regional wine[58]. Moreover, several studies are currently being carried out to discover the complexities of quorum sensing mechanisms that will support future innovations in winemaking.
-
Mainly two approaches are used to detect the quorum sensing mechanism in yeasts. They are chemical approaches and genetic approaches. However, combining these two approaches may lead to an optimal result that may favor the final product's quality.
Chemical approaches
-
In chemical approaches, the morphology of QSMs is confirmed by using X-ray crystallography, nuclear magnetic resonance, and high-resolution liquid chromatography/mass spectrometry (LC/MS) and facilitating the researchers to analyze the spectra of purified molecules from the conditioned medium to synthetic molecules[46]. The morphology of many quorum sensing molecules, their respective cognate synthases, and receptor compounds have been identified by X-ray diffraction analysis or nuclear magnetic resonance (NMR) methods. The chromatographic methods, such as thin-layer chromatography, autoradiography, and high-performance liquid chromatography (HPLC) coupled with diode array detectors, are used to identify the QSMs tyrosol, 2-phenylethanol, and tryptophol in S. cerevisiae. Samples should be concentrated by using distillation due to the low sensitivity of diode array detectors. The effectiveness of detection can be improved by using more expensive and advanced instruments like gas chromatography. Various techniques have been used to separate an analyte, the specific aromatic alcohols (QSMs) from the sample. However, these chemical approaches are not that practicable physically and economically. Even if the initial and final concentrations of aromatic alcohols (QSMs) and microbial cells in the culture medium are known, the QSM feedback of a microbe cannot be clearly stated, as the QSMs synthesis curves and the growth curves differ from each other. To rectify these limitations, several techniques have evolved. They are miniature fermentation models, fluorescent detector systems, and modern high-resolution phenyl columns coupled with HPLC and are considered the best QSM monitoring approaches that provide more detailed, specific, and rapid information at low cost[58].
Double fermenters and microfluidic devices are better approaches for studying cell to cell contact and QS interactions in yeast and bacteria. Microfluidic devices play a significant role in the analysis of biofilm formation and cell surface attachments. Among S. cerevisiae, flor yeasts carry out the biofilm formation on top of the wine surface during the aging process and enhance the quality of the wine. During this process, the cell-to-cell contacts can be studied by using microfluidic devices. There are not many works related to QS analysis during winemaking by using these devices. It has several specific properties, such as large-scale fluid flow systems, and these multiplex devices have advanced image processing and analyzing technologies like confocal microscopy. Figure 6 shows a simple microfluidic setup consisting of external pumps, a bubble trap, and a sewage collection tube. As these devices are made up of small micro-sized channels, it has become easy to alter the fluid flow rate, transfer samples, manipulate reagents, and carry out reactions in microliter volumes. The benefits of this device are low reagent consumption, a short reaction period, and detailed monitoring[76].
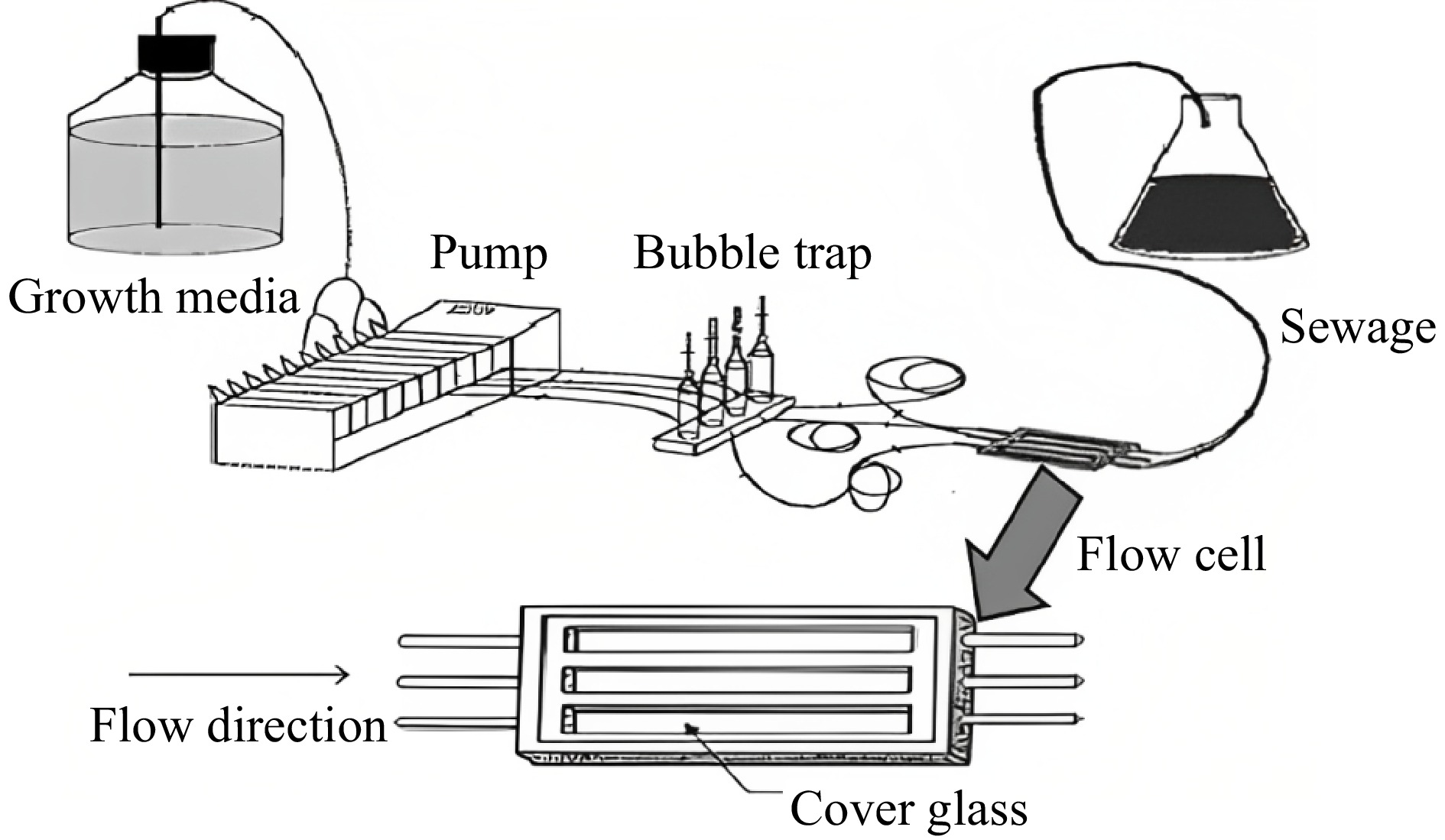
Figure 6.
A simple microfluidic setup[76].
Genetic approaches
-
In genetic approaches, the gene knockout technique is used to analyze QSMs and mechanisms[77]. Real-time PCR (RT-PCR) is an effective method for such analysis. In recent years quantitative RT-PCR has become a significantly common technique for analyzing gene expression in various samples. Generally, the expression of the target gene is studied with the expression of a reference gene to normalize the quantity of the PCR template and, thus, to evaluate the extent of relative expression of the target gene (i.e., normalized gene expression). Instead of analyzing a standard plot, target gene expression values are evaluated relative to the reference gene. The identity of synthetic and natural (purified) QSMs can be analyzed by mixing synthetic molecules into a mutant culture that cannot create its signals[46]. In a study carried out by the Fink laboratory, ARO genes are primarily responsible for the production of QSMs such as tyrosol, 2-phenylethanol, and tryptophol[59]. TaqMan chemistry and S. cerevisiae ARO knock-out strains are used to study the expression of the FLO11, ARO9, and ARO10 genes. Their results connected the environmental sensing, that is QS mechanism, and cell morphology of S. cerevisiae depending on the nutritional level and specific cell density in the medium. Furthermore, they used whole-genome microarray analysis to confirm the outputs of the RT-PCR approach, discovering genes involved in the coordination of aromatic alcohol synthesis induced by ammonium limitation. Later they carried out kinetic studies on ARO gene expression and synthesis of QSMs collectively during wine fermentation. The understanding of the kinetics of QSM is an essential component for the compelling study of QSMs. This study revealed the correlation between the peak expression of specific genes ARO8, ARO9, and ARO10 responsible for the peak synthesis of QSMs like tyrosol, 2-phenylethanol, and tryptophol[55].
-
Currently, omics technologies have been increasingly applied to wine production. Genomics, transcriptomics, metagenomics, metabolomics, and proteomics provide immense data into the metabolomics, genetic and molecular aspects of microbial consortium involved in winemaking[78]. Several multi–omics technologies were applied in winemaking that created new avenues in the wine industry. Among them, metagenomics, metatranscriptomics, metaproteomics, and metabolomics are significant techniques[78]. In winemaking, metagenomics is used to observe the changes in the microbial community during wine production and strain level detection. This application mainly targets DNA molecules. NGS or DNA-seq are the analytical techniques used in metagenomics[79]. Metatranscriptomics is used to analyze microbial functions, microbial pathways during wine production, and proactive spoilage detections. Total RNA and mRNA are the target molecules for this application. NGS or RNA-seq, and stable isotope probing (SIP) are the analytical techniques used in metatranscriptomics[79]. Metaproteomics targets protein molecules and is used to observe microbial proteins during fermentation and the infectious impact of unwanted microorganisms. Mass spectrometry is the analytical technique used for this analysis[80]. Finally, metabolomics targets volatile and non-volatile substances in wine. Mainly metabolomics characterizes metabolic pathways and precursors, identification of metabolites during fermentation, and analyze interaction among microorganisms. Gas chromatography – Mass Spectrometry (GC/MS) and Nuclear Magnetic Resonance (NMR) are the analytical techniques used in metabolomics[80]. Furthermore, in silico approaches are also applied to some extent to study quorum sensing during wine fermentation. It comprises computational and modeling techniques to analyze the mechanism of quorum sensing and its impact on wine fermentation. These approaches unveil the mechanism of microbial communications and regulation of gene expression during fermentation. There were not many in silico approach applications in grape wine fermentation. In a previous study related to quorum sensing in cocoa bean fermentation through in silico perspective, QS and Quorum quenching (QQ) related effectors in bacteria that were involved in cocoa bean fermentation were identified[80]. This study facilitates the selection of strains with QS or QQ potential to enhance the fermentation and swap the ineffective microbial consortium. Several studies must be carried out in wine fermentation to find out the best WMC which results in better sensorial properties[81].
-
Yeast, lactic acid, and acetic acid bacteria are the major microbial species involved in QS during winemaking. Quorum sensing (QS) is an intercellular communication process in which wine microbial consortium collectively adapts their metabolism by secreting quorum sensing molecules (QSM) into their environment. These QSMs continuously diffuse into the medium until approaching the threshold level, which stimulates the microbial cell population. Moreover, these molecules bind with their target sensory proteins and stimulate the transcription and translation of genes responsible for aromatic alcohol production. The research findings revealed that ARO genes regulate the synthesis of quorum sensing molecules like tyrosol, 2-phenylethanol, and tryptophol. For instance, ARO8, ARO9, and ARO10 present in S. cerevisiae are the significant genes regulating the above QSMs and other aromatic alcohols, which determine the organoleptic qualities of wine. Another essential gene that affects the quality of wine is FLO11. Hence, different grape cultivars harbor different types of wine fermenting microbes with unique quorum sensing systems, leading to the unique organoleptic qualities in regional wine. Based on the studies of yeast and lactic acid bacterial species, it is postulated that the microbial consortium is responsible for the organoleptic properties of wine. In several food industries, aroma production and wine quality analysis are conducted using biotechnological applications on aromatic alcohols such as 2-phenylethanol, tryptophol, and tyrosol. The perfect selection of suitable species and strains with specific combinations favors various aromatic profiles of wine. According to the current findings, and if further molecular engineering level techniques are developed, we predict the potential of the QS technique to be used in regulating the wine fermentation process within the next decade. Although scientists are familiar with trophic interactions and cell signaling interactions, there is more knowledge to be unveiled on the roles of the specific interactions of significant microbial species connected to wine fermentation. Currently, most intellectuals have shown interest in mixed culture fermentation over the specific single yeast strain. CRISPR-Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats) technology also can be used to alter the DNA sequences and modify gene function. After selecting the best QS genes from various WMCs, responsible regions of their DNA sequences can be cut and merged as the most effective wine starter culture by using highly advanced CRISPR-Cas9 technology. Furthermore, current studies on the kinetics of QSMs and their regulation in wine fermentation facilitate a colossal platform for future developments. Even now, public perception remains a significant barrier to the commercialization and application of genetically modified microbial strains in winemaking. Therefore, to achieve novel qualities in wine, intellectuals need to enhance awareness of the significant advantages of recombinant DNA technology in food industries. The debate on the utilization of genetically modified organisms (GMOs) in food production continues. Most of the countries banned the imports and production of foods using GMOs, including Sri Lanka. Among these challenges, the novel focuses on QSM stimulating, and coding genes leading to new biotechnological approaches to develop high-quality, region-specific wine varieties.
-
The authors confirm contribution to the paper as follows: data collection and analysis, draft manuscript preparation and editing: Thivijan S; conceptualization, resources, validation: Undugoda LJS; guidance: Undugoda LJS, Thambulugala KM; supervision, manuscript review and editing: Undugoda LJS, Manage PM, Nugara RN, Thambulugala KM, Kannangara SD. All authors reviewed the results and approved the final version of the manuscript.
-
These data were derived from the following resources in the reference section and were analyzed during this study.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Thivijan S, Undugoda LJS, Nugara RN, Manage PM, Thambulugala KM, et al. 2023. Quorum sensing capability of wine microbial consortium involved in spontaneous fermentation of regional wine production. Studies in Fungi 8:20 doi: 10.48130/SIF-2023-0020
Quorum sensing capability of wine microbial consortium involved in spontaneous fermentation of regional wine production
- Received: 26 July 2023
- Accepted: 29 November 2023
- Published online: 21 December 2023
Abstract: Quorum sensing (QS) is an intercellular communication process in which wine microbial consortium collectively adapts their metabolism by secreting quorum sensing molecules (QSM) into their environment. These QSMs continuously diffuse into the medium until approaching the threshold level, which stimulates the microbial cell population. Moreover, these molecules bind with their target sensory proteins and stimulate the transcription and translation of genes responsible for aromatic alcohol production. The research findings revealed that ARO genes regulate the synthesis of quorum sensing molecules like tyrosol, 2-phenylethanol, and tryptophol. For instance, ARO8, ARO9, and ARO10 present in Saccharomyces cerevisiae are the significant genes regulating the above QSMs and other aromatic alcohols, which determine the organoleptic qualities of wine. Another essential gene that affects the quality of wine is FLO11. Hence, different grape cultivars harbor different types of wine fermenting microbes with unique quorum sensing systems, leading to the unique organoleptic qualities in regional wine. Since we could still find the quorum sensing system of S. cerevisiae, this may open avenues to conduct much research to discover the unique quorum sensing systems of different wine microbes. These findings will lead to novel wine starter cultures with many specific genes developed through recombinant DNA technology. Therefore, this review focuses on quorum sensing of wine microbial consortium involved in the fermentation process of spontaneous wine fermentation through the chemistry of QSMs and how these signaling processes are genetically manipulated. Furthermore, this focus reviews the organoleptic quality development of regional wine products due to different quorum sensing abilities.
-
Key words:
- Wine /
- Yeast /
- Fermentation /
- Recombinant DNA technology /
- Microbial consortium /
- Quorum sensing