-
Vegetables are important food sources for humans, providing fiber, vitamins, carotenes and other nutrients[1]. However, due to limited arable land, many vegetables are cultivated in unfavorable conditions, such as drought and salinity. In addition, some vegetables are repeatedly grown in the same areas for several years in succession, which often causes serious soil-borne diseases. These stresses significantly reduce the yield and the quality of vegetables[2].
AM (arbuscular mycorrhizal) symbiosis is an important way to enhance the stress tolerance in host plants. More than 72% of terrestrial plants, including many vegetables, can form AM symbiosis with AMF (AM fungi), which belongs to the sub-phylum Glomeromycota[3]. Modern agricultural activities often cause serious environmental pollution partly because farmers tend to over-apply fertilizers to achieve high yield. The application of AM fungi is a good strategy to improve the agricultural environment and ecology during vegetable cultivation, and in particular it has positive effects on vegetable growth under low Pi condition as summarized by two previous reviews[4,5]. However, establishment of AM symbiosis is not always successful in plants and it usually inhibited by high Pi (phosphate) conditions[6−13]. In addition to the optimization of AM fungi application in vegetable cultivation, we can breed new vegetable varieties with stronger ability to establish AM symbiosis. To this end, understanding the molecular mechanism underlying AM symbiosis in different vegetables, as well as in various conditions, will be very helpful. In recent years, the molecular mechanism of AM symbiosis in vegetable studies has emerged, and here we review the important progress in the molecular mechanism of AM symbiosis, with special focus on its application in vegetable growth.
-
The extraradical hyphae of AMF can extend the root surface by more than 10 cm, forming an extensive absorbing network beyond the rhizosphere nutrient-depletion zones around roots[14,15]. This allows host plants to access a larger volume of soil than the roots without AMF colonization. The extraradical hyphae of AMF can transfer many elements, such as nitrogen (N), phosphorus (P), sulfur (S) and potassium (K), and H2O, from soil far away to the host roots, which greatly benefits the plant growth (Fig. 1)[3]. It was reported that AMF colonization promoted the accumulation of biomass of Allium L. genus, such as onion and leek[16]. This study showed that the total biomass of Allium cepa inoculated with R. intraradices was at least twice as much as the uninoculated plants[17]. Interestingly, the Allium cepa inoculated with the mixed AMF (F. monosporus, R. clarum, D. nigra and A. laevis) (Table 1) exhibited increased dry weight of shoots, roots and bulbs by 84%, 110%, and 41.6% respectively[18]. However, the promotion effect by AMF varies in different species of Allium cepa. The fresh weight of Allium cepa cv. Karmen inoculated with R. irregularis was just slightly increased compared with Allium cepa cv. Kuba, Sochaczewska and Wolska, although Allium cepa cv. Karmen could also be well colonized by AMF[19]. The total dry weight of Allium fistulosum inoculated with R. intraradices (Table 1) was increased by nearly 150%[17] and the Allium ampeloprasum var. Porrum inoculated with R. irregularis (Table 1) showed an increased biomass of at least 100%[20]. These results suggest that the establishment of AMF can help to achieve the increased yield in Allium L genus without further fertilizer application.
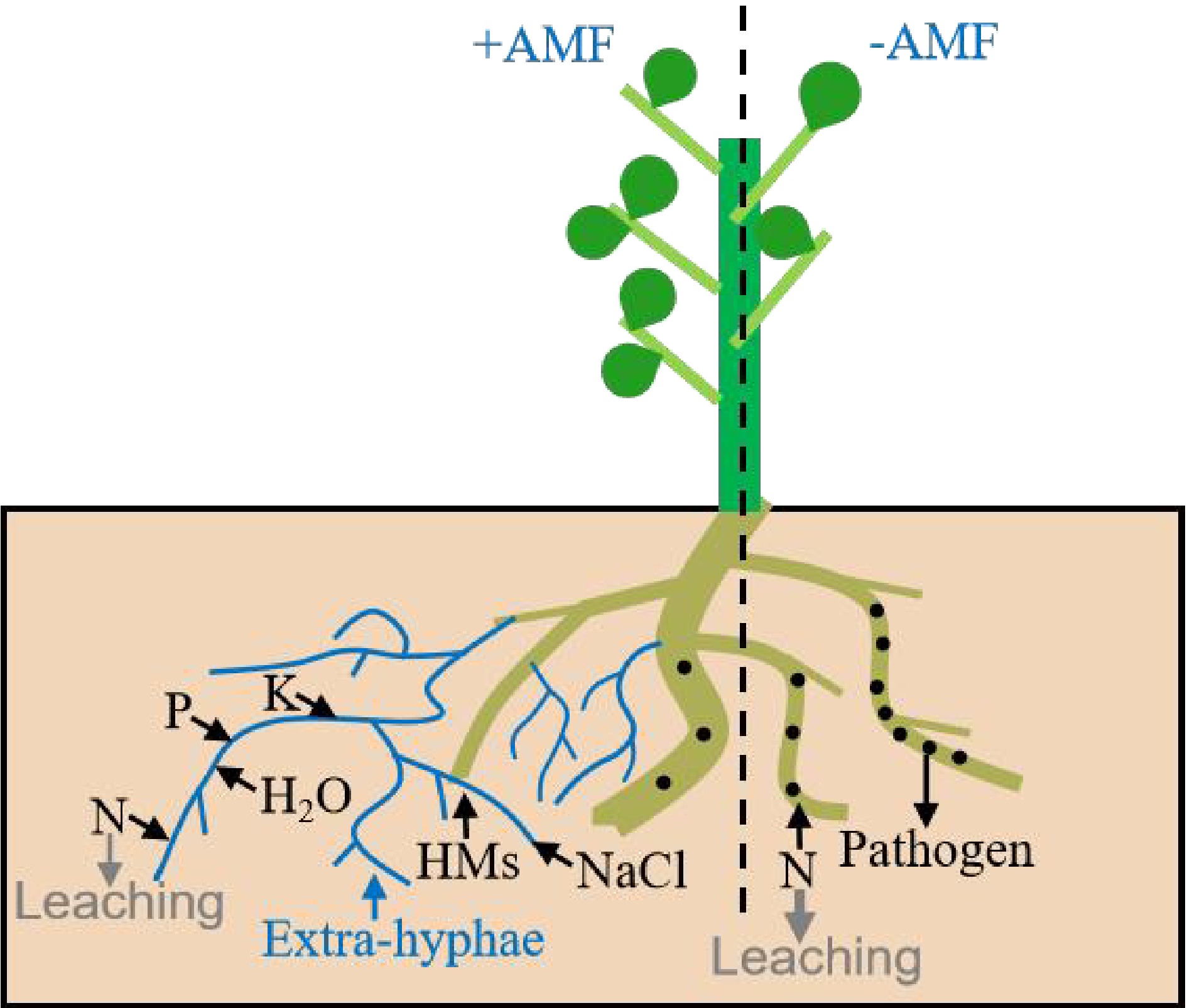
Figure 1.
The formation of AM symbiosis improves the stress tolerance of host plants. The phosphorus (P), nitrogen (N), potassium (K) and other essential nutrients in soil are transferred to the host plants via extraradical hyphae to increase the quality and yield of host plants. The formation of AM symbiosis also enhances the defense of the host plants against various pathogens and the tolerance to HMs (heavy metals) and salt stresses. The thickness of the gray arrow represents the amount of N leaching.
Abbreviated name Full name Number of spore walls Mycorrhizal structures stained
by trypan blueA. laevis Acaulospora laevis 3 V, A, H C. claroideum Claroideoglomus claroideum 1 V, A, H C. etunicatum Claroideoglomus etunicatum 1 V, A, H D. nigra Dentiscutata nigra 3 A, H F. monosporus Funneliformis monosporus 1 V, A, H F. mosseae Funneliformis mosseae 1 V, A, H G. albida Gigaspora albida 1 A, H R. clarum Rhizoglomus clarum 1 V, A, H R. intraradices Rhizoglomus intraradices 1 V, A, H R. irregularis Rhizophagus irregularis 1 V, A, H S. sinuosa Sclerocystis sinuosa 1 V, A, H V, vesicles; A, arbuscules; H, hyphae. The formation of AM symbiosis is significantly inhibited in the plants supplied with high Pi[6−13]. Therefore, most studies examining the promotion effect by AMF were performed under low Pi conditions, which considerably limits the application of AMF in agriculture[5,21]. Interestingly, field trials showed that potato inoculated with R. intraradices could increase yield by 9.5% under conventional agricultural conditions, including the application of pesticides and fertilizers[22,23]. In addition, Solanum lycopersicum cv. Brioso and var. MicroTom inoculated with R. irregularis were also shown to gain the increased yield and enhanced fruit quality under sufficient Pi condition[24,25].
Drought and soil salinity are the frequently occurred stresses that limit crop production in agriculture[26]. The tolerance of host plants against drought and soil salinity can be greatly enhanced by AMF (Fig. 1)[21]. The mixture of AM-fungal spores (R. intraradices, C. etunicatum, F. mosseae, F. geosporum, and C. claroideum) (Table 1) was still shown to increase the tolerance of Solanum lycopersicum cv. Syta seedlings to salt-stress under sufficient Pi conditions[27]. In addition, the inoculation with AMF can boost the plant defense responses to pathogen and disease under low Pi condition (Fig. 1)[20]. Under sufficient Pi condition, the inoculation with S. sinuosa and G. albida (Table 1) enables Phaseolus vulgaris cv. French bean survive the root-rot incidence, resulting in the yield increase by nearly 100%[28]. In Capsicum annuum cv. Charliston Bagci addition, the inoculation with F. mosseae and C. etunicatum (Table 1) alleviates the symptoms of Phytophthora blight[29].
Usually crops can only take up half of the nutrients in the applied chemical fertilizers, while the other half are leached into ground water and surface water, leading to eutrophication and loss of biodiversity[30]. N, a major nutrient in fertilizers, can also be lost from soil as a potent greenhouse gas, including N2O (nitrous oxide) and N2 (dinitrogen gas)[31−34]. Interestingly, the inoculation with R. irregularis in Solanum lycopersicum cv. 76R decreased losses of
$\text{NO}_3^- $ Heavy metal pollution is a worldwide problem. AMF has been emerging as a cost-effective and environmently friendly strategy to assist alleviation of heavy metal phytotoxicity[36]. The previous study showed that Cajanus cajan cv. Millsp inoculated with F. mosseae significantly decreased Cd uptake (by 20%) in the root[37]. This result indicates the potential value of AMF for cultivating vegetables in soil slightly polluted by heavy metal. In addition to the alleviation of heavy metal phytotoxicity, AMF can also enhance the plant tolerance to cold, which has been well summarized in a previous review[5].
-
SLs (Strigolactones) are a group of carotenoid-derived compounds synthesized in plant roots and secreted into the rhizosphere where SLs activate mitochondria and energy metabolism of AMF to promote spore germination and prime the branching of extraradical hyphae[38]. Pea shoots branching mutants (rms (RAMOSUS)1-6) were obtained many years ago by ethylmethanesulfonate mutagenesis[39], but until recently, researchers confirmed the causive genes in the mutants (rms1-5) are involved in the biosynthesis and perception of SLs and the reduced AM symbiosis was observed in these mutants (Fig. 2a, Table 2)[40−43]. In tomato, the function of SlCCD7 (Carotenoid Cleavage Dioxygenase 7), the enzyme involved in the biosynthesis of SLs, is essential for the formation of AM symbiosis[44]. Other genes that affect SLs pathway, such as SlIAA7 was also shown to regulate AM symbiosis (Fig. 2a, Table 2)[45].
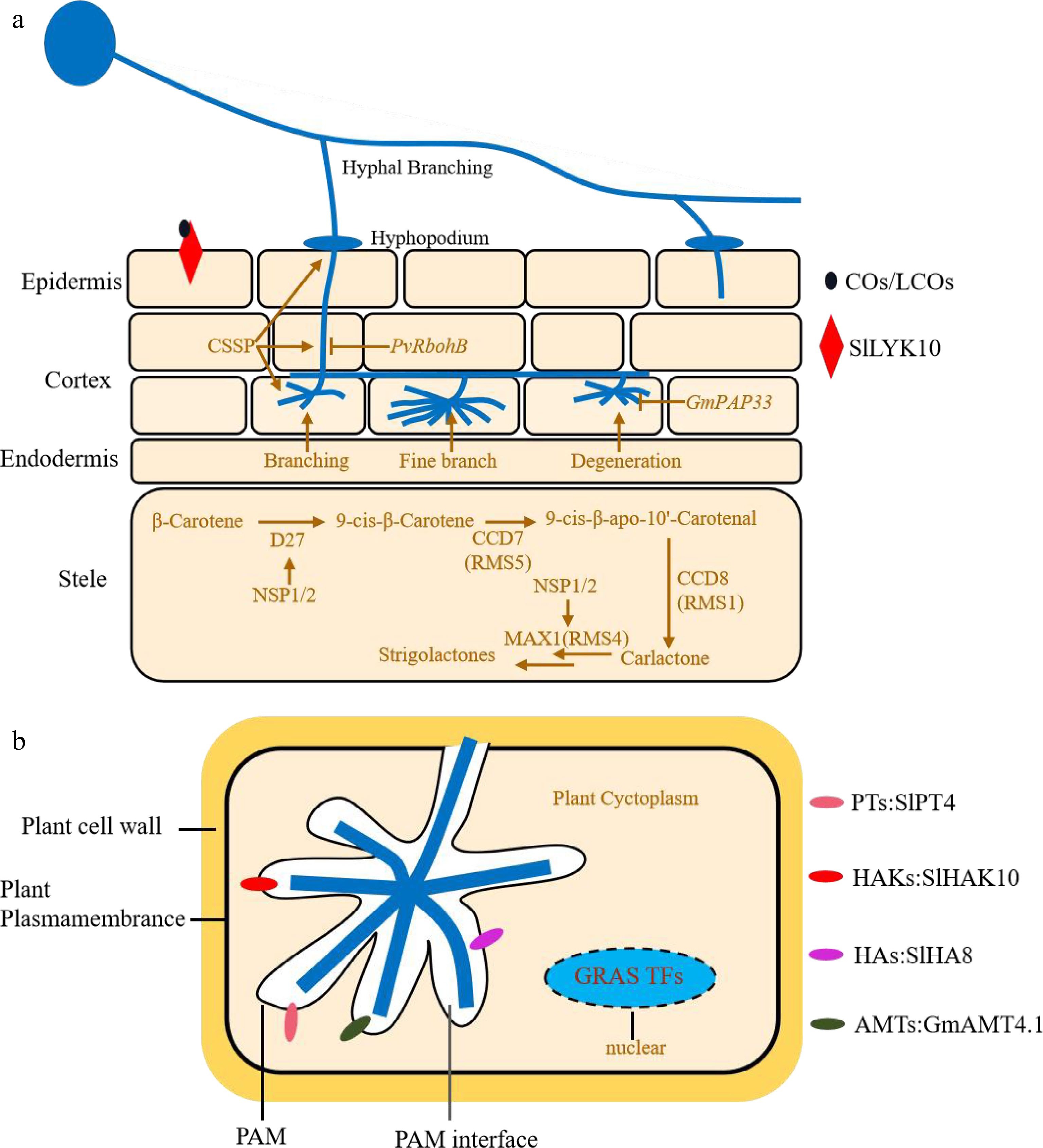
Figure 2.
The molecular mechanism of AM symbiosis in vegetables. (a) RMS1/4/5 are involved in biosynthesis of SLs that promote the spore germination and prime the branching of extraradical hyphae. SlLYK10 binds to COs/LCOs that are secreted by AMF to regulate AM symbiosis. CSSP is required for the early invasion of intra-hyphae and branching of arbuscule. PvRbohB negatively affects the early invasion of intra-hyphae by controlling ROS levels in beans. GmPAP33 negatively regulates the arbuscule degeneration by promoting the hydrolysion of phosphatidylcholine and phosphatidic acid. (b) Many proteins located on PAM, such as SlPT4, SlHA8, SlHAK10 and GmAMT4.1, are required for the exchange of the nutrients and signals between AM and host plants. In addition, GRAS proteins, such as DELLA, RAM1 and RAD1, can regulate the formation of arbuscule.
Table 2. The genes involved in the formation of AM symbiosis in different crop species.
Gene Organism Gene function Mutant phenotype involved in AM symbiosis Reference RMS1 Pea Required for the biosynthesis of SLs Reduced the colonization levels of AM symbiosis [40] RMS4 Pea Required for the biosynthesis of SLs Reduced the colonization levels of AM symbiosis [41] RMS5 Pea Required for the biosynthesis of SLs Reduced the colonization levels of AM symbiosis [40] NA Pea Required for the biosynthesis of GAs Increased the colonization levels of AM symbiosis [46] SlCCD7 Tomato Required for the biosynthesis of SLs Reduced the colonization levels of AM symbiosis [44] SlIAA7 Tomato Required for the biosynthesis of SLs Reduced the colonization levels of AM symbiosis [45] SlLYK10 Tomato Required for the perception of LCOs Reduced the colonization levels of AM symbiosis [48] SlLYK12 Tomato Required for the perception of LCOs Reduced the colonization levels of AM symbiosis [49] SlCCaMK Tomato Required for the calcium ion spiking in the nuclear Reduced the early invasion of intra-hyphae and the branching of arbuscule [48] SlCYCLOPS Tomato Required for the induction of RAM1 Reduced the early invasion of intra-hyphae and the branching of arbuscule [51] GmSYMRKα/β Soybean Required for the calcium ion spiking in the nuclear Reduced the early invasion of intra-hyphae [52] PvRbohB Bean Required for the production of ROS Increased the early invasion of intra-hyphae [53] CRY/LA
(DELLA)Pea Required for the induction of RAM1 and interacted with MYB1 Inhibited the branching and the degeneration of arbuscule [46] SlDLK2 Tomato Remains unknown Increased the colonization levels of AM symbiosis [60] SlPT4 Tomato Required for the transporting of phosphate Reduced the colonization levels of AM symbiosis [54] SlHA8 Tomato Required for the generation of H+ gradient Inhibited the branching of arbuscule [57] SlHAK10 Tomato Required for the transporting of potassium Reduced the colonization levels of AM symbiosis [56] TSB Tomato Microtubules-associated gene Reduced the colonization levels of AM symbiosis [62] GmPAP33 Soybean Required for the promotion of hydrolysis of phosphatidylcholine and phosphatidic acid Increased the percentage of small arbuscule [63] In contrast to the rms1-5 mutants in pea, the Gibberellin acids (GAs)-deficient pea mutant (na-1) showed a significant increase of AM symbiosis (Table 2), suggesting GAs have a negative effect on AM symbiosis[46]. This GA-mediated negative effect on AM symbiosis was also confirmed in other species, including rice, Medicago truncatula and Lotus japonicus[37]. Besides SLs and GAs, other phytohormones can also affect AM symbiosis and their roles in AM symbiosis have been previously reviewed[46].
AMF can produce short chain chitooligosaccharides (COs) and lipo-chitooligosaccharides (LCOs), which are bound by LYSIN MOTIF RECEPTOR KINASE (LYK) to activate Ca2+ spiking and initiate AM symbiosis[47]. In tomato, SlLYK10 and SlLYK12 are required for AM symbiosis and SlLYK10 can bind to LCOs (Fig. 2a, Table 2)[48−50]. Besides the perception of signals from AMF, the common symbiosis signaling pathway (CSSP) is activated in the host plants to initiate the formation of AM symbiosis and nodulation in legume (Fig. 2a, Table 2)[3]. In tomato, CCaMK (Calcium/Calmodulin-dependent protein Kinase) and CYCLOPS were shown to be indispensible for the early invasion of intra-hyphae (Fig. 2a, Table 2)[48,51]. In soybean, the ancestor of SYMRK (Symbiosis Receptor-like Kinase), also belonging to CSSP, gave rise to two paralogous genes GmSYMRKα and GmSYMRKβ, both of which are required for the early invasion of intra-hyphae (Fig. 2a, Table 2)[52]. In addition, the influencing factors also include ROS (reactive oxygen species) as PvRbohB has been shown to negatively regulate the early invasion of intra-hyphae by affecting ROS levels in beans (Fig. 2a, Table 2)[53].
The intra-hyphae enter into the cortical cells of the host plants to form arbuscule, which is then enveloped by the cell membrane called PAM (Periarbuscular Membrane) (Fig. 2b)[3]. Many PAM locating proteins, such as SlPT4 (Phosphate Transporter 4), SlHA8 (H+-ATPase 8), SlHAK10 (High-affinity K+ Transporter 10) and GmAMT4.1 (Ammonium Transporter 4.1), are required for the exchange of the nutrients and signals and AM symbiosis (Fig. 2b, Table 2)[54−57]. The arbuscule is also regulated by many TFs (Transcriptional Factors), such as RAM1 (Reduced Arbuscular Mycorrhiza 1) and RAD1 (Required for Arbuscule Development 1), both of which belong to GRAS family[57]. The loss-of-function of DELLA, another GRAS family gene, caused impaired AM symbiosis in pea[46]. The follow-up studies further showed that DELLA interacted with CYCLOPS to enhance the expression of RAM1[58]. Interestingly, SlDELLA was also shown to interact with SlDLK2 (DWARF14-Like 2), a negative regulator of AM symbiosis, but the function of SlDLK2-SlDELLA complex is unclear yet (Table 2)[59]. The study in tomato suggested that many GRAS family TFs function redundantly to regulate AM symbiosis (Fig. 2b, Table 2)[60].
The formation of arbuscule is also accompanied by the morphological and physiological changes of the arbuscule-containing cortical cells, partly due to the affected cytoskeletons[47]. A recent study showed that tsb (tomato similar to SB401) gene clustered into the same group as microtubules-associated proteins of Solanaceae species, providing further evidence supporting the roles of cytoskeletons in the arbuscule formation (Table 2)[61]. The arbuscule can usually survive for 7−14 d before the final degeneration, which is accompanied by the gradual disappearance of the PAM and matrix[58]. Previous studies showed that MYB1 induced the expression of hydrolase genes by forming a complex with GRAS family transcription factors DELLA and NSP1, and GmPAP33 (Purple Acid Phosphatase 33) inhibited the arbuscule degeneration by promoting the hydrolysis of phosphatidylcholine and phosphatidic acid (Fig. 2b, Table 2)[62,63].
The formation of AM symbiosis is influenced by environmental conditions. Previous studies showed that high N and P both inhibited the formation of AM symbiosis[6−13, 64−68]. Recently, a study in tomato found that the FAB (fasciated and branched) and FIN (fasciated inflorescence), homologous genes to PsNARK (Nodule Autoregulation Receptor Kinase) and PsRDN1 (ROOT DETERMINED NODULATION 1) that are involved in autoregulation of nodulation, were required for the suppression of AM symbiosis by high N[68]. We found that conseved SPX-PHR module regulated the formation of AM symbiosis in response to different phoaphate conditions[69]. SlPHR1/4/10/11/12 were localized in the arbuscule-containing cells to transactivate the expression of AM maker genes. SlSPX1 interacted with them to inhibit their transactivity and negatively regulate the formation of AM symbiosis under medium and replete phoaphate conditions. The elevated atmospheric carbon dioxide (CO2) concentration, which causes problems to the global climate, has increasingly raised concerns. A recent study showed that the rise of atmospheric carbon dioxide could promote the formation of AM symbiosis by triggering the redox-auxin-strigolactone systemic signaling cascade[70].
-
Accumulating evidence has shown that AMF can enhance the tolerance of host plants (including vegetables and crops) under abiotic and biotic stresses. However, the mechanism behind this AMF-mediated stress tolerance is not entirely clear. Microbiome has been emerging as an important way to dissect the underlying mechanism[71]. A few studies showed that AM symbiosis shaped the rhizosphere microbiota and some bacteria such as Streptomyces were beneficial to plants under different stresses[72−74]. In future, recently developed microbiome approaches will drive the progress in understanding whether AM symbiosis recruits the beneficial microbe during the defense against the abiotic and biotic stresses.
Another major issue in the study of AM symbiosis is the effect of external environmental conditions, such as high N and P, which limit the application of AMF in vegetable cultivation[6−13,64−68]. The future studies of the molecular mechanism that limits AM symbiosis under different environmental conditions will benefit the breeding for cultivars with stronger stress resistance. Recently, we uncovered the mechanism behind the inhibition of AM symbiosis by repleted Pi condition in tomato and the key role of SlSPX1 in this process[69]. It is interesting to further examine how AM fungi help Slspx1 mutants to cope with abiotic and biotic stresses under replete Pi conditions, and this will provide the basis for the application of AM fungi in modern agriculture.
The application of AM fungi in vegetables can also benefit from the studies in other crops as many genes involved in the formation of AM symbiosis are conserved in different species. In rice, it has been shown that the natural variation of OsCERK1 enhanced the colonization of AM symbiosis[75]. It is possible to modify the amino acid sequence of such genes, including the homologs of OsCERK1 in vegetables, to enhance the AM colonization in vegetable growth. Technical advancement can also facilitate this purpose. A recent study showed that the expression of the target gene can be up-regulated by a donor-DNA-free CRISPR/Cas-based approach[76], so the specific increase of the expresssion of target genes, such as SlPT4, SlHAK10 and SlHA8 that localize at PAM, to enhance the absorption of N, P and K form AM fungi is theoretically feasible.
This work is supported by the Natural Science Foundation of Fujian Province (2019J01418) and the Prominent Subject Foundation of Fujian Province (712018011) to Dehua Liao.
-
Shuang Wu is the Editorial Board member of journal Vegetable Research. He is blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of this Editorial Board member and his research groups.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Liao D, Sun C, Li S, Tauqeer A, Bian X, et al. 2023. The utilization and molecular mechanism of arbuscular mycorrhizal symbiosis in vegetables. Vegetable Research 3:1 doi: 10.48130/VR-2023-0001
The utilization and molecular mechanism of arbuscular mycorrhizal symbiosis in vegetables
- Received: 25 May 2022
- Accepted: 22 November 2022
- Published online: 17 January 2023
Abstract: Vegetables are an important food and many of them can form AM (arbuscular mycorrhiza) symbiosis with AMF (AM fungi) that belongs to the sub-phylum Glomeromycota. The symbiosis with AM in vegetables enhances their tolerance to various stresses, such as low Pi (phosphate), salinity and soil-borne diseases. In the past decades, the molecular mechanism of AM symbiosis in vegetables has begun to emerge. Here, we review the key studies characterizing the molecular mechanism of AM symbiosis and highlights the huge potential of AMF in vegetable cultivation.
-
Key words:
- Vegetables /
- Utilization /
- Molecular mechanism /
- AM symbiosis












