-
Coconut is a perennial evergreen tree in the palm family Arecaceae and is an important woody oil and food crop in tropical regions. Currently, the global coconut cultivation area is about 12 million hectares, with a total production of approximately 61 million metric tons in 2020[1]. The top coconut-producing countries are Indonesia, the Philippines, and India, which collectively account for over 75% of the world's coconut production[2]. By 2020, China's coconut cultivation area reached 58,000 hectares, with a yield of 220,000 tones, mostly concentrated in Hainan Province.
The structure of a coconut fruit from outside to inside is the fruit peel, seed coat, coconut meat (solid endosperm) and coconut water (liquid endosperm). The endosperm of the coconut has two forms: one is the white solid endosperm (coconut meat), and the other is coconut water, which is a sterile liquid endosperm in the fruit cavity[3]. It is a natural and organic beverage that is nutritious and has no side effects, with various uses in food manufacturing and is widely enjoyed by people. In clinical applications, it can be used for intravenous infusion, treating diarrhea, dehydration, preserving periodontal ligament cells, protecting cardiovascular, liver, kidney, and other functions[4]. Some scholars believe that coconut water has a complex chemical composition, containing several active substances that can stimulate the rapid and irregular division of mature cells in higher plants, thus promoting plant growth[5]. Therefore, it can be attempted to use 'liquid endosperm' containing multiple beneficial components as a liquid culture medium, or add coconut water to the culture medium to provide nutrient components for the cultured plant tissue[6].
A protoplast is a 'naked cell' formed by removing the cell wall by mechanical or enzymatic methods and then enveloped by a plasma membrane[7]. The yield and activity of protoplasts obtained by mechanical separation are low, while those obtained by enzymatic separation are numerous, with good integrity and strong activity, which is the choice of most researchers[8]. Due to the absence of cell wall obstruction, various molecular biology studies can be performed by artificial means. Protoplasts have important application value and are currently successfully applied in many fields, including obtaining regenerated plants through protoplast culture, transient expression of genes, protein interactions, and gene editing[9]. The use of plant protoplasts has provided valuable insights into plant developmental processes, stress responses, hormone signaling, and gene regulation. They have been extensively employed in the study of gene function, protein-protein interactions, promoter analysis, and subcellular localization of proteins[10,11]. Moreover, protoplast-based approaches have facilitated the production of transgenic plants, including genome editing using CRISPR/Cas9 technology[12,13].
Currently, research on coconut water mainly focuses on its special uses, chemical composition, and processing utilization, but there is relatively little research on protoplast isolation and identification. In recent years, there have been reports on protoplast preparation and transient transformation of Areca catechu and oil palm, which are also palm plants. This study referred to the experimental methods of these two plants to establish the optimal experimental condition combination for isolating coconut protoplasts.
-
Coconut leaves and coconut water were selected to isolate protoplasts of different quantities and qualities. The observation results showed that the background of the leaves was contaminated with impurities, and there were not only protoplast cells but also long needle-like substances in the field of view. The cell size was not uniform, and the interior of the cells was relatively transparent (Fig. 1a, b). The largest number of protoplasts was obtained from leaves after 2.5 h of enzyme digestion, which was about 7.6 × 106 cells/mL, and the activity reached 88.7%. In comparison, the background of the coconut water was relatively clean compared to the leaves, and the cells contained more inclusions. When the enzyme digestion time was 0.75 h, the number of protoplasts was the highest, reaching 7.1 × 107/mL, and the cell morphology was large and round (Fig. 1d, e), with a strong cell viability of 89.6%. The majority of cells from both materials had a regular circular shape. Compared with the leaves, the diameter of the liquid endosperm cells was about 2−3 times larger.
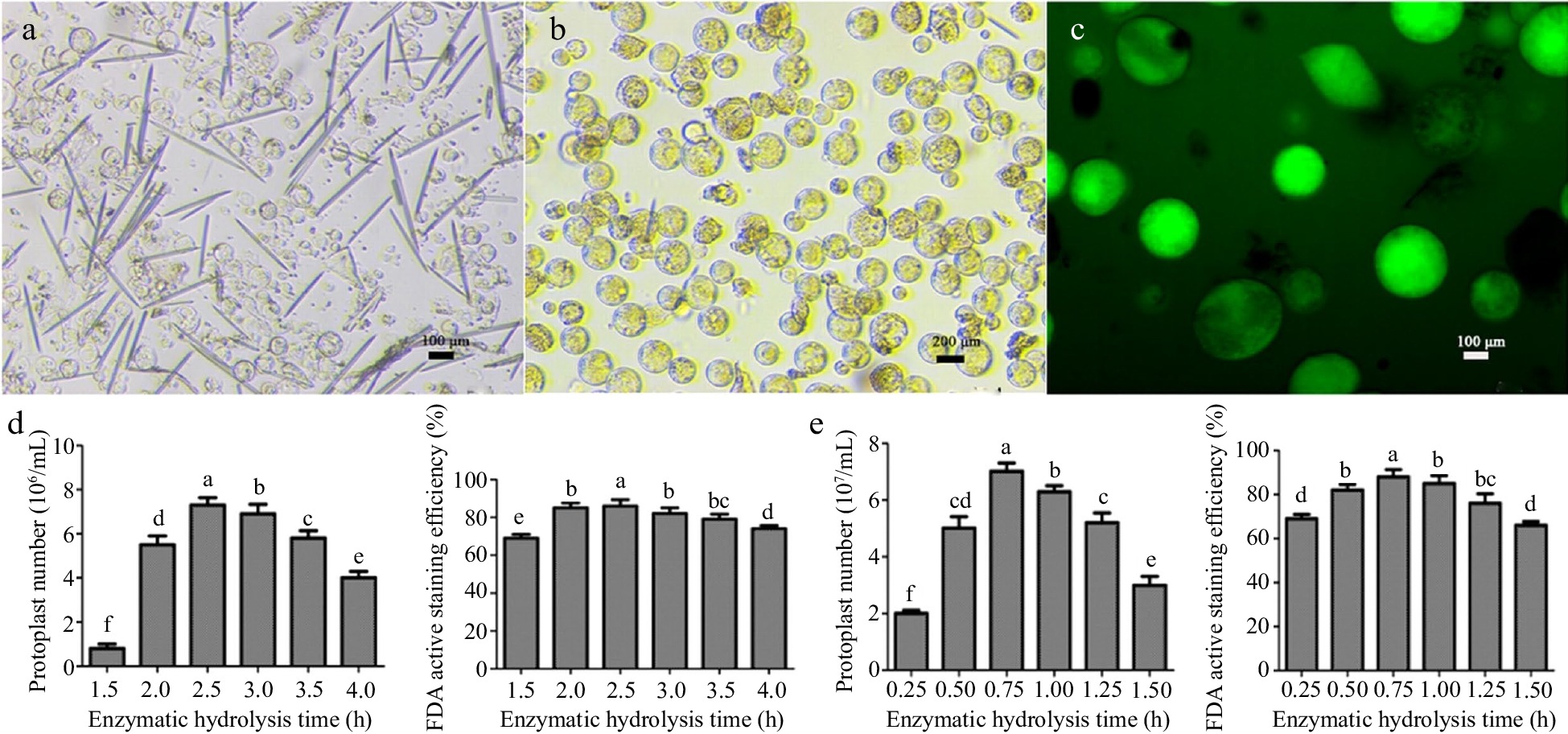
Figure 1.
Isolation of coconut protoplasts, determination of FDA activity, and effects of enzymatic hydrolysis time on the number and activity of protoplasts. (a) Young leaves of coconut, (b) coconut liquid endosperm, (c) FDA staining of protoplasts, (d) leaf enzymatic hydrolysis time, (e) enzymatic hydrolysis time of endosperm. Different lowercase letters indicate significant differences (p < 0.05).
Ploidy analysis of leaf and endosperm protoplasts
-
To determine the ploidy of coconut endosperm protoplasts, physical cutting was performed on freshly picked young coconut leaves, and flow cytometry was used for ploidy detection. Coconut leaf with diploid chromosome number was used as the control. The peak of leaf cell appeared at around 12,250, which could accurately determine that the coconut leaf protoplast was diploid. The peak of endosperm cell appeared at around 19,500. By using the formula: sample ploidy = sample G1 peak fluorescence intensity/control G1 peak fluorescence intensity × control ploidy, 19500/12250 × 2 ≈ 3.18, indicating that the coconut endosperm protoplast was triploid (Fig. 2a, b).
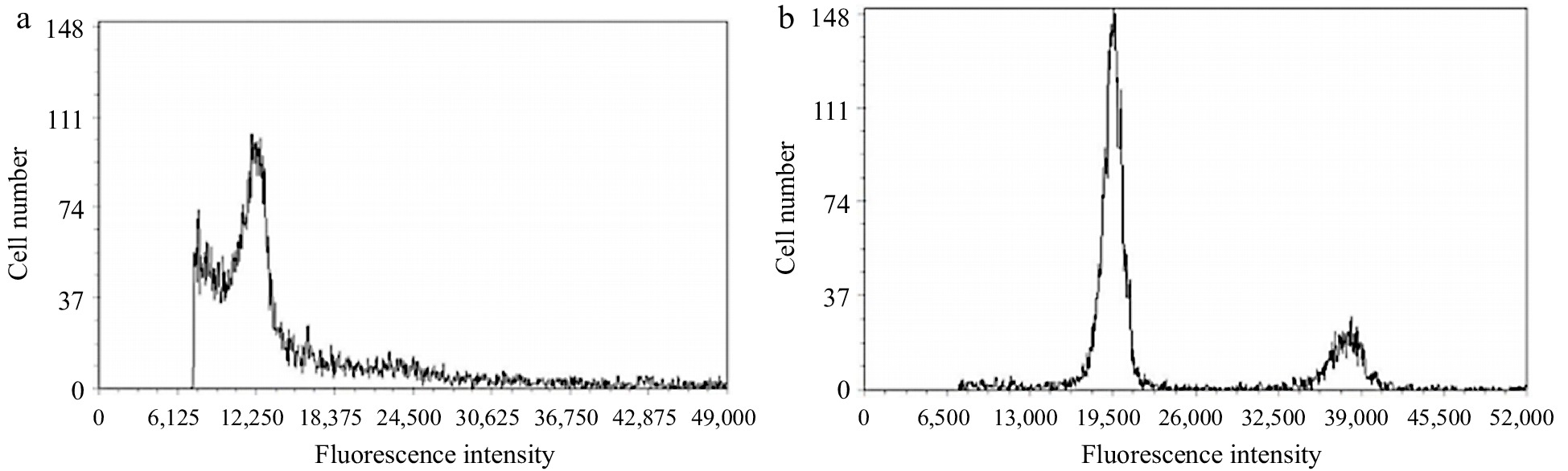
Figure 2.
Ploidy analysis of coconut leaf and liquid endosperm protoplast cells. (a) Leaf protoplasts, (b) endosperm protoplast.
Nucleus localization of leaf and endosperm protoplasts
-
DAPI was used to stain the nuclei of intact protoplasts from both leaves and endosperm. The results showed that the nuclei of both leaf and endosperm protoplasts emitted bright blue fluorescence under weak blue light excitation, but the nuclei of these two types of cells were distinct. The diameter of leaf cells was about 160 μm (Fig. 3a), and a bright blue fluorescent spot in the lower right corner occupied a very small part of the entire cell, about 17% of the cell area, and the cytoplasm was colorless. The diameter of liquid endosperm cells was about 200 μm (Fig. 3b), and bright blue fluorescence almost occupied the entire interior of the cell, about 95%. Random observation of endosperm cells with confocal microscopy with a diameter of about 29 μm also showed that a large area of blue fluorescence was emitted from the middle part of the cell, about 85%. It was inferred that there is a large nucleus in the liquid endosperm cells, which is the main component of the cell interior (Fig. 3c).
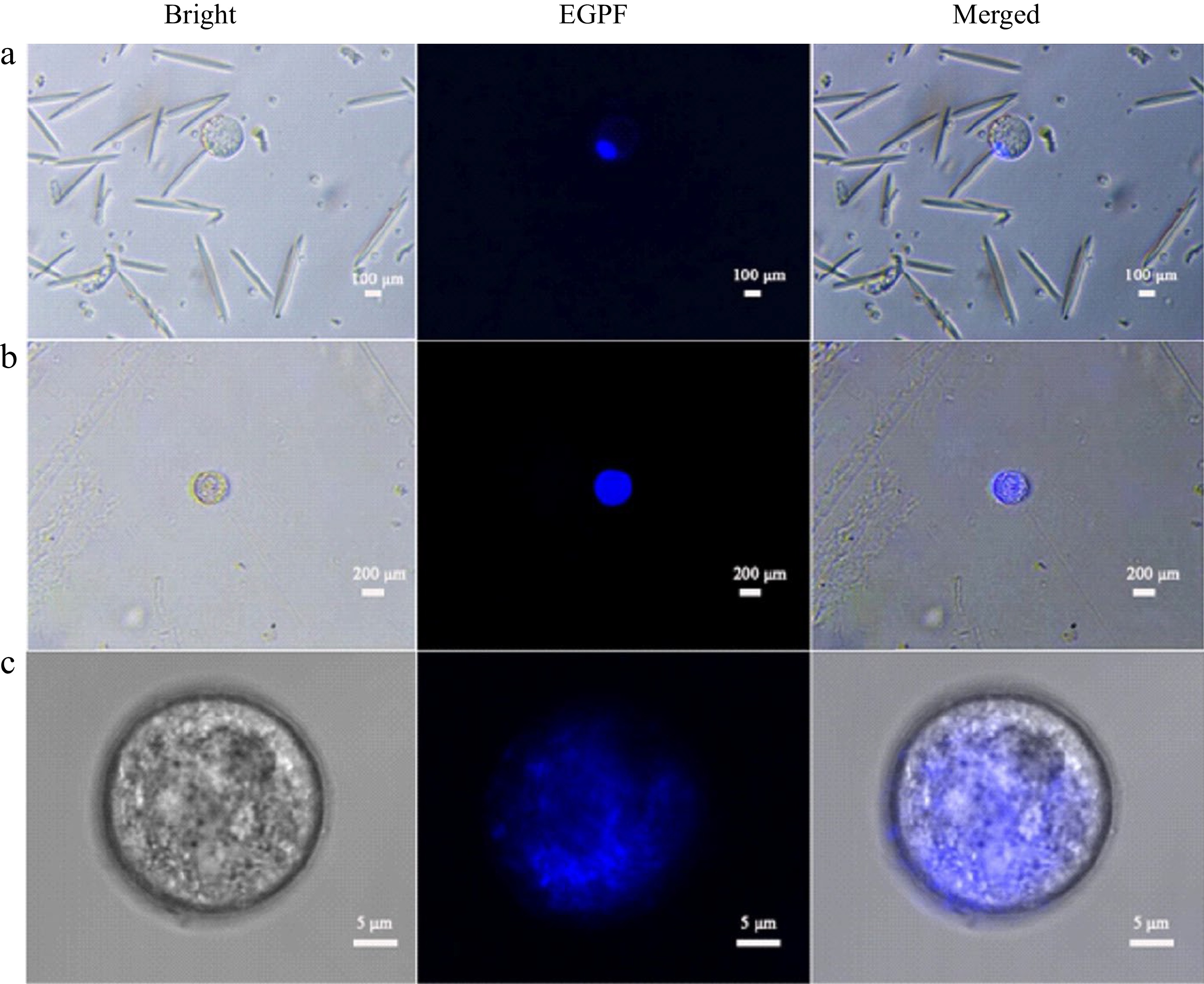
Figure 3.
DAPI stained coconut protoplast. (a) Leaf protoplasts, (b) endosperm protoplast, (c) endosperm protoplasts under confocal microscopy.
Optimization of PEG-mediated transient transformation of protoplasts
-
In order to improve the transient transformation system of coconut protoplasts, the transformation method of endosperm protoplasts was optimized based on the transformation of leaf protoplasts. The results showed that both low and high PEG concentrations would affect the transformation efficiency. The highest transformation efficiency of 14% was achieved when the final PEG concentration was 25% and the incubation time was 25 min (Fig. 4a, b). The optimized transformation method was used to transfect P35-EGFP into endosperm protoplasts, and the bright field, dark field, and overlay images under this transformation method are shown in Fig. 4c.
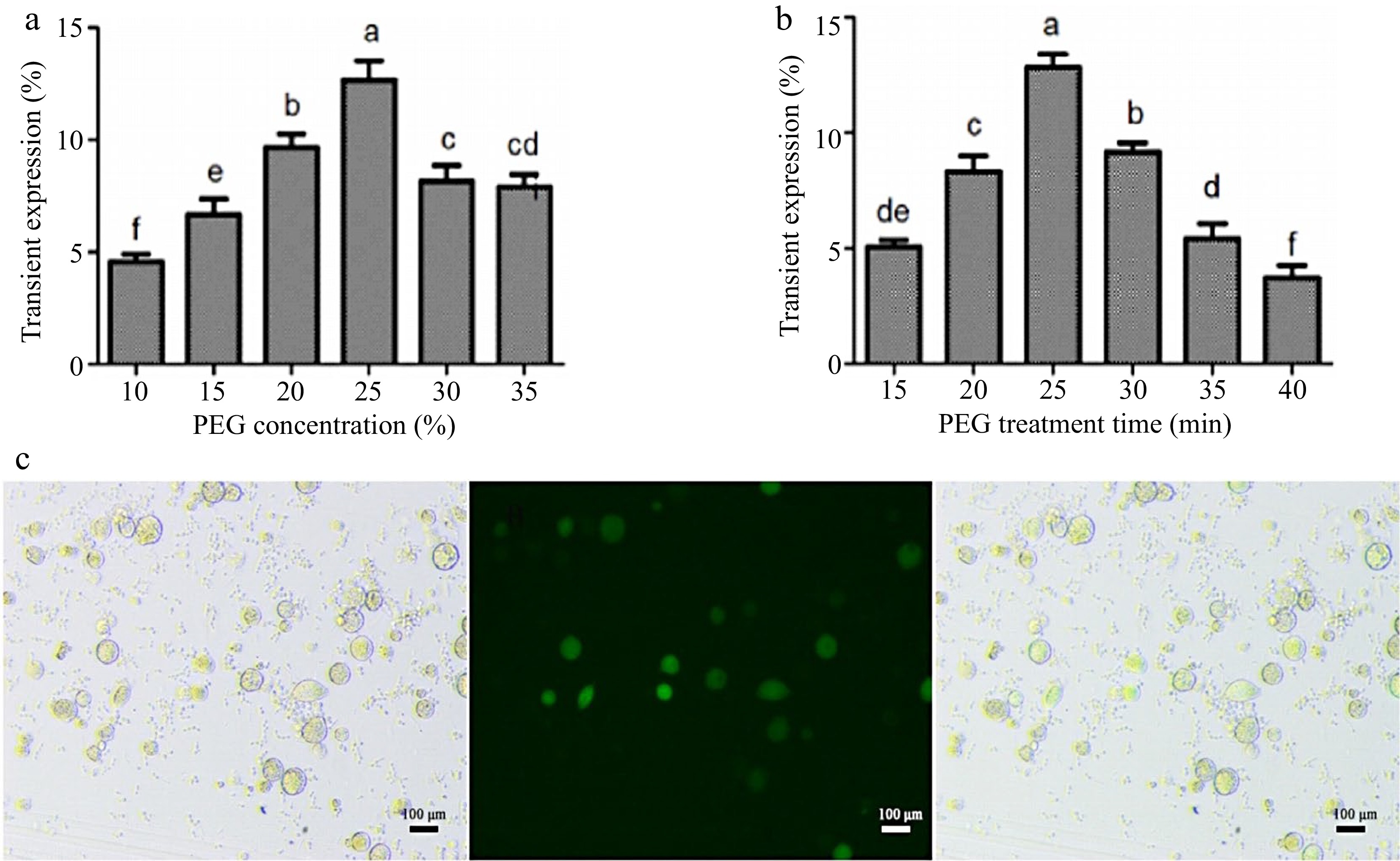
Figure 4.
Optimization of the instantaneous transformation system for coconut endosperm protoplasts. (a) PEG final concentration treatment, (b) PEG incubation time, (c) effect diagram of endosperm protoplast transformation. Different lowercase letters indicate significant differences (p < 0.05).
Subcellular localization of proteins in coconut protoplasts
-
To verify the feasibility of the protoplast transient expression system and to confirm and determine the presence and size of the nucleus using subcellular localization, p35-eGFP and p35-MYB108-eGFP transient expression vectors were introduced into coconut young leaves and liquid endosperm protoplasts, respectively. The transformation results showed that in the young leaves, eGFP was distributed in the nucleus and cytoplasm, while the fusion protein MYB108-eGFP was distributed on the nucleus, with a green fluorescent area of approximately 3 μm (Fig. 5a, b). When introduced into endosperm protoplasts, eGFP was evenly distributed throughout the cell, and the fusion protein MYB108-eGFP was also evenly distributed throughout the cell, covering the entire cell with green fluorescence, with the green fluorescent area accounting for approximately 95% of the cell area (Fig. 6a, b).
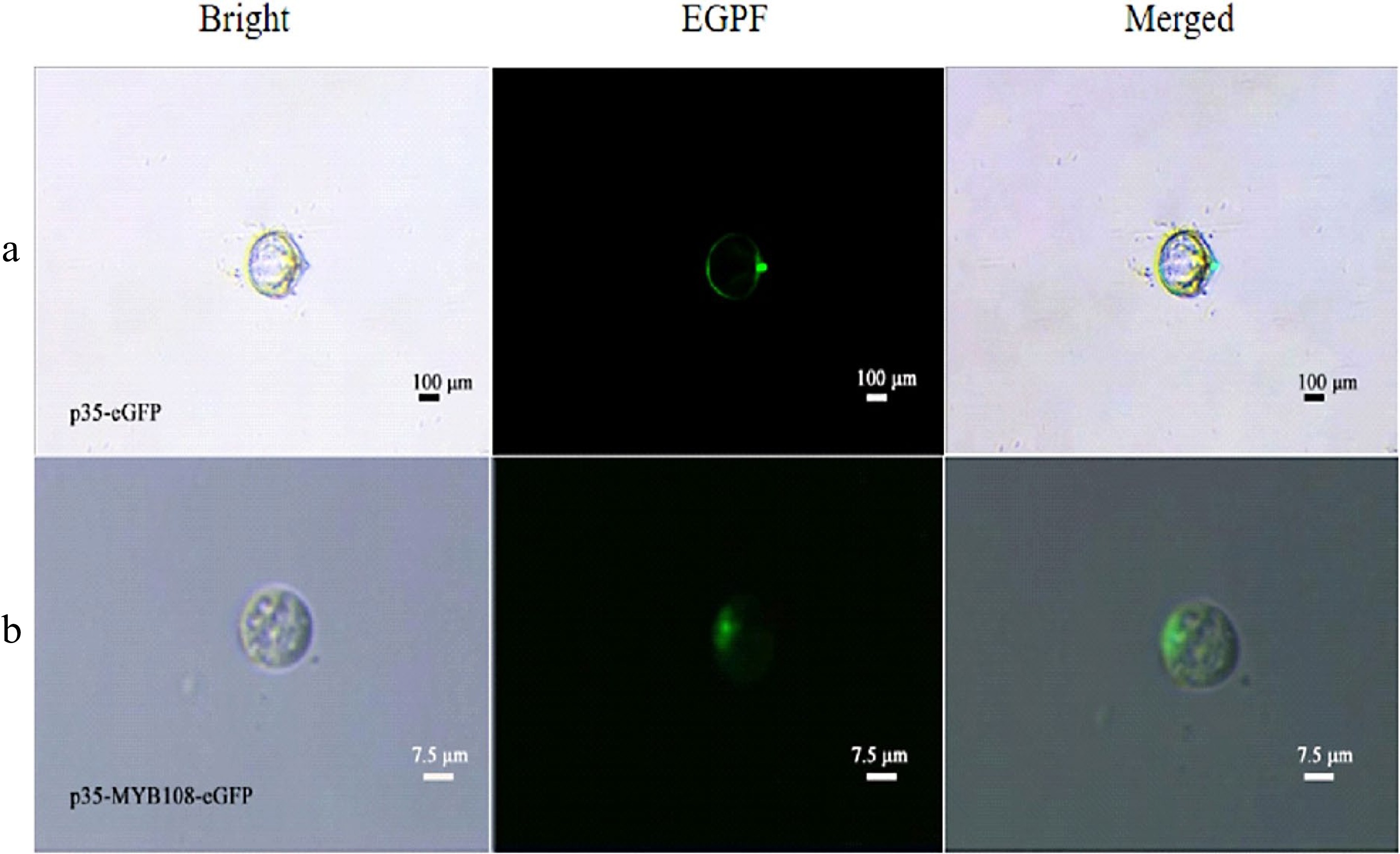
Figure 5.
Coconut leaf protoplasts. (a) p35-eGFP fluorescence localization, (b) subcellular localization of p35-MYB108-eGFP in leaf protoplasts.
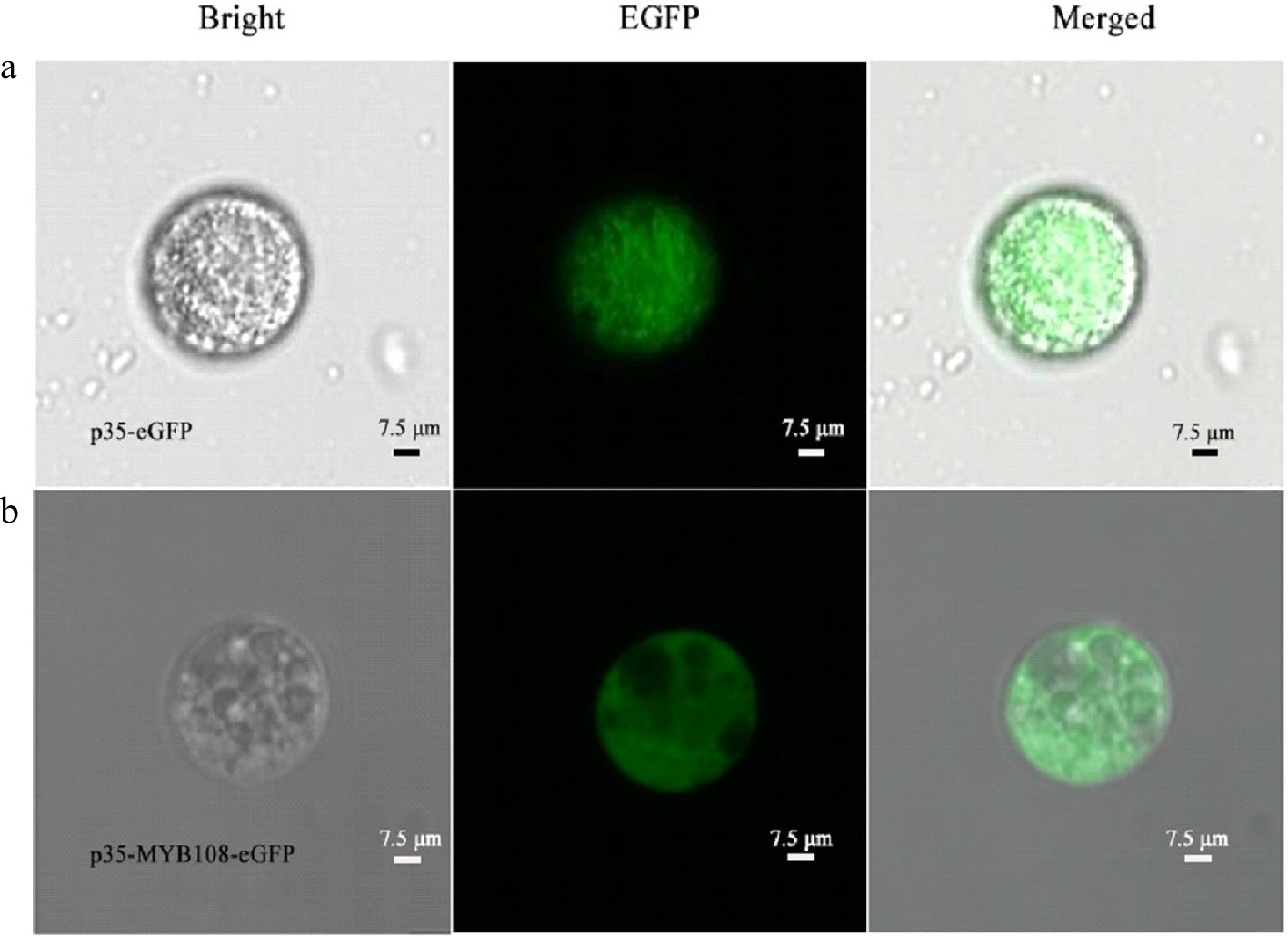
Figure 6.
Coconut liquid endosperm protoplasts. (a) p35-eGFP fluorescence localization, (b) subcellular localization of p35-MYB108-eGFP in endosperm protoplasts.
Transient expression of genes in coconut protoplasts
-
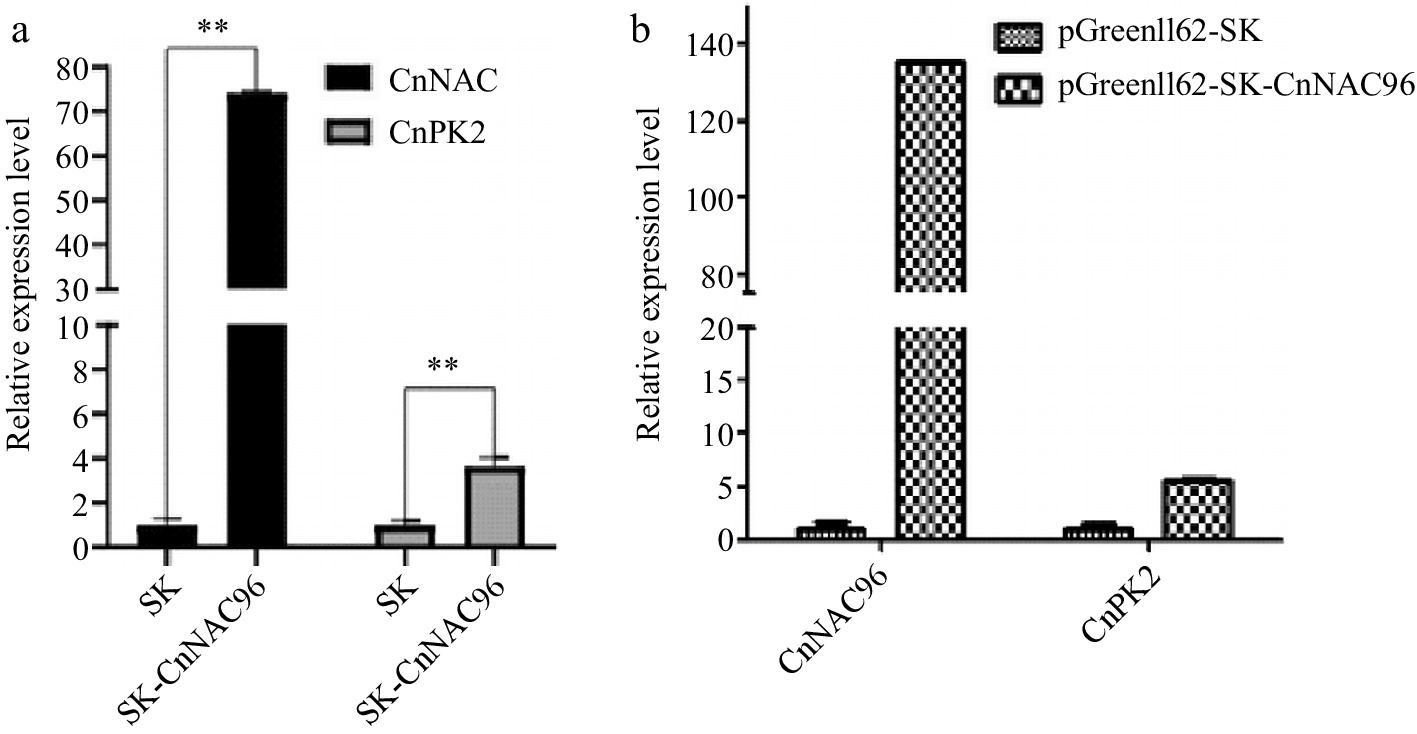
The transient expression system in plant protoplasts is commonly used for gene function analysis and validation. In this study, we investigated the capability of the established transient transformation system in coconuts to explore the potential gene expression and its regulatory relationships. The expression of genes in the transformed coconut protoplasts was assessed using RT-qPCR. The results demonstrated successful expression of CnNAC96 in leaf and endosperm protoplasts, leading to the upregulation of CnPK2 (Coconut pyruvate kinase2). In the endosperm protoplasts, CnNAC96 exhibited approximately 135-fold overexpression, resulting in a five-fold upregulation of CnPK2 (Fig. 7). These findings affirm the feasibility of the optimized transient transformation system in coconut protoplasts and highlight its applicability in investigating potential gene expression and regulatory mechanisms in coconuts.
-
The protoplast transient expression system (PTES) is a necessary tool in molecular biology and is of great significance for determining the function of plant genes[14]. At the same time, by isolating stable and efficient plant protoplasts, homogeneous experimental materials can be provided for subsequent research[15]. Currently, protoplast isolation from various tissues of many species of plants has been achieved. Plant leaves, callus tissue, petioles, and stems can all be used as materials for protoplast isolation and preparation, and protoplasts have been successfully isolated from leaf flesh of Populus tomentosa, papaya seeds, petal of Clerodendrum indicum, wheat leaves, oil palm callus tissue, and Areca catechu inflorescences[15−17]. However, the optimal conditions for isolating protoplasts from different plants vary greatly, so there are still many plants for which a stable protoplast isolation system has not been established.
This study successfully established a mature and efficient system for the isolation of coconut endosperm protoplasts. Optimization was carried out for the important parameter of enzymatic hydrolysis time. Coconut endosperm protoplasts are fragile and have thin cell walls, so the enzymatic hydrolysis time should not be too long. The optimal enzymatic hydrolysis time was found to be 45 min. For coconut leaves, the highest quantity and vitality of protoplasts were obtained after enzymatic hydrolysis for 2.5 h. The diameter of endosperm cells was found to be 2−3 times larger than that of leaf cells. Using flow cytometry, the ploidy levels of coconut leaf and water cells were determined to be diploid and triploid, respectively.
A stable PEG-mediated system for the transient transformation of coconut protoplasts was established. In endosperm protoplasts, the optimal transformation efficiency was achieved with a final PEG concentration of 25% and incubation time of 25 min. The exogenous gene NAC96 was successfully expressed in coconut protoplasts, and CnPK2 was significantly upregulated. Similarly, P35-YMB108-EGFP was successfully transferred into both young coconut leaves and liquid endosperm protoplasts. The subcellular localization results, combined with DAPI nuclear staining, revealed that the endosperm protoplasts had a huge nucleus occupying the entire cell.
In angiosperms, the endosperm is derived from the development of the fertilized polar nuclei and is mostly triploid. Its main function is to provide nutrients for the development of the embryo or seedling, and endosperm development has important significance in plant development[18]. For cereal crops, the endosperm is also the main part of the grain. Currently, most studies on protoplast isolation from plant endosperm have been conducted on monocotyledonous plants, such as isolating rice endosperm protoplasts to study the mechanism of events during rice endosperm development[19]. Similarly, the isolation of maize endosperm protoplasts laid the foundation for research on maize grain starch formation and endosperm cell growth and development regulation[18]. The coconut has a relatively long development cycle, and the protoplasts isolated from the endosperm can provide a homogeneous and stable receptor for the study of molecular regulation and cell expression analysis in coconut growth and development. The establishment of an endosperm protoplast transient transformation system can also validate related research analyses on coconut gene expression regulation and has important platform value for studying coconut endosperm development and fruit formation.
-
The experimental materials were leaves and fruits of Hainan tall coconuts. The leaves were collected from the Hainan University campus, specifically from the white part of the endosperm. The liquid endosperm was taken from green coconuts of Hainan tall coconuts that were 7−8 months post-flowering. Cellulase, pectinase, 2-(N-morpholino)ethanesulfonic acid (MES), bovine serum albumin (BSA), 4',6-diamidino-2-phenylindole (DAPI) were purchased from Beijing Solarbio Science & Technology Co., Ltd. (Beijing, China), while fluorescein diacetate (FDA) was obtained from Shanghai Macklin Biochemical Co., Ltd. (Shanghai, China) Mannitol, PEG-4000, magnesium chloride, and others were purchased from Guangdong Xilong Scientific Co., Ltd (Guangdong, China).
Collection of coconut liquid endosperm and protoplast isolation
-
The protoplast isolation method was based on the oil palm leaf protoplast isolation method with appropriate modifications[20]. Specifically, coconut water was placed in a 50 mL centrifuge tube and centrifuged at 1000 rpm for 5 min. The supernatant was removed and the white flocculent material was added to 15 mL of enzyme solution (0.45 g cellulase, 0.12 g pectinase, 7.5 mL of 0.8 M mannitol, 0.15 mL of 2 M KCl, 0.15 mL of 1 M CaCl2, 0.015 g BSA, and 5.7 mL of ddH2O). The mixture was shaken at 50 rpm at room temperature for 45 min. The resulting mixture was filtered through a 200-mesh cell sieve into a centrifuge tube, and an equal amount of wash solution (30 g/L KCl, 5 g/L CaCl2·2H2O, 36 g/L mannitol, pH = 5.6) was added. The mixture was centrifuged at 1000 rpm for 4 min, and the supernatant was discarded. The process was repeated by adding an equal amount of wash solution and centrifuging. Finally, an appropriate amount of suspension solution (30 g/L KCl, 36 g/L mannitol, pH = 5.6) was added to suspend the coconut liquid endosperm protoplasts.
Isolation of coconut leaf protoplasts
-
Young coconut leaves were cut into 0.5 mm wide strips and completely immersed in 15 mL of enzyme solution. The mixture was placed on a shaker at 60 rpm at room temperature for 3.5 h under dark conditions. The resulting mixture was filtered through a 200-mesh cell sieve into a centrifuge tube, and 15 mL of wash solution was added. The mixture was centrifuged at 2,000 rpm for 5 min at 4 °C, and the supernatant was discarded. The process was repeated twice. Finally, an appropriate amount of suspension solution was added to suspend the coconut leaf protoplasts.
Ploidy analysis of coconut liquid endosperm cells
-
Flow cytometry was used to detect the ploidy of coconut liquid endosperm cells. The leaves of coconut plant (diploid, 2n = 32) were selected as control[21]. In the flow cytometric analysis, fluorescence intensity was plotted on the x-axis and the number of cells was plotted on the y-axis. Fluorescence intensity reflected the amount of DNA content in the cells[22]. The formula for determining the ploidy of the sample was as follows: Ploidy of the sample = fluorescence intensity of G1 peak of the sample/fluorescence intensity of G1 peak of the control × ploidy of the control.
Staining of cell nuclei in coconut leaves and endosperm protoplasts using DAPI
-
DAPI was used to stain the cell nuclei of coconut leaves and endosperm protoplasts. DAPI binds to double-stranded DNA in the cell nuclei, penetrating the cell membrane and producing fluorescence that is over 20 times stronger than DAPI alone. The staining was performed by adding 3 times the volume of staining solution to the sample, mixing well, and incubating at room temperature in the dark for 7 minutes. The stained samples were observed and photographed under a fluorescence microscope.
Expression of GFP gene in coconut leaves and endosperm protoplasts
-
The transformation of coconut endosperm protoplasts was optimized using the method of oil palm protoplast transformation[23]. Five hundred μL of protoplasts were dropped into a six-well plate to form a droplet, and 10%, 15%, 20%, 25%, 30%, and 35% PEG/MgCl2 solutions were dropped onto adjacent positions of the protoplast droplet. 5−25 μg of plasmid DNA was added and gently mixed. The mixture was incubated in the dark at room temperature for 15, 20, 25, 30, 35, and 40 min. The protoplast droplet and PEG droplet were then mixed well and incubated under weak light for 30 min. Four mL of wash solution (KCl 30 g/L, CaCl2·2H2O 5 g/L, sorbitol 36 g/L, pH = 5.6) was added, and the mixture was incubated overnight at room temperature under weak light. The mixture was centrifuged at 1000 r/min for 5 minu at 4 °C, and the supernatant was discarded. Two hundred μL of solution was added to resuspend the protoplasts. The transformation efficiency was calculated by inverted fluorescence microscopy.
Expression and localization of transcription factors in protoplasts
-
Protoplasts from a large number of coconuts were collected, and RNA was extracted using the Novozymes plant RNA extraction kit according to the instructions. cDNA was obtained by reverse transcription. RT-qPCR was used to detect the expression level of CnNAC96 in coconut protoplasts. Subcellular localization analysis was performed using EgMYB108-encoded oil palm protein. The full-length coding sequence (CDSs) of MYB108 was cloned into the P35-eGFP vector to obtain a transient expression vector. The fluorescence of the MYB108-eGFP fusion protein was observed under confocal laser microscopy.
Statistical analysis
-
All samples were reproducible through three biological replicates. Graphs were created using GraphPad Prism 5. The error bars represent the standard deviation. Statistical significance was determined using independent sample T-tests in SPSS 20.0 software. P values of less than 0.05 (*), less than 0.01 (**), and less than 0.001 (***) indicate statistically significant differences.
-
The authors confirm contribution to the paper as follows: Guo Q: Data curation, Writing – original draft; Wang Y: Visualization, Investigation; Huang J: Software, Validation; Zou J: Conceptualization, Methodology; Li D: Supervision, Writing – review & editing. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analyzed during this study are included in this published article.
This research was supported by the Hainan Province Science and Technology Special Fund (No. ZDYF2022XDNY148) and the National Natural Science Foundation of China (NSFC) (No. 32260064).
-
The authors declare that they have no conflict of interest. Dongdong Li is the Editorial Board member of Tropical Plants who was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of this Editorial Board member and his research groups.
-
Received 20 April 2023; Accepted 1 September 2023; Published online 7 October 2023
-
A rapid and efficient method for isolating coconut protoplasts from liquid endosperm and leaves was established.
Optimal conditions for high protoplast yield were determined: 45 minutes of enzyme digestion for endosperm and 2.5 hours for leaves.
Endosperm cells were significantly larger, triploid, and featured enlarged nuclei, making them distinct from leaf cells.
The exogenous gene NAC96 was successfully overexpressed in both leaf and endosperm protoplasts using PEG-mediated transformation, enabling gene function verification and expression regulation research.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press on behalf of Hainan University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Guo Q, Wang Y, Zou J, Jing H, Li D. 2023. Efficient isolation and transformation of protoplasts in coconut endosperm and leaves for gene function studies. Tropical Plants 2:16 doi: 10.48130/TP-2023-0016
Efficient isolation and transformation of protoplasts in coconut endosperm and leaves for gene function studies
- Received: 20 April 2023
- Accepted: 01 September 2023
- Published online: 07 October 2023
Abstract: To establish a rapid and efficient method for isolating and transforming protoplasts in coconut endosperm, coconut liquid endosperm and young leaves were utilized as starting materials. By comparing factors such as enzyme digestion time, quantity, activity, cell and nucleus morphology, ploidy, and subcellular localization, it was discovered that the optimal conditions for achieving the highest protoplast yield and activity were 45 min of enzyme digestion for liquid endosperm and 2.5 h for leaves. Furthermore, the endosperm cells were found to be 2−3 times larger than leaf cells and exhibited triploid characteristics. Moreover, DAPI staining and subcellular localization revealed the presence of significantly enlarged nuclei in protoplasts from coconut liquid endosperm, occupying nearly the entire cell. Using PEG-mediated transformation, the exogenous gene NAC96 was successfully overexpressed in protoplasts derived from both coconut leaves and endosperm. This study has established an efficient method for isolating and transforming protoplasts in coconut while identifying the distinct characteristics of protoplasts in liquid endosperm. These findings provide a crucial experimental platform for verifying coconut gene function and exploring gene expression regulation.
-
Key words:
- Isolation /
- Transformation /
- Protoplasts /
- Coconuts /
- Endosperms