-
As the main component of grain yield, grain size is one of the most critical agronomic characteristics of rice. Although grain size may be affected by the growth environment, it is predominantly controlled by intrinsic signals determined by the interplay of genetic regulators. Therefore, grain size is a goal of genetic improvement in rice breeding, and studies are focusing on the identification and characterization of rice grain-size regulators[1−3]. With the development and application of breeding technologies, it is of great value to understand the mechanisms of grain-size control in breeding high-quality, high-yield rice varieties to ensure food security.
A rice grain is comprised of the embryo and the endosperm, which are covered by an aleurone layer and a thin seed coat. In a mature rice grain, the endosperm occupies most of the volume and determines grain size. The grain is enclosed by the spikelet hull, which consists of a palea and a lemma. The size and shape of the spikelet hull limits the growth and development of the embryo and endosperm, and therefore influences final grain size and shape. Recent studies have revealed that several signaling pathways participate in rice grain-size control[1−5].
The mitogen‐activated protein kinase (MAPK) cascade is comprised of three types of serine-threonine protein kinases, namely MAPK kinase kinase (MAP kinase kinase kinase, MKKK), MAPK kinase (MAP kinase kinase, MKK), and MAPK. They are activated by different upstream receptors on the plasma membrane upon external stimuli, leading to the sequential phosphorylation of MKK and MAPK and the activation of MAPK, which phosphorylate specific downstream substrates to regulate diverse biological processes[6,7]. In plants, receptor‐like kinases (RLKs) and receptor‐like proteins function as upstream receptors of the MAPK cascade to recognize and transmit external and internal signals[8]. RLKs are characterized by an extracellular domain, a single transmembrane domain, and a cytoplasmic kinase domain. They compose a superfamily in plants and have evolved to mediate the communication needed to regulate growth, immunity, and development, as well as final seed/grain size[9−13]. Signaling from RLK to MAPK also requires intermediate components such as receptor‐like cytoplasmic kinase (RLCK)[14,15]. The identified phosphorylation substrates of the plant MAPKs include kinases, enzymes, transcription factors, and other proteins. Phosphorylation may alter their activity, subcellular localization, and/or protein stability to mediate different downstream events[16,17].
The rice genome encodes approximately 75 MAPKKKs, 8 MAPKKs, and 15 MAPKs[18,19], which have been reported to function in plant development, phytohormone biosynthesis and signaling, immune response, and abiotic stress[16,17]. Studies indicate that MAPK signaling controls grain size in rice. Here, we discuss the recent findings on the role of MAPK signaling in this process.
-
OsMKKK10/SMALL GRAIN 2 (SMG2), OsMKK4/SMG1, and OsMPK6 /DARWF AND SMALL GRAIN 1 (DSG1) are part of a cascade regulating grain size and panicle architecture in rice by promoting cell proliferation[20−23]. Sequential phosphorylation of OsMKK4 and OsMPK6 by OsMKKK10 activates OsMPK6, and OsMPK6 activity positively associates with grain size[20,23]. Loss-of-function of OsMKKK10, OsMKK4, or OsMPK6 decreases cell proliferation, resulting in dwarfism, dense panicles, and small grains but increased spikelet number per panicle[20−23]. On the contrary, constitutive activation of OsMKKK10 (CA-OsMKKK10) or OsMKK4 (OsMKK4-DD) increases grain size and plant height[20−23]. In addition, the osmkk4 mutant (large11-1D), in which OsMKK4 activity is enhanced due to the replacement of alanine227 by threonine, produces larger grains and shows increased grain weight[20]. Genetic analysis has shown that OsMKKK10, OsMKK4, and OsMPK6 function in a same pathway to regulate grain growth[20].
OsERECTA1 (OsER1), a leucine-rich repeat (LRR)-RLK, positively regulates grain size and negatively regulates spikelet number per panicle[24]. Genetic and biochemical analyses have suggested that the OsMKKK10–OsMKK4–OsMPK6 module acts downstream of OsER1 to regulate spikelet formation. Furthermore, OsMPK6 interacts with and phosphorylates the zinc finger transcription factor DROUGHT AND SALT TOLERANCE (DST), and it can enhance the transcriptional activity of DST, leading to increased CYTOKININ OXIDASE2 (OsCKX2) expression. This study demonstrates that the OsER1–OsMKKK10–OsMKK4–OsMPK6 module negatively regulates the number of spikelets per panicle by affecting cytokinin metabolism[24]. Attention has also been given to identify the ligands recognized by the OsER1 receptor. REGULATOR OF AWN ELONGATION2 (RAE2) or GRAIN NUMBER, GRAIN LENGTH AND AWN DEVELOPMENT1 (GAD1), an EPIDERMAL PATTERNING FACTOR (EPF)/EPF-LIKE (EPFL) peptide family member, has been shown to regulate rice grain number and size[25]. A similar panicle morphology in the GAD1 loss-of-function mutant and oser1 suggests that GAD1 may be one of the peptides activating the OsMKKK10–OsMKK4–OsMPK6 cascade to control grain size. More recently, the EPF/EPFL small secretory peptides (SSPs), including OsEPFL6, OsEPFL7, OsEPFL8, and OsEPFL9, were reported to be the ligands of the OsER1 receptor and to activate the MAPK cascade during panicle morphogenesis[26]. Notably, OsEPFL8, but not other SSPs of OsER1, is involved in spikelet fertility, and suppression of ligand (OsEPFL6/7/9)-receptor (OsER1) pairs optimizes panicle architecture and enhances rice yield[26]. However, the molecular mechanism between OsER1 and OsMKKK10 is still unclear.
In Arabidopsis, the MKK4/MKK5–MPK3/MPK6 cascade functions downstream of the receptor-like protein kinase ERECTA (ER) to control inflorescence architecture and stomatal development[27−29]. This regulatory pathway of inflorescence architecture appears to be conserved between monocots and dicots. In addition, it has been reported that ER regulates cell proliferation in the outer integument, thereby controlling seed size by the MKK4/5–MPK3/6–DA1–UBIQUITIN-SPECIFIC PROTEASE15 (UBP15) module in Arabidopsis[30]. The ubiquitin-activated protease DA1 cleaves and inactivates the positive seed-size regulator UBP15 to negatively regulate seed size[31−33]. MPK3/6 can phosphorylate DA1, which inactivates and destabilizes DA1 and increases UBP15 accumulation, thereby promoting seed growth[30]. Interestingly, ER regulates seed size independently of its intracellular domain, which is essential for the function of ER in inflorescence morphogenesis and stomatal development[27,30,34]. ER may interact with other membrane-located receptor(s) to recognize the ligands for seed-size regulation. In stomatal development and plant immune responses, ER binds to the co-receptor TOO MANY MOUTHS (TMM) and/or main co-receptors, BRI1-ASSOCIATED RECEPTOR KINASE 1/SOMATIC EMBRYOGENESIS RECEPTOR KINASE (BAK1/SERK) family LRR-RLKs[34−36]. Future studies should identify and characterize the ligands of ER, as well as the potential ER-interacting receptor(s) in seed-size control.
-
A recent study has shown that OsMKKK70 function is redundant with that of its homologs OsMKKK62 and OsMKKK55, which control grain size and leaf angle by OsMKK4–OsMPK6–OsWRKY53 signaling[37]. OsMKKK70 overexpression increases grain length and leaf angle. The osmkkk70 mutant shows no significant differences in grain size and leaf angle compared with wild type; however, the osmkkk62/70 and osmkkk55/62/70 mutants show smaller grains, erect leaves, and reduced brassinosteroid (BR) sensitivity, suggesting that OsMKKK70, OsMKKK62, and OsMKKK55 functions are redundant in the control of grain size and leaf angle. OsMKKK70-overexpressing plants showed an increase in lemma cell number, indicating that OsMKKK70 enhances cell proliferation in spikelets and promotes grain growth. Although OsMKK4 phosphorylation by OsMKKK70 was not observed, OsMKKK70 can interact with OsMKK4, and OsMKKK70 can promote OsMPK6 phosphorylation in vivo. In addition, overexpression or constitutive activation of OsMKK4 or OsMPK6 partially rescues the grain size, leaf angle, and BR hyposensitivity phenotypes of the osmkkk62/70 double mutant, suggesting that OsMKKK70, OsMKK4, and OsMPK6 function through a common pathway to regulate grain size and leaf angle.
OsMKKK55/62/70 are involved in diverse developmental processes and stress responses. OsMKKK70 overexpression increases grain size but reduces pollen fertility and seed setting percentage, indicating its role in reproductive development[37]. OsMKKK55/62/70 also modulate the gibberellin (GA) content in anthers to regulate cold tolerance at the booting stage[38]. In normal temperatures, the seed setting rate of osmkkk62/70 and osmkkk55/62/70 is similar to that of wild type, whereas in cold conditions the mutants show an increased seed setting rate compared to wild type. In addition, OsMKKK62 functions upstream of the OsMKK3–OsMAPK7/14 module to control seed dormancy[39], and OsMKKK55/70 may also be involved in this process[38].
OsWRKY53 positively regulates grain size and BR signaling[40]. OsWRKY53 overexpression increases grain size, leaf angle, and exogenous BR sensitivity, whereas the oswrky53 mutant produces small grains and shows BR-deficient phenotypes such as decreased leaf inclination, dwarfism, and low sensitivity to exogenous BR. OsWRKY53 can be phosphorylated by OsMPK6 in an OsMKK4-dependent manner, which is critical for the positive regulation of BR signaling by OsWRKY53[40]. Phosphor-mimicking OsWRKY53 can partially rescue the grain size and BR hyposensitivity phenotypes of the osmkkk62/70 mutant[37], indicating that both OsWRKY53 and OsMKKK70/62 control grain size and BR signaling. OsWRKY53 overexpression can also suppress the small-grain phenotype of osmpk6 and osmkkk10, suggesting that OsWRKY53 functions downstream of the OsMKKK10–OsMKK4–OsMPK6 module to control grain growth[41]. Notably, WRKY53 mainly affects cell size and slightly affects cell number in the spikelet hull[41], whereas OsMKKK70–OsMKK4–OsMPK6 and OsMKKK10–OsMKK4–OsMPK6 modules regulate grain size mainly through cell proliferation, indicating that there are unknown components downstream of the MAPK cascade that enhance cell division to promote grain growth. In addition, OsWRKY53-overexpressing and OsMKKK70-overexpressing plants display dwarfism, whereas OsMKKK10 overexpression increases plant height, indicating that OsMKKK10 and OsMKKK70 function through different downstream signaling pathways to regulate plant height[37].
Similar to OsMKKK70 and its homologs, OsWRKY53 negatively regulates cold tolerance at the booting stage by modulating the GA content in anthers. The oswrky53 mutant shows a higher fertile pollen ratio and a higher seed setting rate compared to the wild type in cold conditions. By contrast, OsWRKY53 overexpression leads to a decreased fertile pollen ratio and seed setting rate, consistent with the findings of osmkkk62/70. These findings suggest that the OsMKKK70–OsMKK4–OsMPK6–OsWRKY53 cascade may mediate a trade-off between grain size and seed setting under cold stress[38]. In addition, OsWRKY53 functions downstream of OsMKK4–OsMPK6 in defense responses to wounding, pathogens, and herbivores[42−44]. Interestingly, it acts as a positive regulator in pathogen defense[42] but as a negative regulator in herbivore-induced defense[44,45].
-
OsMKK3 affects cell proliferation in spikelet hulls to regulate grain size[46]. Loss-of-function of OsMKK3 reduces grain length, grain width, and chalkiness, whereas overexpression of OsMKK3 increases grain size. Interestingly, natural variation in OsMKK3 influences grain size and chalkiness in rice. Four OsMKK3 haplotypes have been identified in wild rice accessions, and it is believed that the OsMKK3 haplotype present in cultivated rice originated from different wild rice accessions. Furthermore, OsMKK3 underwent strong selection during the domestication of indica and japonica, and polymerization of OsMKK3-Hap1 with other beneficial alleles increased grain length and quality.
OsMKK3 overexpression in rice has been reported to contribute to increased resistance to brown planthopper (Nilaparvata lugens) and leaf blight disease (Xanthomonas oryzae)[47,48]. OsMKK3 phosphorylates OsMPK7 and activates OsWRKY30 to enhance the defense response against X. oryzae, which causes leaf blight disease[48]. In Arabidopsis, MKK3 is involved in several hormone signaling pathways and stress responses[16]. It functions in the MAPKKK14–MKK3–MPK1/MPK2/MPK7 cascade, which is activated by wound-induced jasmonic acid (JA) production, and in the MAPKKK17/18–MKK3–MPK1/2/7/14 cascade, which is triggered by ABA signaling[49−51]. It can also act upstream of MPK6 to regulate JA signaling[52] and blue light-induced seedling development[53]. However, the components acting upstream and downstream of OsMKK3 in grain-size control are still unclear.
-
MITOGEN-ACTIVATED PROTEIN KINASE PHOSPHATASE1 (OsMKP1)/GRAIN SIZE AND NUMBER1 (GSN1) negatively regulates grain size and weight, but positively regulates grain number per panicle[21,54]. Loss-of-function of OsMKP1/GSN1 results in large grains and sparse panicles; however, OsMKP1 expression is positively correlated with the grain number per panicle. OsMKP1 directly interacts with OsMPK6 and inactivates it via dephosphorylation. Furthermore, the large-grain, sparse-panicle phenotype of the gsn1 mutant is rescued by suppression of OsMKKK10, OsMKK4, or OsMPK6, suggesting that OsMKP1/GSN1 and the OsMKKK10–OsMKK4–OsMPK6 module employ a common pathway to regulate panicle morphogenesis and grain size[21]. Therefore, OsMKP1 coordinates the trade-off between grain size and grain number per panicle by regulating the OsMKKK10–OsMKK4–OsMPK6 module.
In Arabidopsis, MKP1 participates in stomatal development and plant immunity by modulating the MAPK cascade[55−58]. MKP1 inhibits the activity of MAPKs in early stomatal lineage cells, thereby positively regulating stomatal development[55]. MKP1 also negatively regulates plant immunity. For instance, the mkp1 mutant displays enhanced activation of MPK3 and MPK6 as well as defense responses[56−58]. These findings reveal that the phosphatase MKP1 is essential for modulating MAPK signaling in various developmental processes, as well as plant immunity.
GSK2 modulates the OsMKK4–OsMPK6 cascade to mediate crosstalk between MAPK signaling and BR signaling
-
The GSK3/SHAGGY-like kinase GSK2 is part of the BR signaling pathway. GSK2 interacts with and phosphorylates OsMKK4 to inhibit OsMKK4-mediated phosphorylation of its substrate OsMPK6, thereby negatively regulating OsMPK6 activity[41]. GSK2 overexpression leads to short grains and typical BR-deficient phenotypes, whereas knockdown of GSK2 by RNA interference increases grain size and enhances BR sensitivity[59]. In Arabidopsis, the GSK2 ortholog BIN2 phosphorylates YDA and MKK4/5 and reduces YODA(YDA)–MKK4/5–MPK3/6 activity to regulate BR-mediated stomatal development[60,61], which is suggestive of crosstalk between BR signaling and MAPK signaling.
Notably, GSK2 modulates the activity of several transcription activators involved in grain-size control, including DWARF AND LOW-TILLERING (DLT/OsGRAS-32/D62/GS6) that positively regulates the BR response and negatively regulates cell division in grain growth[59,62], GS2/GROWTH-REGULATING FACTOR 4 (OsGRF4) that increases grain size by enhancing cell elongation[63−65], and GRAIN SHAPE GENE ON CHROMOSOME 9 (GS9) that regulates grain shape by influencing cell division[66]. GSK2 can also phosphorylate OsWRKY53, the downstream target of OsMPK6, to reduce its stability[41]. Knockout of OsWRKY53 rescues the large grain size and leaf angle phenotypes caused by the knockdown of GSK2, indicating that OsWRKY53 acts downstream of OsGSK2 to control grain size and BR signaling. In terms of grain growth, OsWRKY53 and GSK2 mainly affect cell elongation and slightly affect cell number, whereas OsMKKK70, OsMKK4, and OsMPK6 mainly affect cell number[41]. Given that BR promotes grain growth by regulating cell expansion in spikelet hulls[1], it is likely that OsWRKY53 plays a significant role in BR-mediated seed-size control and an insignificant role in MAPK-regulated seed-size control[41].
OsRac1 controls rice grain size by influencing the phosphorylation level of OsMPK6
-
OsRac1 is a member of the highly conserved ROP/Rac small GTPase family. ROP GTPases function as molecular switches in plant development processes and stress responses[67−70]. OsRac1 positively regulates grain size and yield by promoting cell proliferation[71]. OsRac1 overexpression increases the grain filling rate, as well as grain width, grain weight, and grain yield. OsRac1 interacts with OsMPK6 and influences its phosphorylation. Both OsMPK6 and OsRac1 affect cell division to control grain size, and OsMPK6 functions downstream of OsRac1 in grain-size control. OsRac1 also positively regulates disease resistance through OsMPK6[72,73]. Therefore, OsRac1 is a promising target for rice breeding.
FLR1 regulates grain size possibly through the OsRac1–OsMPK6 module
-
The Catharanthus roseus RLK (CrRLK1L) family FERONIA-like receptor 1 (FLR1) negatively regulates grain size in rice[74]. FLR1 interacts with OsRac1 via its kinase domain, indicating that FLR1 may function through the OsRac1–OsMPK6 module to control grain growth. However, the interaction of FLR1 with the OsRac1–OsMPK6 module in grain-size control is unclear. FLR1 influences both cell expansion and cell division in spikelets, suggesting that FLR1 regulates cell division through OsRac1–OsMPK6 but regulates cell elongation through other downstream components. Interestingly, FLR1 tends to bind to activated OsRac1 over inactivated OsRac1, and it has been proposed that inactive OsRac1 is liberated from the cell membrane to trigger downstream effectors. In addition, the flr1 mutant produces large and wide grains but displays an increased chalkiness percentage, indicating that FLR1 negatively regulates grain size but positively regulates grain quality[74].
Among the rice FLRs, FLR1 and FLR2 are most homologous to Arabidopsis FERONIA (FER). The Arabidopsis fer mutant shows large seeds caused by increased cell elongation in the integuments, but small leaves, short root hairs, and few epidermal hairs caused by decreased cell elongation, indicating that FER inhibits cell elongation during seed growth but enhances cell elongation in certain vegetative tissues[75−78]. FER establishes a signaling complex with RopGEF (Rop guanine nucleotide exchange factor) and Rop/Rac GTPase to mediate auxin-induced root hair growth[75]. GEF1 overexpression in Arabidopsis limits seed growth[77], suggesting that FER may function through Rac1–MPK6 to regulate seed growth. Moreover, FER can recognize different RAPID ALKALINIZATION FACTOR (RALF) peptides to regulate growth, immunity, and development[79−81]. Therefore, it is possible that FLR1 regulates grain growth by recognizing specific RALF peptides. In addition, FER-RALF1 activates TOR signaling in response to low nutrient levels[82]. It would be interesting to investigate whether FLRs can increase grain size under nitrogen-deficient conditions.
-
Recent studies have identified two MAPK signaling pathways and several regulatory components that play key roles in grain-size control (Fig. 1). However, there are still many gaps to be filled. For instance, OsMPK6 regulates grain growth mainly through cell proliferation, whereas OsWRKY53 plays a minor role in cell proliferation and a major role in cell elongation[41]. However, the main downstream regulators of OsMPK6 in grain-size control remain elusive. Genetic screening of the modifiers of the osmpk6 mutant or phosphor-proteomic searches may be key in identifying the OsMPK6 substrates that mediate grain-size control. Considering that DA1 functions downstream of the MKK4/5–MPK3/6 module to control seed size in Arabidopsis[30], the rice ortholog of DA1 is also likely to be a target of the OsMKK4–OsMPK6 module in grain-growth control.
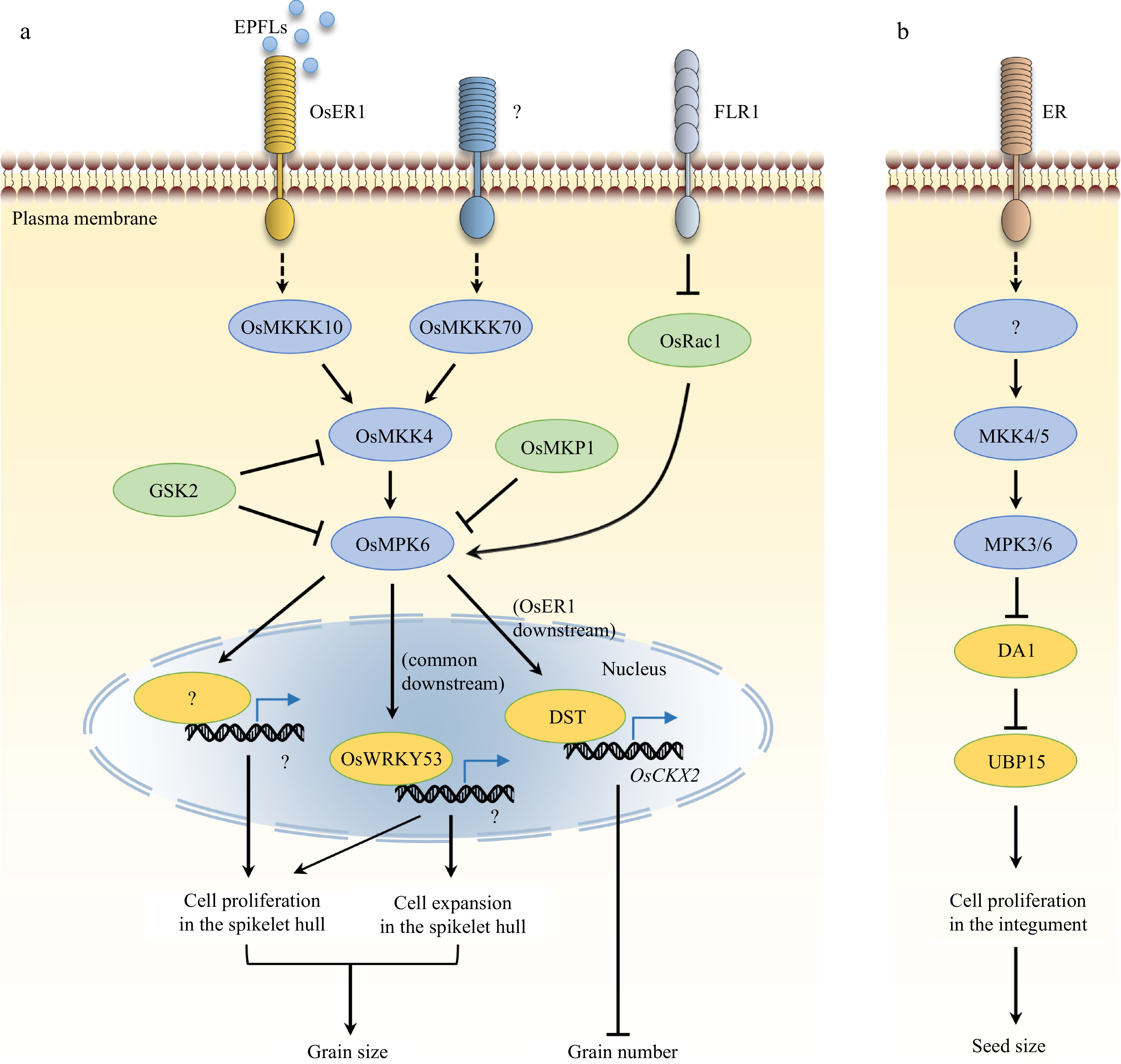
Figure 1.
Control of grain size and number by the MAPK signaling in rice and a comparison to that in Arabidopsis. (a) OsMKKK10–OsMKK4–OsMPK6 and OsMKKK70–OsMKK4–OsMAPK6 ascades play key roles in grain-size control in rice. OsWRKY53 acts downstream of OsMAPK6 to regulate grain size by mainly promoting cell expansion and slightly promoting cell proliferation in the spikelet, whereas the major downstream targets of OsMAPK6 that facilitate cell proliferation in the spikelet hull remain elusive. OsER1 functions upstream of the OsMKKK10–OsMKK4–OsMPK6 module and regulates the morphology of panicles and the number of spikelets per panicle by influencing the metabolism of cytokinin. EPFL peptides act as ligands of OsER1 in this signaling pathway. GSK2, OsMKP1, and OsRac1 influence grain size by modulating the MAPK cascade, whereas FLR1 regulates grain size probably through the OsRac1-OsMAPK6 module. OsMKK3 is not included in the illustration as the upstream and downstream components are unknown. (b) In Arabidopsis, ER functions upstream of the MKK4/5–MPK3/6 cascade to control seed size by regulating DA1-UBP15 activity.
Although the plant genome encodes multiple MKKKs, MKKs, and MAPKs that can form countless MAPK-cascade combinations, different signaling pathways sometimes use a common MAPK module to regulate diverse cell processes[6,7,16,17]. For instance, the OsMKKK10–OsMKK4–OsMPK6 cascade regulates both grain size and grain number per panicle, whereas the OsMKK4–OsMPK6 module controls many other developmental processes and immune responses[16,17]. How different signaling pathways are activated during panicle and grain development to balance grain number, grain size, and other traits remains to be investigated. To achieve signaling specificity, a MAPK cascade can be activated by different upstream signals, or it can target different substrates to participate in different signaling pathways. Signal specificity may lie in the spatiotemporal expression of upstream and downstream components of a certain MAPK cascade[16,17]. Thus far, the expression patterns of different receptors and downstream targets of MAPK signaling that are involved in grain-size control remain elusive. Notably, Arabidopsis ER is expressed in different tissues where it participates in different developmental processes. It is likely that the activation of OsER1–OsMKKK10–OsMKK4–OsMPK6–cytokinin signaling relies on the cellular or temporal-specific expression of EPFLs during panicle morphogenesis. For instance, OsER1 recognizes specifically expressed EPFLs and activates downstream OsMKKK10–OsMKK4–OsMPK6 signaling. DST phosphorylation by OsMPK6 upregulates its activity, resulting in increased OsCKX2 transcription and decreased cytokinin levels, which limits grain number[24,26]. Meanwhile, OsMPK6 activates OsWRKY53 and other unknown targets to enhance cell proliferation and cell expansion in the spikelet hull, thereby increasing grain size. Antagonistic to this signaling cascade, OsMKP1 balances grain number and grain size by affecting OsMPK6 activity[24]. Activation of the OsMKKK70–OsMKK4–OsMPK6 module by unknown ligands and receptors can also trigger downstream signaling to promote grain growth. However, it is unclear whether this occurs at the same time and in the same space as OsMKKK10–OsMKK4–OsMPK6 module activation. Identification of the ligands and substrates, as well as an investigation of the spatiotemporally expressed components, would help answer these questions.
The scaffold proteins mediating interactions between MAPK components also contribute to signaling specificity[16,17]. By assembling the required components, they can increase the efficiency of the interaction or control the spatiotemporal specificity of the cascade. For instance, BREAKING OF ASYMMETRY IN THE STOMATAL LINEAGE (BASL) functions as a scaffold for YDA and MPK3/6 to regulate the asymmetric division of stomatal lineage cells[83]. Phosphorylation of BASL by MPK3/6 leads to its polar localization and the recruitment of MPK3/6 and YDA, which enhances spatial YDA–MPK3/6 signaling and specifies cell fate. However, it is unclear whether the scaffold proteins recruit specific MAPK signaling components to regulate grain development and other downstream events.
The mechanisms by which MAPK cascades coordinately regulate grain development processes and stress/immune responses are unclear. A study has shown that membrane receptors for development- and immune-related signaling pathways can be spatially separated within nanodomains of their associated signaling components, thereby forming specific pools of signaling components within a cell[84]. Alternatively, MKKKs of different pathways can compete with common downstream MKKs and MAPKs to antagonize the interactions between a development-related MAPK pathway and an immune-related MAPK pathway[85]. At this point, we wonder whether different MAPK signaling pathways are compartmentalized for signal specificity or antagonistic with each other in the regulation of grain growth and other traits. Future studies are expected to answer these questions.
We sincerely apologize to our colleagues whose work could not be discussed due to space limitations. This work was supported by the National Natural Science Foundation of China (31871219), the Starting Grant from Hebei Agricultural University, China (YJ201920), the State Key Laboratory of North China Crop Improvement and Regulation (NCCIR2021KF-7), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA24010101, XDB27010102), and AgroST Project (NK2022050103).
-
The authors declare that they have no conflict of interest. Yunhai Li is the Editorial Board member of Seed Biology who was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of this Editorial Board member and his research groups.
-
# These authors contributed equally: Na Li, Liangliang Chen
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press on behalf of Hainan Yazhou Bay Seed Laboratory. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Li N, Chen L, Li Y. 2023. Control of grain size and number by MAPK signaling in rice. Seed Biology 2:15 doi: 10.48130/SeedBio-2023-0015
Control of grain size and number by MAPK signaling in rice
- Received: 27 June 2023
- Accepted: 11 August 2023
- Published online: 07 October 2023
Abstract: Grain size, a main component of grain yield, is regulated by a complex network. The mitogen-activated protein kinase (MAPK) cascade participates in multiple signaling pathways to regulate various biological processes. Recent studies indicate that MAPK signaling plays key roles in regulating grain size. For instance, OsERECTA1(OsER1)–OsMKKK10–OsMKK4–OsMPK6 signaling regulates grain size and grain number per panicle. Grain size is also affected by the OsMKKK70–OsMKK4–OsMPK6 module, which functions upstream of OsWRKY53. In addition, MITOGEN-ACTIVATED PROTEIN KINASE PHOSPHATASE1 (OsMKP1), the GSK3/SHAGGY-like kinase GSK2, and the Rho-family GTPase OsRac1 controls grain size in rice by modulating MAPK signaling. Here, we discuss recent findings on the importance of MAPK signaling in rice grain-size control and examine mechanisms by which MAPK signaling coordinates grain size, grain number and stress responses.
-
Key words:
- MAPK signaling /
- Grain size /
- Grain number /
- Receptor-like kinases













