-
Rice yield and quality have always been of concern as it is one of the main food crops worldwide. The rice yield greatly improved after the two green revolutions. However, research and breeding practices related to rice quality lag far behind those focused on rice yield. This may reflect the neglect of rice quality in the past, complexity of rice quality research, and lack of a consensus definition and evaluation criteria for rice quality. Rice quality is a complex character based on several evaluation indices that reflect both internal and external attributes, such as appearance, taste, and flavor. Rice quality encompasses various qualities, such as appearance, milling, nutritional value, cooking and eating, and sanitary qualities[1]. Rice quality, especially appearance, cooking, and eating quality, plays a crucial role in determining consumer preference and commercial value. Undoubtedly, rice with excellent appearance and taste is more likely to be favored by consumers.
Rice quality is mainly determined by the synthesis, composition, distribution, and accumulation of nutrients and storage products. Cooking and eating quality are important indicators of many quality traits. Currently, in breeding practice, improving the taste quality of rice relies primarily on allelic variations in the waxy gene Wx, encoding granule-bound starch synthase I (GBSSⅠ). This allows the cultivation of rice varieties with varying amylose contents to meet the needs of different consumer groups[2−9].
Wx is known to control amylose synthesis in rice endosperm. In addition to its direct function on amylose synthesis, the Wx gene interacts with many other genes, both directly and indirectly. These interactions regulate the expression level of Wx and ultimately impact the amylose content and quality of rice. The regulation occurs at the transcriptional and post-transcriptional levels, and also involves interactions with GBSSI proteins, as well as some unknown forms of interaction. Certain genes or QTL (Quantitative trait loci), such as Du1[10], Du3[11], Du13[12], FLO2[13], qAC2[14], qSAC3[15], and LowAC1[16], have been reported to manipulate Wx mRNA splicing efficiency, directly controlling the amylose content in rice grains. Additionally, certain transcription factors like OsNAC24[17], OsNAC20[18], OsNAC26[18], OsNF-YB1[19], OsNF-YC12[9], bHLH144[15], OsMADS7[20], OsMADS14[21], OsBP-5[22], OsEBP-89[22], REB[23], and OsbZIP60[24] have been found to regulate the expression of Wx gene in rice endosperm, either directly or indirectly. There are also some quality genes, such as OsAAP6[25], Chalk5[26], WCR1[27], FLO19[28], that affect the expression level of Wx genes in unknown ways. Furthermore, OsGBP[29] and FLO6[30] have been discovered to be involved in starch synthesis through direct interaction with GBSSI. Exploring natural variations of these genes that interact with Wx, or modifying these genes themselves through genetic engineering, are potential strategies for enhancing the quality of cooked rice.
The Wx gene not only affects the synthesis of endosperm starch and the physicochemical properties of starch[31], but also has a powerful pleiotropic function that impacts the quality of rice from multiple aspects. Tan et al. identified a major protein content QTL in the Wx gene region, which is also responsible for flour color[32]. Chen et al. investigated the total protein content and four storage protein contents of 527 rice germplasms using a genome-wide association study, and discovered that the distance between an association site with a phenotypic variation rate greater than 10% and the quality gene Wx was less than 20 Kb[33]. By conducting haplotype analysis and endosperm expression analysis of the Wx gene, they observed that the 2.3 kb mRNA level of Wx was inversely proportional to the albumin content[33]. This finding further supports the conclusion that Wx is a gene responsible for endosperm protein content. In 2020, two different research groups almost simultaneously reported that Wx also regulates grain transparency and eating quality[8,9]. According to Xia et al.'s genome-wide association studies, it was discovered that the Wx gene acts as a negative regulator for both crude fatty acid content and rice quality[34,35]. Qiu et al. showed that Wx gene is highly correlated with cooked rice elongation[36]. Deng et al.'s research highlighted the significance of the Wx gene in regulating grain fissure resistance, and its genetic variation conferred different levels of tolerance to fissuring in grains as well as head rice yield[37]. Thus, Wx does not affect rice quality solely by controlling amylose synthesis, but rather depends on its pleiotropic effects, which determine the overall quality of rice from various perspectives (Fig. 1).
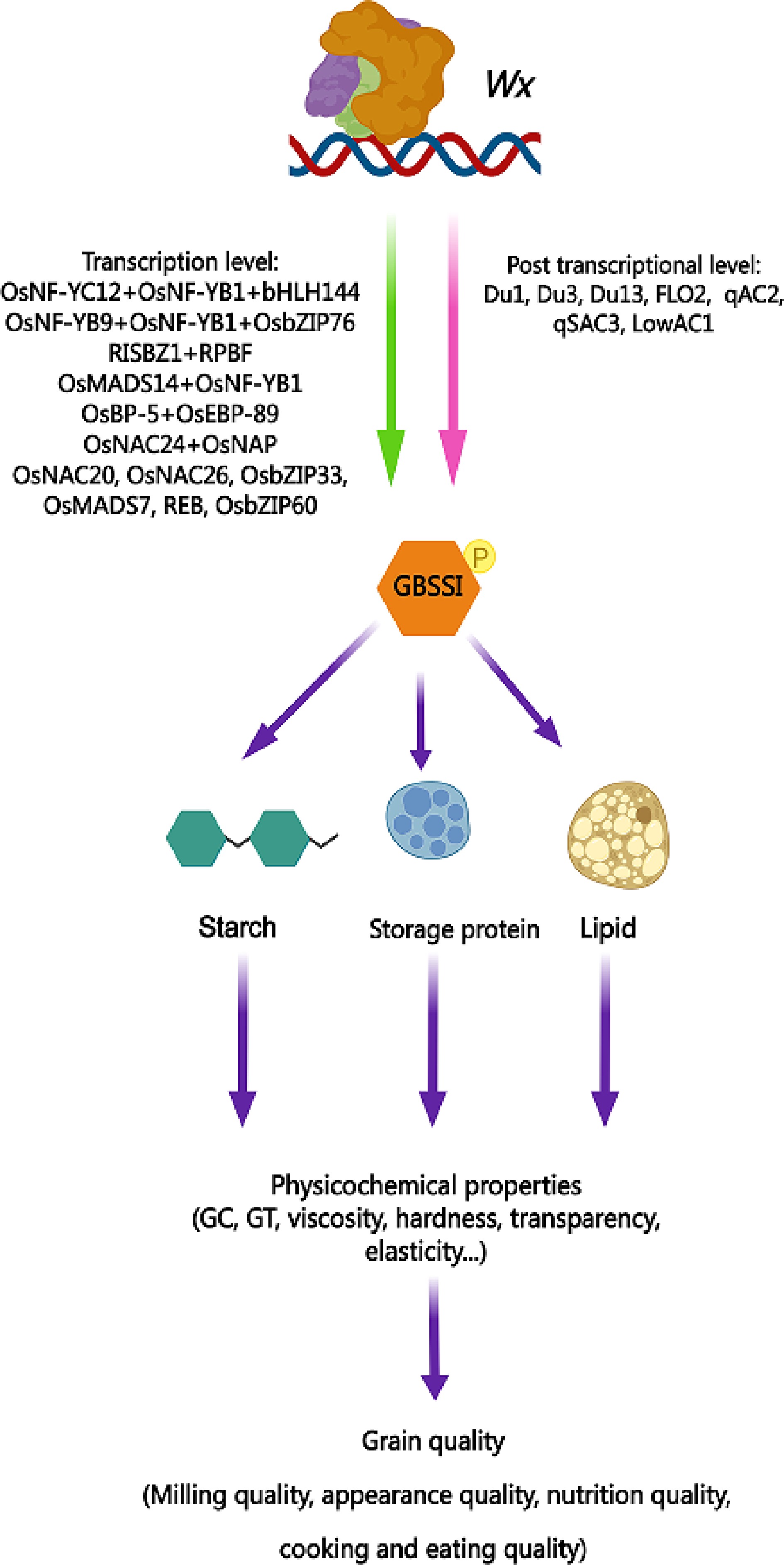
Figure 1.
Pleiotropy of Wx gene in rice quality regulation. This pattern diagram presents the regulatory factors of the Wx gene at both the transcriptional and post-transcriptional levels that have been reported thus far. It also highlights the encoding product GBSSI of the Wx gene, which plays a role in regulating the three major components of rice endosperm (starch, protein, and lipids) and subsequently affects various physicochemical properties of rice grains. The pleiotropic effect of the Wx gene ultimately determines the overall quality of rice from multiple perspectives. GC, gel consistency; GT, gelatinization temperature. Yellow spheres with the letter P indicate phosphorylation.
Although significant progress has been made in improving the quality of rice with regard to starch, the quality of the existing rice still fails to meet the diverse needs of consumers. Proteins have attracted the attention of rice quality researchers because they are the second-largest storage substances in rice, after starch. Proteins in rice can be divided into two categories according to their functions: storage and structural proteins. Most of the proteins in rice seeds are storage proteins, whereas structural proteins are responsible for maintaining normal cell metabolism, mainly hormones, enzymes, and enzyme inhibitors, and their total content is relatively small[38]. Therefore, rice proteins are generally referred to as storage proteins. Studies have shown that rice seed storage proteins are mainly distributed in the aleurone layer and embryo[39]. Albumin and globulin are more abundant in the aleurone and glume layers. Glutelin is the most abundant protein in the endosperm, and albumin is evenly distributed in rice bran, fine bran, and milled rice[40]. The storage proteins in the endosperm are mainly filled between starch granules in the form of independent proteomes (PBs), which can be divided into two types: spherical type I proteomes (PB-I) with concentric lamellar structures and elliptical type II proteomes (PB-II) without lamellar structures. PB-I mainly contains prolamin, accounting for approximately 20%–30% of the total endosperm storage protein[41]. PB-II is mainly composed of glutelin with a small amount of globulin, accounting for approximately 65% of the total stored protein in endosperm[42]. Glutelin is the first major component of rice grain storage protein[43]. To date, research on the biosynthesis and genetics on glutelin has been the most common. In this regard, there have been two excellent reviews[40,44] on the progress in rice glutelin biosynthesis and genetics, which will not be repeated here.
The protein content and composition of rice also seriously affect various quality traits, particularly the cooking and eating quality.
Singh et al. found that the gelatinization characteristics of rice flour and starch from the same variety were different, suggesting that factors other than starch affect the gelatinization process of rice[45]. Martin & Fitzgerald found that during the early stage of cooking, protein reduced the water absorption of rice by binding water and increased the concentration of starch gel in both dispersed and viscous phases through a disulfide-linked protein network, resulting in an increase in the peak viscosity of RVA spectra[46]. Hamaker & Griffin used the reducing agent dithiothreitol (DTT) to break the protein disulfide bond and observed significant swelling of the starch granules and an increase in peak viscosity[47,48]. After adding protease or DTT to the rice flour and non-glutinous rice flour, Xie et al. found that the peak viscosity, disintegration value, and recovery value of glutinous rice decreased significantly, indicating that the protein network formed by disulfide bonds enhances the gelatinization rigidity of glutinous rice flour. In contrast, the disintegration value of non-glutinous rice increases without disulfide bonds, and the expansion process of starch particles became more gelatinized[49]. Chavez-murillo et al. reported a significant negative correlation between rice protein, peak viscosity, and disintegration value in RVA, suggesting that proteins affect these indices by binding to water[50]. Baxter et al. added four protein components to pure starch and found that the addition of glutenin and albumin increased the starch gelatinization temperature, whereas the addition of globulin decreased it[51]. The albumin content showed a linear and positive correlation with the hardness of the starch gel and a negative correlation with adhesion. The addition of prolamin to the starch gel results in decreased hardness, adhesion, and adhesive properties. Furthermore, an increase in the prolamin content led to a decrease in both hardness and adhesion[52]. Zhou et al. hydrolyzed the protein in rice with protease and observed that during gelatinization, the number of starch molecules leached and water permeability increased, and the heat absorption temperature of gelatinization decreased, suggesting that protein is an important factor that affects the thermodynamic properties of stored rice[53]. Moreover, many studies have demonstrated that a higher endosperm protein content increases the hardness and roughness of rice and reduces its adhesion and smoothness[54,55]. Existing studies have generally found that the higher the protein content, the worse the cooking and eating quality of rice[56,57]. In summary, the protein content and composition of rice are closely related to its cooking and eating quality.
This review mainly focuses on the evaluation methods of rice cooking and eating quality and starch physicochemical properties, factors affecting rice protein content, the genetic basis of rice protein content, progress made in the genetic improvement of rice protein content, and future prospects in this field.
-
The evaluation methods for rice taste include instrumental, artificial sensory, physical, and chemical index evaluation methods. At present, the most widely used instrument in rice taste evaluation is the rice taste value analyzer produced by the SATAKE Company in Japan[58]. The instrument can calculate physical parameters, such as viscosity, elasticity, hardness, and balance, by measuring the light characteristics of rice. Based on these measurements, the instrument calculates the taste value of rice using a formula[59]. Sensory evaluation is the most accurate and direct manifestation of the taste quality of rice. After the rice is steamed and cooked under standardized and unified conditions, the evaluators comprehensively evaluate the color, odor, taste, viscosity, softness, hardness, and overall palatability of the tested rice using visual, nasal, and oral methods.
In addition to these two methods, physical and chemical index evaluation methods are widely used in the evaluation of rice quality and breeding of high-quality rice. The main physical and chemical indices involved include the amylose content, fine structure of amylopectin, gelatinization properties, texture characteristics, and thermodynamic properties (Fig. 2). Amylose content is considered the most important factor in determining rice quality[31,60−62]. Therefore, the effects of amylose on rice quality have been extensively and comprehensively studied, which has been summarized in detail in some excellent reviews[1,62−64] and will not be repeated here.
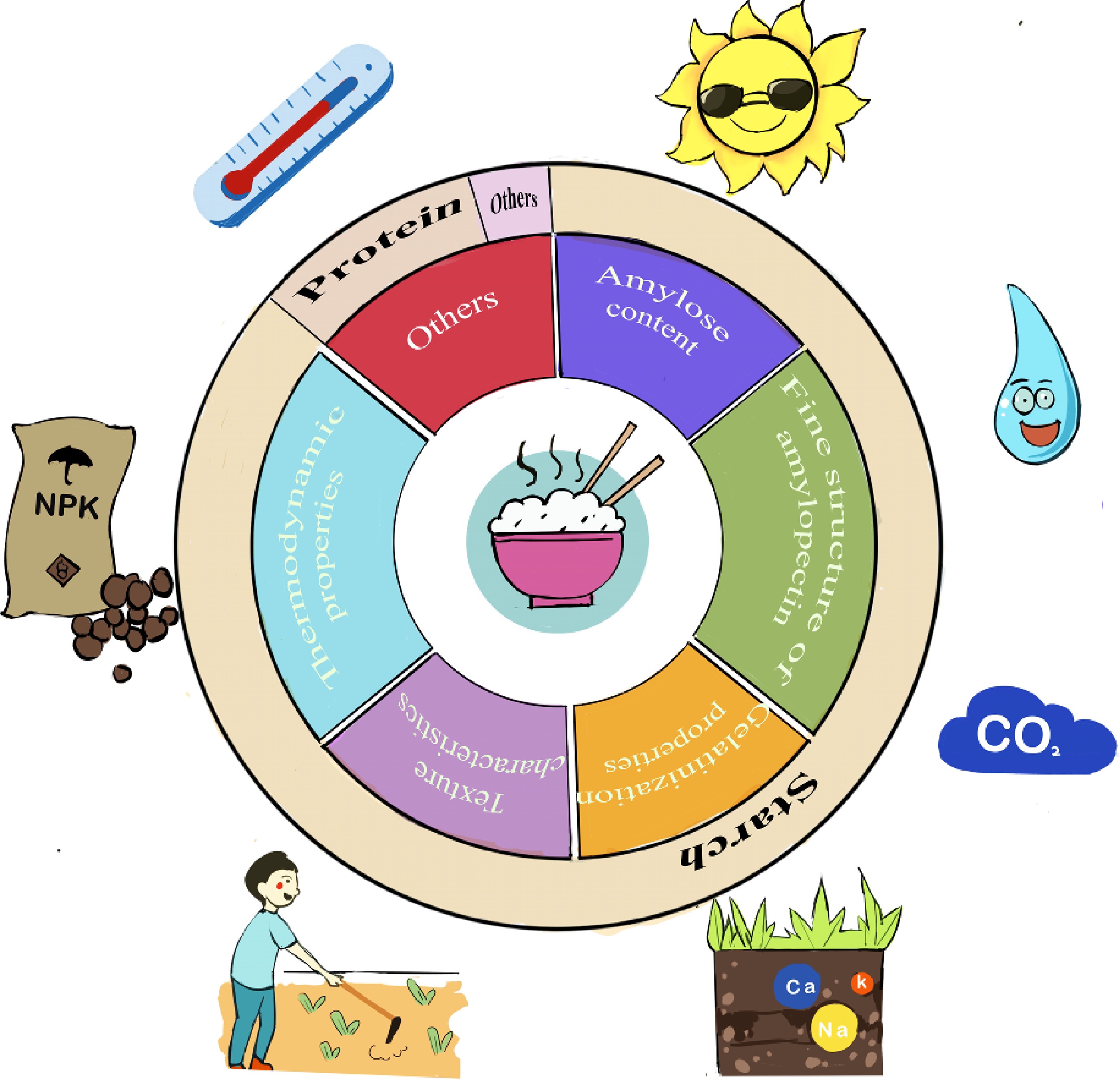
Figure 2.
Determinants of cooking and eating quality of rice and external factors affecting protein content of rice grains. The six parts in the inner circle represent the physical and chemical characteristics or indicators that determine the cooking and eating quality of rice; the three parts in the outer circle represent the material composition that affects the physical and chemical characteristics of rice; the cartoon diagram in the outermost circle represents the ecological factors and environmental factors that affect the grain protein content, from clockwise from 1 o'clock to indicate light, moisture, carbon dioxide, salinity, cultivation, fertilizer and temperature.
Amylose is a very small, linear, or slightly branched structure, whereas amylopectin is a highly branched component of starch. The difference in the eating quality of rice with the same amylose content is generally considered to be closely related to the fine structure of amylopectin. Amylopectin accounts for a large proportion of the starch in the rice endosperm, approximately 70%–90%, and its molecular weight is relatively large. It is generally composed of thousands of glucose residues. There are many non-reducing ends in the molecule, but only one reducing end. The fine complex structure of amylopectin is initially evident over a wide range of chain lengths. The amylopectin chain is divided into A, B, and C chains based on the branch point and degree of polymerization. The reducing end of the A chain is involved in the formation of α-1,6 glycosidic bonds. The reducing end of the B chain is connected to the C chain or the other B chains, and the C chain is the only macromolecule with a reducing end. Depending on the length of the chain, it can be divided into main short chain (generally 0–35 DP) and secondary long chain (generally greater than 35 DP), A chain (6 ≤ DP ≤ 12), B1 chain (13 ≤ DP ≤ 24), B2 chain (25 ≤ DP ≤ 36), B3 chain (DP ≥ 37), and some amylopectin molecules have extra-long chains (EL chains). Second, the fine and complex structure of amylopectin is also evident in the multilevel structure of the starch granules. Amylose and amylopectin aggregate and twist to form a double helix and then aggregate to form different types of crystals. The crystals form amorphous sheets alternately. In addition, crystalline sheets form periodic shell-shaped or ring-shaped growth rings, small starch granules have raised elastic bodies on the surface, and these small starch granules further fused to form composite starch granules.
The gelatinization properties of rice flour can be characterized by measuring the change in the viscosity of the starch suspension during heating and cooling. During heating, the viscosity gradually reaches a maximum value with increasing temperature and then gradually decreases. which is a function of the change in starch granules[65]. During the cooling process, the viscosity of the starch paste increases over time, which indicates that the gelatinized starch was recycled[66]. Currently, rapid viscosity analyzer (RVA) is mainly used to investigate the gelatinization properties of starch. The RVA gelatinization curve provides several important parameters: the gelatinization temperature (PM), which is the temperature at which starch begins to gelatinize; peak viscosity (PV), which represents the maximum viscosity of starch particles before breaking; peak time (PT), which indicates the time when starch reaches peak viscosity; decay value (BD), which indicates the stability of starch hot paste under shear stress; retrogradation value (SB), indicating that the gelatinized starch paste begins to retrograde when cooled; and final viscosity (FV), which represents the final viscosity of the RVA curve[67].
Texture properties, such as hardness, brittleness, stickiness, resilience, elasticity, and gel strength, are important food quality factors. Sensory evaluation is a highly recognized evaluation method; however, it has some limitations, such as strong subjectivity and being greatly influenced by the evaluator's own preferences. In this context, a texture instrument has been developed to replace human sensory evaluation and provide specific index parameters. A texture tester, also known as a physical property tester, is mainly used to simulate the mechanical movement of oral chewing. The most commonly used test is the texture profile analysis (TPA) program, which is also known as the secondary extrusion cycle or twice mastication test (two-bite test, TBT). By simulating the chewing movement of the human cavity, a solid or semi-solid sample is extruded twice to obtain texture parameters.
Starch gelatinization is an irreversible process that includes granule swelling, crystal dissolution, loss of birefringence, and starch dissolution, accompanied by changes in viscosity. The phase transition of starch granules involves the expansion and rupture of starch granules. The gel temperature is the key point of these two stages because it is at this point that the impact on the rupture of starch granules is the greatest. The temperature parameters of the starch gelatinization process can be determined by differential scanning calorimetry, including the degree of crystallization, transition effect, and melting point of starch during the gelatinization reaction, as well as the degree of gelatinization and recovery characteristics of starch.
-
As a typical quantitative trait, phenotypic differences in rice protein content among different genotypes are greatly affected by the environment. Ecological factors, such as temperature, light, and carbon dioxide concentration, and environmental factors, such as cultivation, affect the protein content of rice (Fig. 2).
Rice is a typical temperature-loving crop, and the protein content of the grain is very sensitive to temperature changes during the grain-filling stage. High temperatures during the filling stage usually lead to an increase in grain protein content, decrease in amylose content and taste value, and a decrease in grain quality[68]. The high temperature during the mature stage leads to abnormal rice quality, shape, and color, which may be attributed to a decrease in enzyme activity, respiratory consumption of assimilation products, and reduced sink activity related to grain filling[69,70].
Light, in addition to temperature, is another key factor that affects protein synthesis. It has been found that the main protein components, such as glutelin, and the most important essential amino acids, including lysine and threonine, increased significantly in rice harvested after low-light treatment at the filling stage; however, cooking quality decreased[71].
Some studies have shown that the atmospheric concentration of CO2 has an important effect on rice quality. Goufo et al. demonstrated that elevated carbon dioxide levels result in reduced protein content in plants, which is attributed to the inhibition of nitrate assimilation[72,73]. Furthermore, the researchers observed an increase in peak viscosity, minimum viscosity, breakdown value, final viscosity, and hardness of rice, while noting a decrease in setback value. All these alterations in physical and chemical indicators collectively indicate an enhancement in the cooking and eating quality of rice[72].
Moisture and fertilizer are the two most important factors in cultivation management. In many systems, 5,000 liters of water is required to produce 1 kg of rice, although this can be reduced to approximately 2,000 liters[74]. Soil moisture status has a significant impact on yield and grain quality[75]. Rice is generally grown under submerged conditions. Submerged plastic film mulching (PM) and submerged wheat straw mulching (SM) are emerging water-saving technologies for rice production. Different water management treatments, namely PM, water-saving grouting, and conventional irrigation, significantly affected the percentage of brown rice, milled rice, chalky grain, amylose content, and protein content in a variety- and grain-position-dependent manner. The protein content is the most affected by water management[76]. Nitrogen is crucial for plant growth and development. As nitrogen is still the main component of proteins, applying nitrogen fertilizer can also significantly affect the quality of grains[35,77]. The application of nitrogen fertilizer at different stages of panicle differentiation, heading, flowering, and grain filling could significantly increase the content of grain storage protein[78]. In addition to nitrogen, potassium is another fertilizer necessary for rice production. Some studies have shown that the application of potassium fertilizer increased gel consistency and grain protein content but had no significant effect on gelatinization temperature and amylose content[79].
Salinity is another important factor that significantly influences crop quality. By comparing rice varieties grown in low-and high-salinity areas, Siscar-Lee et al.[80] found that salt-tolerant and salt-sensitive varieties grown in saline-alkali soil had higher storage protein content than those grown in normal soil; however, these varieties also had less translucent grains and lower starch and amylose contents[80]. Owing to the significant influence of the environment, the interaction between the protein content genotype and the environment is large, and the heritability is relatively small. Some studies have shown that the heritability of the phenotype of protein content in rice is only 13.0% and 37.2%[81]; therefore, the phenotype with high protein content has little effect in the early generation. However, it is also reported that its heritability can reach 58.8%[82]. Therefore, it is feasible to screen materials with high protein content in the lower generation. Several external factors aggravate the complexity and challenge of rice protein research.
-
Rice grain protein content is a quality trait controlled by multiple genetic factors with a complex genetic basis. There are large differences in the protein content among the different varieties[83]. Chen et al. determined the grain protein content of 527 cultivated rice core germplasm and showed that the protein content ranged from 44.06 to 106.71 mg/g in 2014 and 32.64 to 80.08 mg/g in 2015[33]. Similarly, Yang et al. used near-infrared spectroscopy to measure the protein content of 402 core germplasms. The two-year phenotypic data showed that the protein content of rice varied from 5.33% to 14.83%, and the protein content of most varieties was distributed between 7.5% and 11.5%[83]. Liu et al. measured the protein content of 24 japonica rice varieties collected from different regions in China and found that the protein content ranged from 6.45% to 11.1%, with an average of 8.26%[84]. Webb et al. analyzed the protein content of approximately 4,000 rice varieties from 57 countries and found that the protein content ranged from 5.3% to 13.6%[85].
Genetic loci for protein
-
To date, numerous studies have mapped quantitative trait loci (QTL) that control the protein content of rice. Tan et al. used a set of recombinant inbred line populations to detect a QTL related to protein content on chromosomes 6 and 7 respectively, one of these QTLs was located near Wx on chromosome 6 and had a large effect, explaining 13.0% of the phenotypic variation[32]. Aluko et al. used DH populations derived from BC3F1 (O. sativa × O. glaberrima) to detect QTL controlling protein content on chromosomes 1, 2, 6, and 11, respectively[86]. Similar to the results obtained by Tan et al., the QTL on chromosome 6 had the largest effect and was located near the Wx gene[86]. Hu et al. also used a DH population (Gui630×02428) and identified five QTLs for rice protein content, among which the QTL effect on chromosome 5 in RG435-RG172a was found to be the largest[87]. Kepiro et al. used an RIL population to map QTLs related to protein content in both brown rice and milled rice respectively[88]. They observed that the two loci were located in brown rice and three loci were located in milled rice. The loci on chromosomes 1 and 4 simultaneously control the protein content of brown rice and milled rice[88]. Wang et al. used the RIL population (Zhenshan97/Nanyangzhan) to map QTLs for different amino acid contents and found that there was a QTL with a large effect at the end of the long arm of rice chromosome 1 that simultaneously controlled the contents of multiple amino acids[89]. Using the RIL population constructed by Chuan 7 and Nanyangzhan, Luo et al. detected two QTLs affecting protein content; however, the effects of these QTLs were small, and phenotypic variation explained only 7.2%[90]. Ye et al. detected at least 15 fragments related to protein content using two-year data of chromosome segment substitution line (CSSL) populations at four sites, among which CSSL-48 on chromosome 8 was detected in all eight environments[91]. Liu et al. mapped nine protein content-related QTL on chromosomes 1, 2, 3, 6, 8, and 11 using a CSSL (Asominori × IR24) population[92]. Bruno et al. identified a minor QTL for brown rice protein content on chromosome 7, using a set of DH populations[93]. Kashiwagi & Munakata used a set of single-segment substitution line populations obtained from crosses between Koshihikari and NonaBokra to identify a stably inherited QTL, TGP12, which controls the protein content of rice across three years of different environments[94]. This QTL can specifically reduce protein content of rice without affecting its cooking and eating quality[94]. Park et al. used a set of RIL populations obtained from the crossing parents with large differences in rice quality, and identified a stable genetic QTL qPro9 on chromosome 9 through 2 years of repeated identification, and fine-mapped it to a specific region of 34 Kb[95]. Zhang et al. studied the inheritance of crude protein and its components in rice using 71 recombinant inbred lines (RIL) obtained from crossing the japonica rice variety Asominori with indica rice variety IR24[96]. They identified a total of 16 QTLs located on eight chromosomes[96]. To investigate the genetic relationship between rice yield and rice nutrient content, Yu et al. used 209 recombinant inbred lines obtained from crossing XieqingzaoB with Milyang46 to map the QTL that affects brown rice yield and two main nutrient contents. Five QTLs related to protein content were detected on chromosomes 3, 4, 5, 6, and 10. Among them, a major QTL qPC-6 was located near the Wx marker RM190 on the short arm of rice chromosome 6, which explains 19.3% of the phenotypic variation with an additive effect of 0.471%[97]. Zheng et al. used 71 lines derived from 'Asominori/IR24' to analyze the developmental behavior of protein content and protein index (PI) through unconditional and conditional QTL mapping. Ten unconditional QTLs, six conditional QTLs of proteins, 11 unconditional QTLs, and nine conditional QTLs of PI have been identified at four stages of grain filling[98].
Advanced molecular breeding methods, such as genome-wide association analysis (GWAS), supported by next-generation sequencing and omics technology, can effectively identify genomic regions related to rice quality traits[99−102]. Thus, in addition to using parental-derived genetic populations to identify QTLs related to rice protein content, researchers have also attempted to use natural populations of rice to conduct GWAS to find genes/QTLs related to protein content of rice. Chen et al. conducted GWAS of total protein content and four stored protein contents of 527 rice varieties in the overall population, as well as in the indica rice subpopulation and japonica rice subpopulation. They detected a total of 107 significant association loci, of which 28 loci overlapped with reported QTLs or intervals known to control rice protein content. Sixteen loci were detected in different populations and nine of them were simultaneously detected in different phenotypes. Based on the analysis of the associated loci that explained a phenotypic variation rate of more than 10%, 13 loci were found to be co-located with genes related to quality, and the distances between 5 loci and quality-related genes (PG5a, Wx, AGPS2a, RP6, and RM1) were less than 20Kb[33]. Similarly, 135 significant loci related to grain protein content have been identified through genome-wide association studies[103], among these loci, six leading SNPs are located near the known genes involved in the biosynthesis and accumulation of storage proteins (less than 150 Kb), including Sar1a, GluB6, OsTudor-SN, and Glb1. In addition, two genes (Susy2 and Flo5), which have been shown to play a key role in rice starch synthesis, are less than 50 Kb from the leading SNPs, and the chalkiness rate and protein content of mutants obtained by editing Flo5 are significantly higher than those of the wild type[103]. Yang et al. used a population of CSSLs obtained by crossing the indica rice variety Habataki with japonica rice variety Sasanishiki for QTL mapping and identified 18 QTLs related to grain protein content across three environmental conditions. Among these, qGPC-1 and qGPC-10 were repeatedly identified in all three environmental conditions, whereas qGPC-3, qGPC-8, and qGPC-12 were detected under both environmental conditions, and the others could only be detected under one environmental condition[83]. Huang et al. conducted a genome-wide association study on heading date and ten grain-related traits, of 950 rice varieties worldwide using a high-density haplotype map to identify five candidate genes for grain protein content on chromosomes 6, 7, and 11, in combination with expression profile data and gene annotation information[104]. Verma et al. used 42,446 SNP markers to perform a genome-wide association study on five grain quality traits of 103 rice varieties and identified multiple grain protein QTLs on chromosomes 1, 2, 6, 7, 10, and 11[105]. Moreover, a novel grain protein QTL, qPRO_1.12 was identified on chromosome 12[105]. Using different sets of 258 germplasm from the 3 K Rice Genome Project, Wang et al. conducted an association study on apparent amylopectin content (AAC), gel consistency (GC), gelatinization temperature (GT), and PC in two environments, and detected three QTLs affecting protein content[106].
Key genes for protein content
-
Although many QTLs have been identified and reported, cloning QTLs related to protein content has become extremely difficult owing to various influencing factors. To date, only a limited number of genes have been cloned from natural populations, which can be used for quality improvement. Peng et al.[25] cloned the first protein content gene, OsAAP6, in rice, which encodes an amino acid transporter and functions as a positive regulator of the storage protein content. Upregulation of its expression can increase the contents of the four storage proteins. Notably, the OsAAP6 allele from Nanyangzhan reduced the content of the four storage proteins while improving the cooking quality of rice. In 2019, Yang et al.[83] cloned a major glutelin gene, OsGluA2, which acts as a positive regulator of rice protein content. The higher the expression level, the higher the glutelin content and the larger the volume of protein body II. In addition, a large number of floury endosperm genes may also affect grain protein content and (Table 1), but previous researchers did not pay enough attention.
Table 1. Reported floury endosperm genes that may affect grain protein content.
Classification Name Gene ID Effect of gene knockout
or knockdown on grain
protein contentAnnotation Biochemical metabolism flo4/OsPPDKB/OsC4PPDK LOC_Os05g33570 Increased[130] Pyruvate, phosphate dikinase OsSSIIIa/Flo5/SS3a LOC_Os08g09230 Increased[103] Starch synthase III FLO8/OsUgp1 LOC_Os09g38030 Increased[131] UTP--glucose-1-phosphate uridylyltransferase FLO12/OsAlaAT1 LOC_Os10g25130 Increased[132,133] Aminotransferase FLO15/OsGLYI7 LOC_Os05g14194 Increased[134] Glyoxalase family protein FLO16 LOC_Os10g33800 Increased[135] lactate/malate dehydrogenase FSE1 LOC_Os08g01920 Increased[136] Phospholipase-like protein OsAGPL2/OsAPL2/shr1/GIF2/osagpl2-3 LOC_Os01g44220 Decreased[137] ADP-glucose pyrophosphorylase large subunit 2 OsBEIIb/be2b LOC_Os02g32660 Unknown[138] Starch branching enzyme IIb Pho1 LOC_Os03g55090 Unknown[139] Plastidial phosphorylase OsGINT1/FSE6 LOC_Os05g46260 Increased[140] Glycosyltransferase OsPK2/OsPKpα1 LOC_Os07g08340 Increased[141] Plastidic pyruvate kinase GIF1/OsCIN2 LOC_Os04g33740 Unknown[142] Glycosyl hydrolases PDIL1-1 LOC_Os11g09280 Increased[143] Protein disulphide isomerase-like enzyme OsACS6/SSG6 LOC_Os06g03990 Unknown[144] Aminotransferase PFPβ/PFP1 LOC_Os06g13810 Unknown[145] Pyrophosphate-fructose 6-phosphate
1-phosphotransferase subunit betaFLO19 LOC_Os03g48060 Increased[28] Class I glutamine amidotransferase FLO19 LOC_Os04g02900 Decreased[146] Plastid-localized pyruvate dehydrogenase complex E1 component subunit α1 FLO23/OsF2KP2 LOC_Os03g18310 Decreased[147] Fructose-6-phosphate-2-kinase/fructose-2, 6-bisphosphatase OsDPE1 LOC_Os07g43390 Unknown[148] Disproportionating enzyme Transcriptional regulation and protein interaction OsNF-YC12 LOC_Os10g11580 Increased[19,149] CCAAT-box-binding transcription factor OsNF-YB1/OsHAP3K/OsEnS-41 LOC_Os02g49410 Increased[19] Nuclear transcription factor Y subunit B bHLH144 LOC_Os04g35010 Increased[19] Helix-loop-helix DNA-binding domain containing protein RISBZ1/OsbZIP58 LOC_Os07g08420 Decreased[150] bZIP transcription factor REB/OsbZIP33/RISBZ2 LOC_Os03g58250 Unknown[151] bZIP transcription factor RPBF/OsDof3/OsDof-10/OsDof7 LOC_Os02g15350 Decreased[150,152] Dof transcription factor FLO2 LOC_Os04g55230 Unknown[153] Tetratricopeptide repeat domain containing protein FLO6 LOC_Os03g48170 Increased[154] CBM48 domain-containing protein OsGBP LOC_Os02g04330 No change[29] GBSS-binding protein FLO7 LOC_Os10g32680 Unknown[155] DUF1388 domain protein OsHsp70cp-2/cpHSP70-2/ flo11-2/ FLO11 LOC_Os12g14070 Unknown[156] Plastid heat shock protein 70 RSR1 LOC_Os05g03040 Unknown[157] Transcription factor of the AP2/EREBP family OsNAC20; OsNAC26 LOC_Os01g01470; LOC_Os01g29840 Decreased when knock out together[18] NAC transcription factor OsNAC23/ONAC023 LOC_Os02g12310 Decreased when gene knock out and increased when gene overexpression[158] NAC transcription factor OsNAC24/OsNAC024 LOC_Os05g34310 Unknown[17] NAC transcription factor OsNAC127 LOC_Os11g31340 Unknown[159] NAC transcription factor OsNAC129 LOC_Os11g31380 Unknown[159,160] NAC transcription factor Du13/TL1 LOC_Os06g48530 No change[12] C2H2 zinc finger protein OsbZIP60/O3/OPAQUE3 LOC_Os07g44950 Decreased[24] Basic leucine zipper transcription factor OsMADS6/MFO1 LOC_Os02g45770 Increased[161] MADS-box transcription factor OsMADS29 LOC_Os02g07430 Unknown[162] MADS-box transcription factor OsMADS14 LOC_Os03g54160 Unknown[21] MADS-box transcription factor Epigenetics OsROS1/ROS1a/DNG702 LOC_Os01g11900 Increased[163] DNA demethylase OsCADT1/FLO20/SHMT4 LOC_Os01g65410 Decreased[164] Serine hydroxymethyltransferase Energy supply FLO10 LOC_Os03g07220 Increased[165] Pentatricopeptide repeat protein FLO14/OsNPPR3 LOC_Os03g51840 Unknown[166] Pentatricopeptide repeat protein FLO18 LOC_Os07g48850 Decreased[167] Pentatricopeptide repeat protein OGR1 LOC_Os12g17080 Unknown[168] Pentatricopeptide repeat–DYW protein FGR1/OsNPPR1 LOC_Os08g19310 Unknown[169] Pentatricopeptide repeat protein FLO13/OsNDUFA9 LOC_Os02g57180 Unknown[169] Mitochondrial complex I subunit FLO22 LOC_Os07g08180 Unknown[170] P-type pentatricopeptide repeat (PPR) protein Material transport ESG1 LOC_Os04g46700 Decreased[171] Bcterial-type ABC (ATP-binding cassette) lipid transporter OsBT1 LOC_Os02g10800 Unknown[172] ADP-Glucose Transporter OsBip1/BiP3 LOC_Os02g02410 Decreased[173] Endoplasmic riculum caperone OsLTPL36 LOC_Os03g25350 Decreased[174] Lipid transfer protein OsRab5a/gpa1/glup4 LOC_Os12g43550 No change, but pro-glutelin accumulation[175] Small GTPase Sar1a; Sar1b; Sar1c LOC_Os01g23620; LOC_Os12g37360; LOC_Os01g15010 Unknown but pro-glutelin accumulation when three genes knockdown together[176] Small GTPase GPA3 LOC_Os03g61950 Increased and pro-glutelin accumulation[177] Regulator of post-Golgi vesicular Traffic GPA4/GLUP2/GOT1B LOC_Os03g11100 Decreased but pro-glutelin accumulation[178] Golgi Transport 1 GPA5 LOC_Os06g43560 Unknown but pro-glutelin accumulation[179] Rab5a Effector OsVPS9A/GPA2/GLUP6/GEF LOC_Os03g15650 Decreased but pro-glutelin accumulation[180] Guanine nucleotide exchange factor GPA6/OsNHX5 LOC_Os09g11450 No change, but pro-glutelin accumulation[181] Vacuolar Na+/H+ antiporter Function unknown SSG4 LOC_Os01g08420 Unknown[182] Unknown function protein Nitrogen-efficient genes that may affect endosperm protein content
-
Rice grain nitrogen is primarily derived from the nitrogen absorbed from the soil. Therefore, in addition to the external factors, the ability of rice to absorb, transport, assimilate, distribute, and even reuse nitrogen may cause changes in the nitrogen (protein) content of the final grain (Fig. 3). In other words, the nitrogen utilization capacity of rice may also be an internal cause of the difference in grain protein content. Therefore, attention should be paid to genes related to nitrogen use efficiency in rice, as they may also be potential targets for the genetic improvement of rice proteins. In a recent study, Zhang et al. utilized the promoter of the Nhd1 gene to develop rice genetic materials with significantly increased endogenous expression levels[107]. There were no significant changes observed in the entire length of growth duration, nitrogen use efficiency and rice yield of the Nhd1 enhanced line. However, the starch granules in the rice showed a more relaxed arrangement, with noticeable reductions in amylose and protein content. This resulted in a lower gelatinization temperature and increased gel consistency, suggesting that the rice is easier to cook, digest, and has an improved taste. The GRF4 gene is a positive regulator of plant carbon and nitrogen metabolism that can simultaneously promote nitrogen absorption, assimilation, and transport, thereby increasing the total nitrogen level in the grain[108]. LNUE1 gene encodes OsAlaAT1, an alanine aminotransferase that regulates nitrogen use efficiency of rice. lune1 mutant has low nitrogen use efficiency, decreased total protein levels in seeds, and severe chalkiness in the endosperm[109]. THP9[110], the first major gene cloned from wild maize to control the high protein content and NUE of maize, encodes asparagine synthetase 4 (ASN4), which is a key component of nitrogen metabolism and is responsible for asparagine synthesis. Transgenic expression of THP9-teosinte in B73 inbred lines significantly increases seed protein content. The OsDREB1C[111] gene drives a wide range of transcriptional activities that regulate the photosynthetic capacity, nitrogen use efficiency, and heading date of rice. Overexpression of the gene enhanced the ability of rice to absorb and transport nitrogen, allocate more nitrogen to the grain, and increase nitrogen use efficiency by 25.8%–56.6% compared with the control group. In view of the fact that the NUE genes cloned and discovered in many studies currently only have yield data, and relevant researchers have not yet investigated whether these genes also affect grain protein content and quality traits, we collected and summarized the relevant information on the currently known NUE genes (Table 2). This compilation will facilitate further exploration of the effects of these genes on rice grain protein content and quality.
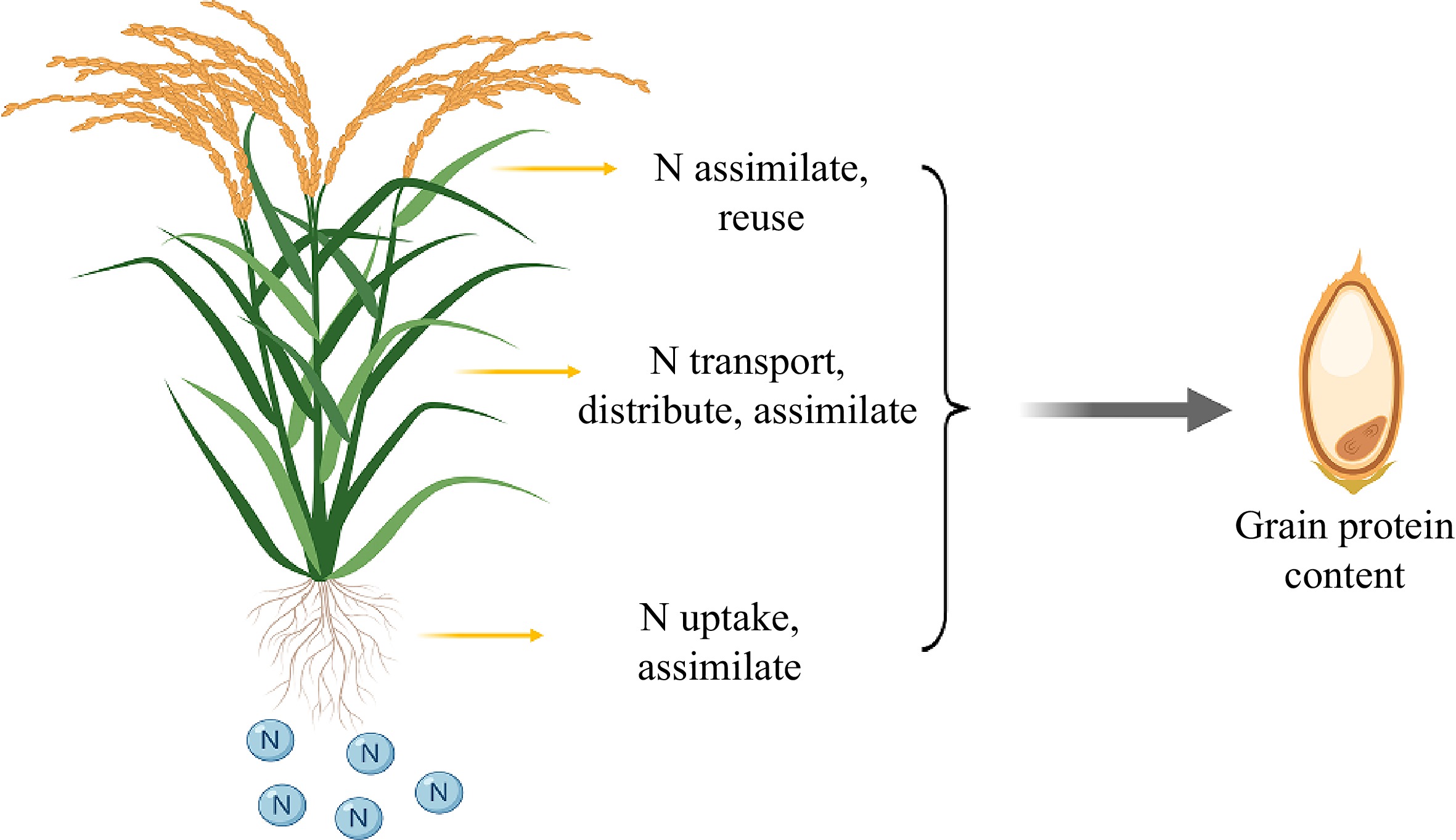
Figure 3.
The nitrogen utilization ability of rice may determine the protein content of grains. Each step of nitrogen utilization in rice, which includes absorption, transportation, assimilation, distribution, and reuse, can potentially impact the final grain protein content.
Table 2. Reported nitrogen use efficiency genes that may also affect grain protein content.
Name Gene ID Effect of altered gene function on grain protein content Annotation MYB61/qNLA1/qCel1 LOC_Os01g18240 Unknown[183] MYB family transcription factor OsGRF4/GS2/GL2/PT2/LGS1/GLW2 LOC_Os02g47280 Increased when gene overexpression[108] Growth-regulating factor TOND1 LOC_Os12g43440 Unknown[184] Unkonwn function protein OsDEP1/DN1/qPE9-1/qNGR9 LOC_Os09g26999 No change when gene overexpression[185] Gγ subunit OsTCP19 LOC_Os06g12230 Unknown[186] Class-I TCP transcription factor SMOS1/shb/RLA1/NGR5 LOC_Os05g32270 Unknown[187] GRAS protein OsNPF6.1 LOC_Os01g01360 Unknown[188] Nitrate transporter OsNAC42 LOC_Os09g32040 Unknown[188] No apical meristem protein OsNR2/qCR2 LOC_Os02g53130 Unknown[189] Nitrate reductase OsNRT1.1B/OsNPF6.5/qCHR-10 LOC_Os10g40600 Unknown[190] Peptide transporter PTR2 Ghd7/E1/Hd4 LOC_Os07g15770 Unknown[191] CCT motif family protein ARE1 LOC_Os08g12780 Unknown[192] Chloroplast envelope membrane protein OsCCA1/OsLHY/Nhd1 LOC_Os08g06110 Decreased when gene expression enhanced[107] MYB transcription factor Ef-cd Unknown[193] Long noncoding RNA OsAtg8/OsATG8a LOC_Os07g32800 Increased when gene overexpression[194] Autophagy-related protein OsPIN9 LOC_Os01g58860 Unknown[195] Auxin efflux transporter OsNLP4 LOC_Os09g37710 Unknown[196,197] NIN-like protein OsNLP3 LOC_Os01g13540 Unknown[198] NIN-like protein OsNLP1 LOC_Os03g03900 Unknown[199] NIN-like protein OsAAP6/qPC1 LOC_Os01g65670 Increased when gene overexpression[25] Amino acid permease OsAAP10 LOC_Os02g49060 Decreased when gene knock out[200] Amino acid permease OsAAP5 LOC_Os01g65660 Unknown[201] Amino acid permease OsAAP3 LOC_Os06g36180 Increased when gene overexpression[202] Amino acid permease OsAAP1 LOC_Os07g04180 Increased when gene overexpression and decreased when gene interference[203] Amino acid permease OsAMT1;1/OsAMT1-1 LOC_Os04g43070 Unknown[204] High-affinity ammonium transporter OsAMT2;1 LOC_Os05g39240 Unknown[205] Ammonium transporter OsGS1/OsGS1;1/OsGLN1;1/λGS28 LOC_Os02g50240 Decreased when OsGS1;1b overexpression[206] Glutamine synthetase OsGS2/OsGLN2/λGS31 LOC_Os04g56400 Increased when concurrent overexpression of OsGS1 and OsGS2[207] Glutamine synthetase OsAMT1;3/OsAMT1.3/OsAMT1;2 LOC_Os02g40710 Increased when gene overexpression[208] Ammonium transporter OsGOGAT1 LOC_Os01g48960 Increased when gene overexpression[208] Glutamate synthetase 1 OsAS1/ OsASN1 LOC_Os03g18130 Increased when gene overexpression[209] Asparagine synthetase OsSHM1/OsSHMT1 LOC_Os03g52840 Unknown[210] Serine hydroxymethyltransferase 1 OsENOD93-1 LOC_Os06g05010 Unknown[211] Early nodulin 93 ENOD93 protein OsNRT2.3/OsNRT2.3a/OsNRT2.3b LOC_Os01g50820 Unknown[212] High-affinity nitrate transporter OsNAR2.1 LOC_Os02g38230 Unknown[213] Partner protein for high-affinity nitrate transport OsNRT2.1 LOC_Os02g02170 Unknown[214] High-affinity nitrate transporter OsNRT1.1A/OsNPF6.3 LOC_Os08g05910 Unknown[215] Nitrate transporter DNR1 LOC_Os01g08270 Decreased when gene knock out[216] Amino transferase qSBM1 LOC_Os01g65120 Unknown[217] Peptide transporter OsRBCS2 LOC_Os12g17600 Unknown[218] Small subunit of Rubisco OSA1 LOC_Os03g48310 Unknown[219] Plasma membrane H+-ATPase OsDREB1C LOC_Os06g03670 Increased when gene overexpression[111] AP2/EREBP transcription factor OsPTR6/OsNPF7.3 LOC_Os04g50950 Unknown[220] Peptide transporter OsMADS1/LHS1/AFO/LGY3/GW3p6 LOC_Os03g11614 Unknown[221,222] MADS-domain transcription factor -
Rice protein content plays a key role in determining the cooking and eating quality of rice. A high protein content tends to make rice grains compact, resulting in poor water absorption and greatly reducing the taste of rice. Therefore, in production practice, reducing the protein content in rice is helpful for improving the cooking and eating qualities of rice. Although research on breeding rice with proper protein content has been conducted for many years, there are currently no effective methods in production practice to regulate rice protein content and improve the cooking and eating qualities of rice. Hybrid breeding is the most commonly used method for quality breeding worldwide[112]. For rice quality traits that exhibit relatively simple genetic behavior, germplasm resources with excellent rice quality traits can be selected and improved by crossbreeding. Some well-known high-quality varieties, such as Koshihikari from Japan, Basmati370 from the United States, IR64 from the International Rice Research Institute, and Daohuaxiang 2 from China, are bred through hybridization. However, for protein content, a typical quantitative trait controlled by multiple genes, the genetic basis is complex and greatly affected by environmental factors, making it challenging to achieve ideal improvement using traditional hybrid breeding.
It may be effective to improve the quality traits of rice using physical and chemical factors to induce variation. Schaeffer & Sharpe performed biochemical mutagenesis by increasing the contents of lysine, threonine, and cysteine in the culture medium and obtained mutant lines with high lysine content in rice through cell culture[113,114].
It is also a good choice for collecting a wide range of germplasm resources, selecting varieties with specific traits, and improving them to meet human needs. Juliano et al. screened 38 rice varieties with lysine content 0.5% higher than the average level of 10,493 rice varieties[115]. Mochizuki & Hara used the low-gluten material NM67 to breed a rice variety LGC-1 with significantly reduced absorbable glutelin, and the content of prolamin in the rice that cannot be absorbed by the human body is high[116]. By crossing LGC-1 with Wuyujing 3, combined with breeding for agronomic traits and molecular marker-assisted selection, Zhang et al. obtained three new rice varieties with excellent agronomic traits and glutelin content close to that of LGC-1[117].
Since the 1990s, the continuous development of molecular biology technology has created good conditions for the genetic improvement of rice quality, and progress has been made in the creation of high-quality rice materials and rice quality breeding using biotechnology. The application of transgenic technology to introduce exogenous special genes and express them efficiently may be an effective strategy for increasing the content of essential amino acids in rice. Lee et al. carried out point mutations in the maize dhps gene and connected the gene to the promoter of CsMV35S and GluB-1, respectively, to transform and obtain transgenic rice, and the lysine content in mature seeds was significantly increased[118]. Liu et al. linked the endosperm-specific expression promoter to lysine-rich foreign proteins, which also increased the lysine content of seeds by 30%[119]. A sulfur-rich storage protein gene from sesame was transferred into rice and expressed, which simultaneously increased cysteine and methionine in the endosperm and total protein content at the same time[120]. The transfer of sulfur-rich genes from sunflower to rice could also increase cysteine and methionine levels in the endosperm; however, the total protein content decreased[121]. Zhou et al. transformed the aspartate aminotransferase gene from Escherichia coli into rice, which increased the content of amino acids and proteins in rice[122]. The OASA1D gene, encoding a feedback-insensitive α-subunit of rice anthranilate synthase, was expressed in rice driven by the maize ubiquitin promoter, which increased the content of tryptophan in seeds[123].
The 3' untranslated region regulates gene expression at the transcriptional and post-transcriptional levels by affecting the accumulation, stability, and translation efficiency of mRNA[124−126]. Li et al. evaluated the 3'-UTRs of nine seed storage protein (SSP) genes as terminators to enhance glutelin GluB-3 promoter-driven β-glucuronidase (gus A) reporter gene expression in stable transgenic rice lines, in which six 3'-UTRs significantly enhanced the activity of the GluB-3 promoter without altering its tissue specificity[127]. Yang et al. found that transgenic seeds using the 3' UTR of GluB-1 as the terminator had more accumulation of the target expression protein than transgenic seeds using the Nos terminator[128]. These results indicate that it is feasible to regulate the key genes of protein content at the transcription and translation levels using genetic engineering.
In recent years, the popularity of gene-editing technology has provided an opportunity to improve the protein content of rice. Yang et al. used Crispr-cas9 technology to edit eight members of the glutelin gene family and obtained seven different homozygous mutation types, including double, triple, quadruple, quintuple, and sextuple mutants. Notably, type II, III, IV, V, and VI mutants with moderately reduced grain protein content significantly increased the rice taste value, improved its appearance, and decreased hardness[57]. Similarly, Chen et al. designed three sgRNAs targeting nine glutelin genes and generated nine T-DNA-free homozygous editing lines that exhibited reduced glutelin content compared with the wild type. These low glutelin lines all showed agronomic traits similar to the wild type, including yield components and viscosity characteristics[129].
-
The cooking and eating qualities of rice may differ among different individuals. Thousands of rice varieties vary in cooking taste, particularly in texture. This highly significant trait in rice has attracted the attention of scientists for nearly three-quarters of a century and has only recently begun to be fully understood. An increasing number of studies have shown that grain protein is the most important factor affecting cooking and eating quality after starch. However, more research is needed to fully understand the relationship between grain protein and rice quality, as well as to develop strategies for optimizing it to meet human dietary needs. Therefore, it can be strengthened in the following aspects:
1. Enhancing the genetic mechanism of protein content in rice.
Rice protein content is a typical quantitative trait. Although hundreds of QTLs affecting rice grain protein content have been identified, only two genes regulating protein content have been cloned, both of which are positive regulators of protein content. Further new genes regulating protein content in rice need to be further explored. The formation of rice storage proteins is a complex process involving nitrogen absorption, transport, assimilation, distribution, reuse, amino acid synthesis, amino acid modification, amino acid transport, protein synthesis, transport, modification, storage, and degradation. The molecular genetic mechanisms of each step need to be further analyzed.
2. Strengthen study on the mechanism of the effect of protein on eating quality.
A series of physical and chemical changes occur in rice during cooking, including water absorption of rice grains, gelatinization of starch, dissolution of starch after endosperm cell breakage, and formation of adhesion layers. In this process, whether the protein itself has an indirect effect on the taste or a direct effect on the water absorption of rice grains, starch gelatinization, and expansion, the type of extract, and the thickness of the adhesive layer, the specific mechanism needs to be further explored. In addition, the interactions among the proteins, starch, and lipid require further clarification. In summary, an in-depth study on the relationship between rice protein, cooking, and eating quality will provide a scientific basis for breeders to select and cultivate varieties with superior tastes.
3. Rational application of nitrogen fertilizer and breeding of varieties with high nitrogen efficiency.
Although nitrogen fertilizer is commonly used to increase rice yield, its inefficient use not only causes environmental pollution but also results to an increase in protein content in rice. This excessive use of nitrogen fertilizer leads to the deterioration of cooking and eating quality of rice. Exploring more nitrogen-efficient genes and analyzing the mechanism of their influence on protein content in rice will help cultivate nitrogen-efficient varieties. This, in turn, will lead to synergistic improvement in rice yield and quality, with minimal nitrogen fertilizer usage.
-
The authors confirm contribution to the paper as follows: study conception and design: Lou G, He Y; data collection: Lou G, Bhat M, Tan X, Wang Y; draft manuscript preparation: Lou G, He Y. All authors reviewed the results and approved the final version of the manuscript.
-
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
This work was supported by grants from the National Natural Science Foundation of China (U21A20211, 31821005), AgroST Project (NK20220501) and China Agriculture Research System (CARS-01-01).
-
The authors declare that they have no conflict of interest.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press on behalf of Hainan Yazhou Bay Seed Laboratory. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Lou G, Bhat MA, Tan X, Wang Y, He Y. 2023. Research progress on the relationship between rice protein content and cooking and eating quality and its influencing factors. Seed Biology 2:16 doi: 10.48130/SeedBio-2023-0016
Research progress on the relationship between rice protein content and cooking and eating quality and its influencing factors
- Received: 25 July 2023
- Accepted: 07 October 2023
- Published online: 27 October 2023
Abstract: Proteins, the second-largest storage substance in rice endosperm, play an important role in determining the cooking and eating qualities of rice. Its contents are influenced by both genetic and environmental factors. This article provides a review of the evaluation methods for cooking and eating qualities of rice and starch physicochemical properties, the factors that affect the protein content of rice, the genetic basis of rice protein content, the research progress made in the genetic improvement of rice protein content, and the prospects for the future, aiming to provide a reference for the genetic improvement of rice protein content and the breeding of rice varieties with excellent taste.
-
Key words:
- Rice /
- Grain protein content /
- Grain quality /
- Cooking and eating quality /
- Nitrogen utilization /
- Genetic basis /
- Genetic improvement













