-
Banana (Musa spp.), the most vital economic crop for both staple food and fruit extensively planted in the tropics and subtropics, are a perennial herbaceous monocotyledonous plant pertaining to the Musaceae family of the order Scitamineae. Based on the data of the United Nations Food and Agriculture Organization, bananas are planted in 138 countries and regions around the world. As a staple food for the largely impoverished continent of Africa, it is the fourth largest staple food crop after rice, wheat and maize. As a fresh fruit, it stands the second largest fruit in the world after citrus, and the consumption and trade volume of fresh fruit rank first in the world[1].
The most vital cultivated banana cultivars globally are triploids, originating from interspecific or intraspecific hybridization of two wild diploid species, Musa acuminata (A genome) and M. balbisiana (B genome)[1]. Owing to their parthenocarpy and polyploidy, it is very hard to cultivate new varieties through conventional breeding[2]. Banana plants are extremely threatened by diverse biotic and abiotic stresses, such as diseases, salt, drought, and cold. Currently, fusarium wilt (commonly known as panama disease) caused by Fusarium oxysporum f. sp. cubense (Foc) seriously threatens global banana production. At present, there are a lack of banana cultivars with both excellent production and Foc-resistance[2].
It is a fundamental way out for the sustainable development of global banana production to improve the new varieties with excellent production and Foc-resistance. Biotechnology involving plant tissue culture is a powerful complementary strategy in conventional plant breeding programs[3]. There are two processes of plant regeneration, namely organogenesis and somatic embryogenesis (SE). In general, organogenesis involves the sequential formation of shoots and roots from tissues, relying on the appropriate culture conditions. On the other hand, SE is a totipotent embryonic stem cell formed by dedifferentiation of plant somatic cells. This new embryo can go on to develop into a complete plant[4]. Currently, there are two different ways to induce explants to form SE: direct SE and indirect SE. In the direct SE pathway, explants directly form somatic embryos without callus formation. SE also can be formed indirectly through a callus stage.
Banana plant regeneration via organogenesis based on meristemic tissue, such as shoot tips and floral apices, are widely used for clonal propagation. Although the regeneration system based on organogenesis has also been applied to genetic transformation, it has the problems of low efficiency of genetic transformation and high proportion of chimeric plants. SE through embryogenic cell suspension (ECS) cultures is an ideal recipient system for genetic transformation in several plants, including banana, due to their oocyte characteristics, strong ability to accept foreign genes and fewer chimeras[5, 6]. Genetic transformation through ECS is a most widely used strategy in different banana varieties[5].
The purpose of this paper is to review the current status of research on the process of SE for Musa spp. Indirect SE from IMFs and scalps are the focus of this review. At the same time, their applications in breeding technologies were also summarized in order to provide reference for the research and practical application of banana SE in the future.
-
The regenerative system of banana somatic embryogenesis based on ECS provides an ideal raw material for mutation breeding, somatic hybridization and genetic transformation[5, 6]. Based on the type of explant, there are four recognized procedures for the establishment of banana EC and proliferation of embryogenic cell suspension (ECS). In majority of the reports, the immature male flowers (IMFs) and/or shoot-tip derived scalps are a preferred choice for developing ECS cultures. Similar to other plants, banana plant regeneration via somatic embryogenesis based on ECS mainly includes callus induction, embryogenic callus selection, embryogenic callus proliferation and initiation of cell suspension culture, development and maturation of somatic embryo, and plant regeneration[7, 8] (Fig. 1).
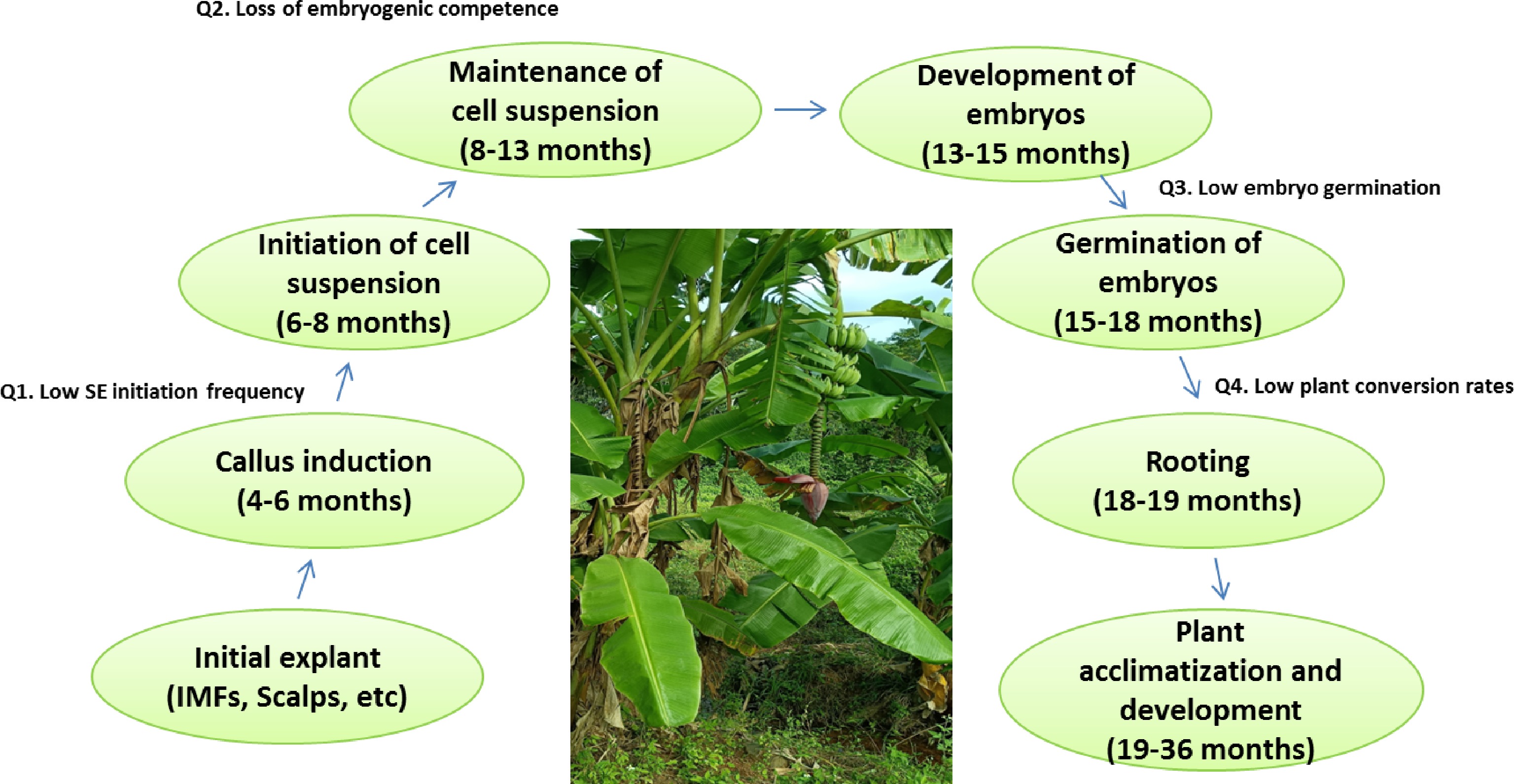
Figure 1.
The main phases and time required for each phase in the somatic embryogenesis of banana and critical questions (Q). (SE) somatic embryo; (IMFs) immature male flowers.
Although somatic embryogenesis in banana is now a well-established method, the initiation of a 'genotype-independent' embryogenic cell culture is still far from routine. There are still some problems that need to be solved in the reported protocols for banana SE. These problems include either all or some of the following: low SE initiation frequency from the explants, reduction or loss of embryogenic competence concomitant with the increased time of subculture, and low embryo germination and plant conversion rates (Fig. 1).
-
In banana, indirect SE was mostly observed. The main culture stages of indirect SE are induction and proliferation of embryogenic callus (EC), maturation and germination of somatic embryos[7, 8]. Therefore, various factors affect the efficiency and quality of EC formation, including the explant type, the genotype of the donor plant, plant growth regulators, and the media and other additives, etc.
Explant
-
The selection of suitable explants is one of the key factors for the success of EC induction. Since early reports in the late 1980s, a series of explants have been successfully used in banana EC. In a word, there are mainly four different types of explants used in banana: immature and mature zygotic embryos[9−14], Rhizome slices and leaf sheaths[15], IMFs and female flowers[5, 16−21], and scalps[22, 23]. Recently, somatic embryos were also successfully induced by secondary explants from male buds and bracts in medium containing TDZ[24, 25]. While, direct somatic embryos developed from split shoot tips under a combination of picloram and 6-benzyladenine (BA)[26]. Despite many options, the most used explants to establish a renewable ECS for seedless banana are still limited to scalps[27−31] and IMFs[32−34].
It is reported that factors such as the developmental and physiological state of the explant and the location of the material can affect SE. Strosse et al.[35] suggested that the immature flowers should be taken from position 8 to 16, which were the most responsive ones in terms of embryogenesis. From the reports, sensitive positions are mainly concentrated in 7-13[17]; 8-15[36], 4-11[37], and 6-11[34]. Interestingly, higher efficiency and taking a short time for EC formation were observed by spraying exogenous 2,4-dichlorophenoxyacetic acid (2,4-D) on immature male flower buds[38].
Genotype
-
SE is highly genome dependent as the efficiency varies with cultivars. Using various explants, SE has been achieved for some genotypes of banana varieties (AA, BB, AB, AAA, ABB, and AAB). Using IMFs as starting materials, three genotypes including six cultivars were tested[17]. The efficiency of EC obtained from different genotypes ranged from 0 to 7%. Even for the different variety of the same genotype, the frequency of EC formation varied differently. Musa AAB cvs. 'French Plantain', 'Mysore' and 'Silk' showed the efficiency of 2%, 3%, and 7%, respectively. As for Musa AAA cv. 'Grande Naine', it had the highest induction rate (37%) of all tested varieties[17]. Among the reported genotypes, two cultivars from Cavendish subgroup (AAA) ranged from 0.7% to 10%, responses[39]. Using the scalps, the mean embryogenic frequency was 6.0%, 3.8%, and 1.8% for cooking bananas (ABB), Cavendish-type bananas (AAA), and plantains (AAB), respectively[30]. Whereas, using inflorescence proliferation for SE induction, the embryogenic frequency was 12.5% and 25% under semisolid and liquid inductive medium, respectively[24].
Plant growth regulators
-
PGRs are crucial in the process of callus formation, proliferation, somatic embryo formation, plant regeneration and rooting. Auxins and cytokinins act a decisive role in somatic embryogenesis in various plant species. At present, the commonly used auxins are 2,4-dichlorophenoxyacetic acid (2,4-D), 1-naphtaleneacetic acid (NAA), indole-3-acetic acid (IAA), Indole-3-butyric acid (IBA), and picloram (4-amino-3,5,6-trichloropicolinic acid). As for CKs, 6-benzylaminopurine (BA), kinetin (KN), and zeatin are the mostly used. 2,4-D is used for EC induction, establishment and proliferation of ECS in most banana cultivars. It is often applied at 1–4 mg L−1, and is combined with low concentrations of cytokinins to control SE. For plant regeneration, BA is often used at concentrations of 0.1–3 mg L−1, and low concentrations of NAA added, or sometimes hormone-free media.
Different concentrations and various combinations of PGRs were required for different explants. For IMFs method, even though a high level of 2,4-D is needed for the EC induction, prolonged exposure will reduce the embryogenic nature of the callus. At the proliferation of EC and initiation of cell suspension cultures, reduction of the concentration of the sole auxin 2.4-D is improtant for proliferation of somatic embryogenic callus and expression of somatic embryos[18, 37, 40]. However, Nandhakumar et al.[34] reported a MS based ECS medium with 10 mg L−1 resulted in the rapid multiplication of embryogenic cells. In addition, picloram also plays a vital role in SE. It was reported that the induction percentage of EC of M. acuminata cv. 'Mas' (AA) reached 15.6% when 2.4-D in the callus induction medium was substituted by 8.28 μM picloram. The induction efficiency of IMFs on medium with picloram was more than twice that of 2.4-D[41]. On the contrary, the opposite results were observed when the effects of different concentrations of 2,4-D and picloram on callus initiation of M. acuminata cv. 'Berangan' (AAA) were studied[40]. It may be induced by the different genotypes of the explants. As for the other explants, both auxin and cytokinin were used and maintained in the medium. Embryogenic callus (17.5%) was induced from scalps of Musa AAB Silk 'Guoshanxiang' on MS medium with 5 μM 2,4-D and 1 μM Zeatin[28]. Using split shoot tips as explants, maximum embryo induction (100%) for M. acuminata AAA cv. 'Grand Naine' occurred in medium with 4.14 μM picloram and 0.22 μM BA. The plant regeneration (2%–3%) occurred in MS medium with NAA (0.53–2.68 μM) and BA (2.22–44.39 μM), or TDZ (4.54 μM) plus glutamine (200 mg/L)[26].
Basal medium and additives
-
Medium is the basic substance for in vitro plant culture. According to the components, it can be divided into Murashige and Skoog (MS) medium[42], Gamborg's B5[43], Woody Plant Medium (WPM)[44], and Schenk and Hildebrandt (SH) medium[45], etc. The basal medium may be solid, semi-solid, or liquid. The commonly used media for SE in Musa spp. include MS and SH. MS is the preferred medium for callus initiation, establishment of ECS, and plant regeneration. SH medium with MS vitamins or 1/2 MS is often used for the development and maturation of somatic embryos of Musa spp.
Medium additives, used along with basal media and PGRs, commonly include carbon source, various amino acids, malt extract (ME), and coconut water (CW), etc. Carbon source plays a major role in plant energy metabolism and regulates the osmotic potential of the cell. The most preferred carbon source for banana was sucrose (2%–4.5%; w/v). In addition, maltose, dextrose, fructose, lactose and galactose are also used as carbon sources in some studies. Adding maltose in the medium promoted the formation of banana ECS[32, 34]. The effect of different amino acids (L-Proline, L-Glutamine and L-Asparagine) on somatic embryo production was compared. Among the tested amino acids, L-Glutamine (400 mg L−1) had a significant strengthening effect on primary and secondary somatic embryos in M3 medium[34]. It was in concert with the early report[46]. The presence of 400 mg L−1 L-Glutamine resulted in optimum somatic embryo development and high regeneration efficiency in banana cv. Berangan (AAA). Although L-Glutamine and L-Proline have been shown to promote the embryo development, high concentrations of proline (400 mg L−1) in liquid media caused abnormal embryo differentiation[46]. CW and ME play a promoting role in banana callus induction[32, 47]. To avoid rapid browning of the explants, anti-oxidant like ascorbate (10 mg L−1), melatonin (50 mg L−1), and L-Glutamine (100 mg L−1) were also added to the medium[34].
-
Exploring the molecular regulatory mechanism of plant SE can not only reveal the process of somatic embryo development, but also afford a basis molecular mechanism for somatic embryo development. In most banana genotypes, the potency of explant to develop EC is highly inefficient. Therefore, it is important to find the molecular regulators that can be explored to enhance the SE potential in banana.
Based on the banana genome database, the differential transcribed fragments between zygotic and somatic embryogenesis were compared by cDNA-AFLP[48]. The role of genes including transcription factors (TFs) was identified in banana SE. The results showed that MaBBM2 and MaWUS2 maybe the prospective candidate TFs and MaPIN1 could be a hopeful gene marker for the embryogenicity in banana[49, 50].
Recently, differentially expressed proteins during the SE in banana were identified by proteome technology[51−53]. Based on comparative proteomics, it is indicated that EC was related to excessive accumulation of ROS scavenging proteins, heat shock proteins (HSP), and growth-regulator related proteins[51]. Furthermore, calcium signaling and PGRs were also involved in the development and germination of somatic embryos. The important role of calcium and PGRs (IAA, BAP, and kinetin) were confirmed by proper induction of five recalcitrant banana cultivars[52]. Based on these results, the medium for optimal SE efficiency in several cultivars could be customized.
-
As the basal materials, ECS is very important for banana germplasm innovation. However, the establishment of banana ECS is very difficult, and after establishment, it needs to be subcultured regularly. Frequent subculture not only consumes a lot of manpower and material resources, but also leads to somatic mutation and the loss of embryogenic characteristics. Furthermore, it is susceptible to bacterial and fungal contamination. Therefore, it is of great significance to study the preservation methods of banana ECS. Cryopreservation is an effective technology that can not only reduce the risk of contamination and gene mutation, but also effectively store plant material for a long time. There are three main methods for cryopreservation of banana germplasm, namely slow-freezing (two-step method), quick-freezing and vitrification. Panis et al.[54] successfully used a two-step method to preserve banana ECS for the first time. In 2010, Li et al. successfully cryopreserved banana ECS by vitrification[55].
Protoplast cultures
-
Protoplast fusion and somatic hybridization offers the potential to produce novel crops and overcome breeding obstacles in polyploid and apomictic banana cultivars. In 1993, the isolation and regeneration of protoplasts from an embryogenic cell suspension culture in banana were successfully received[56, 57]. Science then, a number of banana cultivars including various genotypes (AA, AAA, AAB, ABB) were effectively regenerated through protoplast culture[58].
Plant regeneration via protoplast culture opens up feasible opportunities for somatic hybridization and protoplast transformation, and eventually leads to genetic modification and breeding of new varieties. Somatic hybridization between triploid (Musa spp. AAB group, cv. 'Maçã') and diploid (Musa spp. AA group, cv. 'Lidi') bananas was attempted using protoplast electrofusion and nurse culture techniques. Somatic hybrids showed different ploidy levels by RAPD and flow cytometric ploidy analyses[59]. Assani et al.[60] successfully obtained banana somatic hybrid plants from Musa spp. triploid cv. 'Gros Michel' (AAA) and diploid cv. 'SF265' (AA). By the comparison of chemical (PEG: polyethylene glycol) and electrical fusion technique, it was found that electric fusion was better for mitotic activities, somatic embryogenesis and plantlet, and chemical procedure was better for the frequency of binary fusion. Xiao et al.[58] developed an asymmetric protoplast fusion with 20% (w/v) PEG and obtained somatic hybrids between Musa Silk cv. 'Guoshanxiang' (AAB) and Musa acuminata cv. 'Mas' (AA). Recently, Wu et al.[61] established a PEG-mediated protoplast transformation, which can serve as an effective and rapid tool for transient expression assays and sgRNA validation in banana.
Transgenics
-
As a perennial fruit crop, banana is susceptible to a plethora of abiotic and biotic stresses[6, 62, 63]. The objectives of banana improvement programs are breeding cultivars resistant to abiotic and biotic stressors without adverse effects on yield and fruit quality. Numerous efforts have been made to breed superior banana cultivars with better resistance to abiotic and biotic stresses and optimum yields at the same time using conventional breeding and genetic modification strategies. However, conventional breeding approaches are time-consuming and severely hampered by inherent banana problems (polyploidy and sterility). Therefore, genetic transformation is becoming increasingly popular and can provide rapid solutions.
In summary, the recipients used for banana genetic transformation are usually ECS, apical meristem, corm slices, thin cell layers from shoot tips, multiple shoot clumps etc. Among them, genetic transformation through ECS is a most commonly used method in different cultivars of banana owing to its strong ability to accept foreign genes and the lower frequency of chimeras shoot production. The transformation method is mainly mediated by gene gun and Agrobacterium. The flow chart of banana genetic transformation using ECS is shown in Fig. 2. The transformation efficiency was between 1.25% and 50.00%, with a large range of changes. Except for NPTII, GUS, GFP and other screening genes and reporter genes, the transformed functional genes mainly involved in banana fruit quality, disease resistance, drought tolerance, dwarfing and other traits improvement[62]. In this part, the studies for banana genetic transformation with added value from 2000 on were mainly summarized as below.
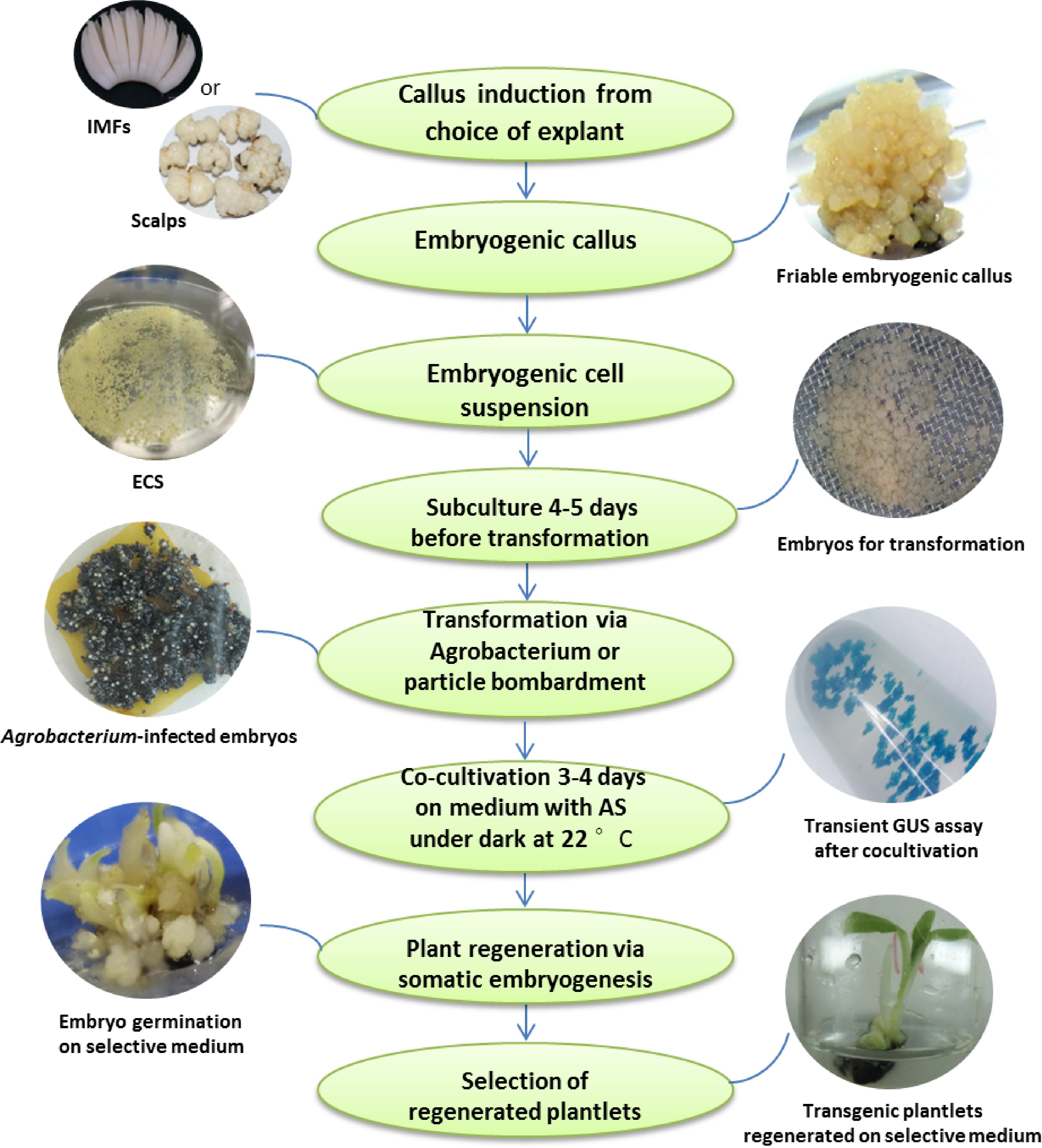
Figure 2.
Schematic representation of genetic transformation steps of banana using embryogenic cell suspension. Scalps and immature male flowers (IMFs) are the most used explants to establish a renewable ECS for seedless banana cultivars. The photos of scalps and friable embryogenic callus are cited from Tripathi et al.[31].
Improvements in disease resistance
-
In banana, the most serious diseases are fungal (Fusarium wilt, black Sigatoka), bacterial (banana Xanthomonas wilt, BXW), and viral (banana bunchy top disease, and banana streak disease)[63, 64]. Researchers have been working to improve disease and pest resistance in bananas using transgenic technology[64−66].
Various transgenes have been used to develop genetically engineered banana and many conferred significant levels of resistance to fungal pathogens (Table 1). Functional genes used to develop Foc-resistance bananas mainly included the antimicrobial peptides belonging to plant or animal origin[67, 68, 70, 72, 75, 78, 80], apoptosis-inhibition-related animal genes (Bcl-xL, Ced-9 and Bcl-2 3' UTR, Ced9)[69, 74, 81], different cell-death-related genes (MusaDAD1, MusaBAG1 and MusaBI1)[72], and defense-related protein[82]. In addition, Foc-resistance has also been conformed using RNAi silencing of key genes of Foc[71, 84]. Although the above studies demonstrate the transgenic plants resistance to Foc in the greenhouse, field evaluation remains to be seen. Recently, transgenic bananas with resistance gene analog 2 (RGA2), isolated from a seedling of Musa acuminata ssp. malaccensis with resistance to TR4, showed promising resistance against Fusarium wilt after a 3-year field trial in Australia[81]. Similarly, two native genes (MaLYK1 and MabHLH) from banana germplasms with Foc resistance were introduced back to Cavendish banana cv. Brazil, had shown increased resistance to TR4[83, 85]. Several studies from the researchers at the International Institute of Tropical Agriculture (IITA) reported transgenic bananas resistant to BXW disease[88−92]. Other studies also dealt with the production of resistance to Black Sigatoka[86, 87] and banana bunchy top disease[93−95] (Table 1).
Table 1. Genetic transformations of banana (Musa spp.).
Trait Gene Sources Transformation method Result Cultivar Reference Fungal Foc Race2 resistance MSI-99 Synthetic Agrobacterium/EHA105/ ECS Improved disease resistance against Foc and black leaf streak disease Rasthali (AAB) [67] Foc Race1 resistance β–1,3–endoglucanase Soybean Agrobacterium/LBA4404/Single buds Increased tolerance to Foc Race 1 Rasthali (AAB) [68] Bcl-xL, Ced-9, Bcl-2 3' UTR Animal Agrobacterium/LBA4404/ECS Apoptosis-inhibition-related genes confer resistance to Foc Race 1 Lady Finger (AAB) [69] PhDef1 and PhDef2 Petunia Agrobacterium/EHA105/ECS Improved fungal resistance with normal growth and no stunting phenotype Rasthali (AAB) [70] ihpRNA-VEL and ihpRNA-FTF1 Agrobacterium/EHA105/ECS Increased resistance to Foc Race 1 Rasthali (AAB) [71] MusaDAD1, MusaBAG1 and MusaBI1 Banana Agrobacterium/EHA105/ECS Increased resistance to Foc Race 1 Rasthali (AAB) [72] Sm-AMP-D1 Stellaria media Agrobacterium/EHA105/ECS Improved resistance against Foc Race 1 and no gross growth abnormalities Rasthali (AAB) [73] Ced9 Synthetic − Increased resistance against Fusarium wilt Sukali Ndiizi (AAB) [74] Ace-AMP1 Onion seeds Agrobacterium/LBA4404/ECS Enhanced resistance to Foc race 1 Rasthali (AAB) [75] Ace-AMP1 + Ca-pflp Allium cepa L; Capsicum annum L. Agrobacterium/AGL1/ ECS Stacked Ace-AMP1 and pflp transgenic plants showed resistance to Foc race 1 Rasthali (AAB) [76] Foc TR4 resistance Human lysozyme (HL) Agrobacterium/EHA105/corm slices Improved resistance to Foc TR4 Taijiao (AAA) [77] Pflp Sweet pepper Agrobacterium/EHA105/
multiple bud clumpsEnhanced resistance to Foc TR4 Pei Chiao (AAA) and Gros Michel (AAA) [78] TLP or PR-5 Rice Biolistics/Single
cauliflower-like bodiesEnhanced resistance to Foc TR4 Pisang Nangka (AAB) [79] ThChit42 Trichoderma harzianum Agrobacterium/EHA105/ECS Enhanced resistance to Foc TR4 Furenzhi (AA) [80] RGA2 or Ced9 Banana Agrobacterium/EHA105/ECS Improved promising resistance against Fusarium wilt Grand Nain (AAA) [81] MaPR-10 banana Agrobacterium/−/ECS Improved tolerance against Fusarium infection Berangan [82] MaLYK1 Banana Agrobacterium/EHA105/ECS Increased resistance to Foc TR4 Cavendish (AAA) [83] Synthesis of ergosterol (ERG6/11) − Agrobacterium/EHA105/ECS strong resistance to Fusarium wilt Brazil (AAA) [84] MpbHLH Banana Agrobacterium/EHA105/ECS Enhanced Foc TR4-resistance of Cavendish banana Brazil (AAA) [85] Sigatoka resistance ThEn-42 + StSy + Cu,Zn-SOD co-transformation Trichoderma harzianum
+ grape + tomatoBiolistics/ECS Enhanced tolerance to Sigatoka Grand Nain (AAA) [86] rcc2 or rcg3 Rice Agrobacterium/EHA105/ ECS Enhanced host plant resistance to black Sigatoka Gros Michel (AAA) [87] Bacterial BXW resistance Hrap Sweet pepper Agrobacterium/AGL1/ ECS About 20% of the Hrap lines showed
100% resistance for both mother and
ratoon crops under field conditionsSukali Ndiizi (AAB) and Mpologoma (AAA) [88, 89] Pflp Sweet pepper Agrobacterium/EHA105/ ECS About 16% of the Pflp lines showed
100% resistance for both mother and
ratoon crops under field conditionsSukali Ndiizi (AAB) and Nakinyika (AAA) [89, 90] Stacked Harp and Pflp Sweet pepper Agrobacterium/AGL1/ ECS Stacked Harp and Pflp transgenic plants
had higher resistance to X cmGonja manjaya (AAB) [91] Xa21 Rice Agrobacterium/EHA105/ ECS 50% of the transgenic lines showed complete resistance to X cm Gonja manjaya (AAB) [92] Viral Rep BBTV − Completely resistant to BBTV infection was found under glasshouse conditions Brazilian (AAA) [93] BBTV-G- cp BBTV Biolistics/apical meristem Williams (AAA) [94] Rep, ProRep BBTV − Rasthali (AAB) [95] Abiotic stress Salt, oxidative stress MusaWRKY71 banana Agrobacterium/EHA105/ ECS Enhanced tolerance towards oxidative and salt stress Rasthali (AAB) [96] Cold, drought, salt MusabZIP53 banana Agrobacterium/EHA105/ ECS Transgenic plants displayed severe growth retardation Rasthali (AAB) [97] Cold MpMYBS3 banana Agrobacterium/EHA105/ ECS The transgenic lines had higher cold tolerance Brazil (AAA) [98] Salt, drought MusaNAC042 banana Agrobacterium/EHA105/ ECS MusaNAC042 is positively associated with drought and salinity tolerance Rasthali (AAB) [99] Drought, salt Musa-DHN-1 banana Agrobacterium/-/ ECS Improved tolerance to drought and salt-stress Rasthali (AAB) [100] Salt, drought AhSIPR10 Arachis hypogaea Agrobacterium/EHA105/
multiple shoot clumpTransgenic plants showed better tolerance of salt and drought conditions Matti (AA) [101] Drought, salt, oxidative stress MusaSAP1 banana Agrobacterium/EHA105/ ECS Transgenic plants displayed better stress endurance characteristics Rasthali (AAB) [102] Cold, salt, drought MusaPIP1;2 banana Agrobacterium/EHA105/ ECS Transgenic plants showed better abiotic stress survival characteristics Rasthali (AAB) [103] Salt MusaPIP2;6 banana Agrobacterium/EHA105/ ECS Transgenic plants showed better tolerance under salt stress Rasthali (AAB) [104] Salt, drought MaPIP1;1 banana Agrobacterium/EHA105/
thin cell layers from shoot tipsImproved tolerance to salt and drought stresses Brazilian (AAA) [105, 106] Drought, cold ,salt MaPIP2-7 banana Agrobacterium/EHA105/
thin cell layers from shoot tipsImproved tolerance to salt, drought, and cold stresses Brazilian (AAA) [107] MaSIP2-1 banana Agrobacterium/EHA105/
thin cell layers from shoot tipsTransgenic plants had a stronger drought and cold tolerance than the control Brazilian (AAA) [108] Salt, drought AhcAPX Arachis hypogea Agrobacterium/EHA105/
multiple shoot clumpEnhanced the tolerance to drought and salt stress Grand naine (AAA) [109] Fruit quality and others Biofortified Iron ferritin soybean Agrobacterium/EHA105/ ECS A 6.32-fold increase in iron accumulation and a 4.58-fold increase in the zinc levels were noted in the leaves of transgenic plants Rasthali (AAB) [110] Biofortified pro-vitamin A MtPsy2a banaan Agrobacterium/AGL1/ ECS A high content of β-CE (75.1 µg/g dw) was found in the fourth generation with no variation in critical agronomical features such as yield and cycle time Dwarf Cavendish (AAA) [111] Fruit ripening MaMADS1 and MaMADS2 banana Agrobacterium/-/ ECS Repression of either MaMADS1 or
MaMADS2, resulted in delayed ethylene
synthesis and maturationGrand Nain (AAA) [112] Sense and anti-sense MaMADS36 banana Agrobacterium/GV3101/
thin cell layers from shoot tipsMaMADS36 represents a central molecular switch in regulating banana fruit ripening Red banana (AAA) [113] Foc, Fusarium oxysporum f. sp. cubense; ECS, embryogenic cell suspension; BBTV, Banana bunchy top virus; NM, not mention. Improvements in the abiotic stresses resistance
-
Many transcription factors (TFs) and downstream genes which respond to abiotic stress have been identified in banana. They mainly include WRKY[96], bZIP[97], MYB[98], NAC[99], dehydrins (DHN)[100], SAP1[102], and aquaporins (AQP)[103−108], and so on (Table 1). In transgenic plants, overexpression of these TFs let them withstand and survive under stress conditions. Because of their important role in plant growth and development, it can cause abnormal growth in transgenic plants by the constitutive overexpression of these TFs, such as bZIP53[97]. Identifying and overexpressing a key gene participated in stress tolerance is a good option. Ectopic expression of stress-related genes has been introduced into banana to enhance the tolerance to some abiotic stresses[100−109]. However, the majority of these studies have been reported from a glasshouse evaluation. Trials in the field are necessary to further prove their worthiness.
Improvements in the fruit quality and others
-
Recently, genetic engineering was also employed to improve fruit nutrient content and control fruit ripening (Table 1). Transgenic bananas with biofortified iron content and pro-vitamin A were tested in the green house and field, respectively. The transgenic banana plants overexpressing soybean ferritin accumulated the higher levels of iron and zinc under in vitro conditions as well as in the green house[110]. PVA-biofortified transgenic Cavendish bananas were also developed[111].
Banana MaMADS transcription factors are necessary for fruit ripening and molecular tools to promote shelf-life. Repression of either MaMADS1 or MaMADS2, resulted in delayed ethylene synthesis and maturation[112]. Similarly, transgenic red bananas were obtained with sense and anti-sense constructs of MaMADS36. Further study demonstrated that MaMADS36 directly binds to the CA/T(r)G box of the MaBAM9b promoter to regulate enzyme activity and starch degradation during ripening[113].
CRISPR/Cas-based genome editing
-
Genome-editing technologies using various site-directed nucleases (SDNs) have become powerful tools for modifying plant genomes. SDNs include meganucleases, ZFNs (zinc finger nucleases), TALENs (transcription activator-like effector nucleases), and CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein)[114]. The CRISPR/Cas system has been widely adopted for plants genetic improvement due to its simplicity and high-efficiency[115].
Although the application of the CRISPR/Cas9 system in banana is still at the preliminary stage, CRISPR/Cas9 mediated genome editing has been applied to improve banana nutrient contents, storage time, disease resistance, and alter the plant architecture[6, 114, 116, 117]. In summary, the CRISPR/Cas9-based genome editing utilized in banana are outline in Table 2.
Table 2. Application of CRISPR/Cas9 gene editing technology in Musa.
Cultivar Explant Strategy of transformation Cas9 promoter sgRNA promoter Target gene Target trait Result Editing efficiency Reference Baxi; AAA ECS Agrobacterium-mediated transformation
EHA105Ubi OsU6a MaPDS Albino and variegated phenotype Murtation in target genes; Albino phenotype in transgenic plants 55% [118] Rasthali; AAB ECS Agrobacterium-mediated transformation
Agl12 × CaMV35S OsU3 MaPDS Albino and variegated phenotype Murtation in target genes; Albino phenotype in transgenic plants 59% [119] Williams; AAA ECS Agrobacterium-mediated transformation
EHA105Ubi;CaMV35s OsU3 MaPDS Albino and dwarf phenotype Murtation in target genes; Albino and dwarfing phenotype in transgenic plants 63% [120] Sukali Ndiizi; AAB;Gonja Manjaya; AAB ECS Agrobacterium-mediated transformation
EHA1052 × CaMV35S OsU6 MaPDS albino and variegated phenotypes Generation of mutants with albino and variegated phenotypes 100% [121] Williams; AAA ECS Agrobacterium-mediated transformation NM NM MaCHAOS Pale-green phenotypes Murtation in target genes; Pale-green phenotypes and normal growth NM [122] Baxi; AAA protoplast PEG-mediated transformation Ubi OsU3 MaPDS - The efficiency of CRISPR/Cas9-mediated mutagenesis was higher than that of CRISPR/Cas12a, and RNP-CRISPR-Cas9 1.04% (Cas9), 0.92% (RNP), 0.39% (Cas12a) [61] Brazilian; AAA protoplast PEG-mediated transformation Ubi MaU6 MaPDS - Increased mutation efficiency of CRISPR/Cas9 genome editing in banana by optimized construct 4-fold [123] Gonja Manjaya; AAB ECS Agrobacterium-mediated transformation
EHA105Ubi OsU6 Viral genes Banana streak virus (BSV) Inactivation of endogenous banana streak virus (eBSV) intergated in host genome and generated resistant banana plants against eBSV 95% [124] Sukali Ndiizi; AAB ECS Agrobacterium-mediated transformation
EHA1052 × CaMV35S OsU6 MusaDMR6 Banana Xanthomonas wilt (BXW) Improved resistance to BXW disease in mutants with normal growth 100% [125] Grand Naine; AAA ECS Agrobacterium-mediated transformation
Agl1CaMV35S OsU3 MaLCYε Regulation synthesis of β-carotene Improved nutritional trait in transgenic plants with normal growth NM [127] Rasthali; AAB protoplasts and ECS electroporation-mediated transformation;
particle bombardment methodCaMV35S OsU3 MaCCD4 Regulatory mechanism of β-carotene homeostasis CCD4 negatively regulates β-carotene biosynthesis NM [128] Brazilian; AAA ECS Agrobacterium-mediated transformation Ubi OsU6a MaACO1 Shelf life More vitamin C and improved shelf life in transgenic plants 98% [129] Gros Michel; AAA ECS Agrobacterium-mediated transformation Ubi OsU6a/
OsU3MaGA20ox2 Semi-dwarf phenotype A lower active GA content and a semi-dwarf phenotype in transgenic plants NM [130] NM, not mentioned. Establishing the editing system
-
Plant albino phenotype is a classic and indicative phenotype for testing and judging whether the CRISPR/Cas9 system is effective. As an indicator gene, phytoene desaturase (PDS) can easily obtain the target albino trait and has been knocked out in most fruit trees. Recently, CRISPR/Cas9-based genome editing in banana has been established using the PDS as a marker gene[118−121]. However, knockout of PDS has adverse effects on plant growth. Optionally, RP43/CHAOS39-edited banana plants were obtained with pale-green phenotype and no negative effects on plant growth[122]. Recently, the transient delivery system by a PEG-mediated protoplast was established[61]. The editing efficiency of the CRISPR/Cas9, CRISPR/Cas12a, and ribonucleoprotein-CRISPR-Cas9 (RNP-CRISPR-Cas9) for targeting the PDS gene in banana protoplasts was compared. The results showed that the efficiency of CRISPR/Cas9-mediated mutagenesis was higher than that of the other two systems. In addition, it was the first report by a RNP-CRISPR-Cas9 system for genome editing in banana[61]. In comparison to the previous report in banana, using endogenous U6promoter and banana codon-optimized Cas9 in CRISPR/Cas9 cassette, the mutagenesis efficiency has a fourfold increase[123].
Enhance disease resistance
-
Banana streak virus (BSV), a double stranded DNA Badnavirus which integrated in the B genome derived from M. balbisiana, is called endogenous BSV (eBSV). It severely affects production of plantain (AAB) in Africa. To inactivate the virus, a multiplexed gRNA strategy targeting all three ORFs of eBSV was constructed and transformed into Gonja Manjaya (AAB). Compared with the controls, the eBSV-edited plants exhibited resistance against eBSV and normal growth. A very high mutation effciency of 95% using three gRNAs were observed[124]. Recently, CRISPR/Cas9 mediated gene editing for banana resistance against bacteria was also reported. To obtain the banana cultivar against Xanthomonas Wilt (BXW), a Musadmr6 gene was edited[125]. The edited plants had a higher resistance to BXW without adverse affecting on plant growth. Researchers are also trying to breed TR4 resistance cultivars by CRISPR[126].
Improve fruit quality and shelf life
-
Fruit quality is an important indicator to measure the value of fruit commodities. Carotenoids are essential for human nutrition. Most Cavendish group cultivars have low β-carotene content in the fruit pulp. Using CRISPR/Cas9 technology, β-carotene-enriched banana plants were created by editing the fifth exon of LCYε gene from A genome, which determines a high α-/β-carotene ratio[127]. Compared with the unedited fruits, the β-carotene count of the fruit pulp of the edited lines increased by 6-fold. More recently, CRISPR/Cas9-mediated editing of CCD4 was conducted in Rasthali. In comparison to the controls, the accumulation of β-carotene in roots was increased in the CCD4-edited plants[128].
The shelf life of post-harvest fruits is an important factor affecting fruit quality. The production of ethylene is closely related to the storage time of banana fruits. Thus, it is the first consideration for developing postharvest preservation technology. MaACO1 encodes for an O2-activating ascorbate-dependent non-heme iron enzyme that catalyzes the last step in ethylene biosynthesis. The MaACO1-editted banana fruit extended shelf life and had more Vitamin C compared with the wild-type fruit[129].
Alter the plant architecture
-
Developing semi-dwarf and dwarf banana varieties is also one of the objectives of banana improvement programs. Gibberellin (GA) is a key gene which determines plant height and the mutations in its biosynthesis genes often leads to dwarf plants. CRISPR/Cas9 technology was applied to generate a semi-dwarf banana cultivar 'Gros Michel' by manipulating the M. acuminata gibberellin 20ox2 (MaGA20ox2) gene, disrupting the gibberellin (GA) pathway[130].
-
At present, extensive advances has been made on banana SE. Reports of banana genetic improvement using ECS in the past five years has increased dramatically. Nevertheless, there are still many problems to be solved in the research on banana SE and genetic modification. Little information is available on the molecular mechanisms of banana SE. The embryogenic capacity and efficiently propagated plantlets are very low. The repeatability of the protocols early reported for SE in banana is poor. So far, SE in bananas is far from being considered a conventional technique and has not even been successfully used in some varieties.
Hence, an important consideration for future work is to explore the basic molecular mechanism of banana embryogenic potency. The gradual application of multi-omics technique in plant SE provides the feasibility to uncover the regulatory mechanism of SE development at the molecular level. Further continuous work is needed for optimizing a highly effcient and versatile transformation and regeneration system which independent on genotype. And the genetic improvement at present only aims at single gene or single trait. More genes associated with disease-resistance, as well as with other important agronomic traits, should be characterized and utilized in target breeding programs. Molecular designing breeding with multi-gene superposition should be carried out to breed new banana varieties with good comprehensive characters.
Despite the rapid progress of banana transgenic, there are no commercial transgenic varieties applied to the production. With the continuous optimization and improvement of CRISPR/Cas and other gene editing technologies, it is possible to obtain an ideal mutant by accurately targeting target sites. In addition, the key technology of modern biotechnology breeding is the delivery system of plant genetic modification. The application of nanocarriers in plant genetic engineering shows a broad application prospect. The introduction of nanotechnology into banana tissue culture showed significant positive effects on callus induction, somatic embryogenesis and other regeneration aspects. More recently, a cut-dip-budding delivery (CBD) system enables genetic modifications in plants without tissue culture[131]. It overcomes the difficulties posed by the traditional technology due to the plant tissue culture process. Therefore, it would be very interesting to explore a simple, fast and efficient method for banana genetic transformation or genome editing without the need for tissue culture.
This research was supported by the specific research fund of The Innovation Platform for Academicians of Hainan Province (YSPTZX202101), the Hainan Provincial Natural Science Foundation (321RC638), the National Natural Science Foundation of China (32172269, 31501043) and the Earmarked Fund for Modern Agro-industry Technology Research System (CARS-31).
-
The authors declare that they have no conflict of interest.
-
Received 30 September 2022; Accepted 29 November 2022; Published online 23 December 2022
-
# These authors contributed equally: Jingyi Wang, Shanshan Gan
- Copyright: © 2022 by the author(s). Published by Maximum Academic Press on behalf of Hainan University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Wang J, Gan S, Zheng Y, Jin Z, Cheng Y, et al. 2022. Banana somatic embryogenesis and biotechnological application. Tropical Plants 1:12 doi: 10.48130/TP-2022-0012
Banana somatic embryogenesis and biotechnological application
- Received: 30 September 2022
- Accepted: 29 November 2022
- Published online: 23 December 2022
Abstract: As one of the most important economic crops for both staple food and fruit widely cultivated in the tropics and subtropics, banana (Musa spp.) is susceptible to a plethora of abiotic and biotic stresses. Breeding cultivars resistant to abiotic and biotic stressors without adverse effects on yield and fruit quality are the objectives of banana improvement programs. However, conventional breeding approaches are time-consuming and severely hampered by inherent banana problems (polyploidy and sterility). Therefore, genetic transformation is becoming increasingly popular and can provide rapid solutions. Numerous efforts have been made to develop superior banana cultivars with better resistance to abiotic and biotic stresses and optimum yields using genetic modification strategies. Somatic embryogenesis (SE) through embryogenic cell suspension (ECS) cultures is an ideal recipient system for genetic transformation in banana. The purpose of this paper is to review the current status of banana somatic embryo research, clarify the process of banana somatic embryo induction and culture, and summarize the main influencing factors in the process of somatic embryogenesis. At the same time, their applications in breeding technologies such as cryopreservation, protoplast culture, genetic transformation and gene editing were also summarized, in order to provide reference for the research and practical application of banana somatic embryogenesis in the future.
-
Key words:
- Banana /
- Somatic embryogenesis /
- Genetic transformation /
- CRISPR












