-
In the current Anthropocene, it is urgent to detect and predict the consequences of ongoing climate change, which is increasingly causing substantial ecosystem imbalances and corresponding biodiversity loss[1]. Given the expected widespread and detrimental effects of rapid climate change, investigating genetic patterns of adaptation and forecasting the magnitude of environmental impacts have been conducted across multiple scales, from individuals to populations, species, and entire communities[2−4]. These efforts have enabled a better understanding of how different species are responding to environmental change and the development of effective strategies for conserving and managing genetic resources.
At the level of single species, signals of local adaptation have been commonly found in plant populations, highlighting the species-wide heterogeneity of adaptive genetic variation[2]. This means that different populations of a single species may have adapted to their local environments in different ways, resulting in unique adaptive divergence history and genomic signatures. Adaptive divergence can result from various factors, including selection imposed by environmental change, restricted gene flow, and genetic drift when subpopulations are small and partially isolated across heterogeneous landscapes, allowing researchers to investigate a variety of ecological and climatic factors that affect neutral and adaptive processes as well as its potentially associated genetic targets[5−8]. Studying adaptive processes and spatial distribution of genetic variation associated with environmental change could contribute to a deep knowledge of species' local adaptation, which is important for elucidating the formation and maintenance of local biodiversity.
In addition, with the current and simulated future climatic data, the adaptation potential of populations under climate change could be evaluated, adding helpful insights into the conservation of endangered plants and informing the management of genetic resources[9]. Compared to main crops and forest trees, however, investigations on the local adaptation of fruit tree across current and future climatic landscapes are still limited[10−12]. Climate change poses critical challenges for fruit production. Fruit are an important component of a healthy human diet. The quality and yield of fruit trees are more directly affected by climate change. The tree-fruit industry is thus particularly vulnerable to climate change. Therefore, a comprehensive understanding of the mechanisms of local adaptation for fruit trees using landscape genomics methods has important significance for the protection and sustainable utilization of fruit tree germplasm. Moreover, fruit trees could be improved for climate tolerance by application of key adaptation genes detected by the methods of landscape genomics. Here, by highlighting the importance of studying local adaptation in fruit trees and introducing the cutting-edge methods (Fig. 1), we call for more efforts to unravel the genomic mechanisms underlying local adaptation and predict the possible maladaptation for fruit trees.
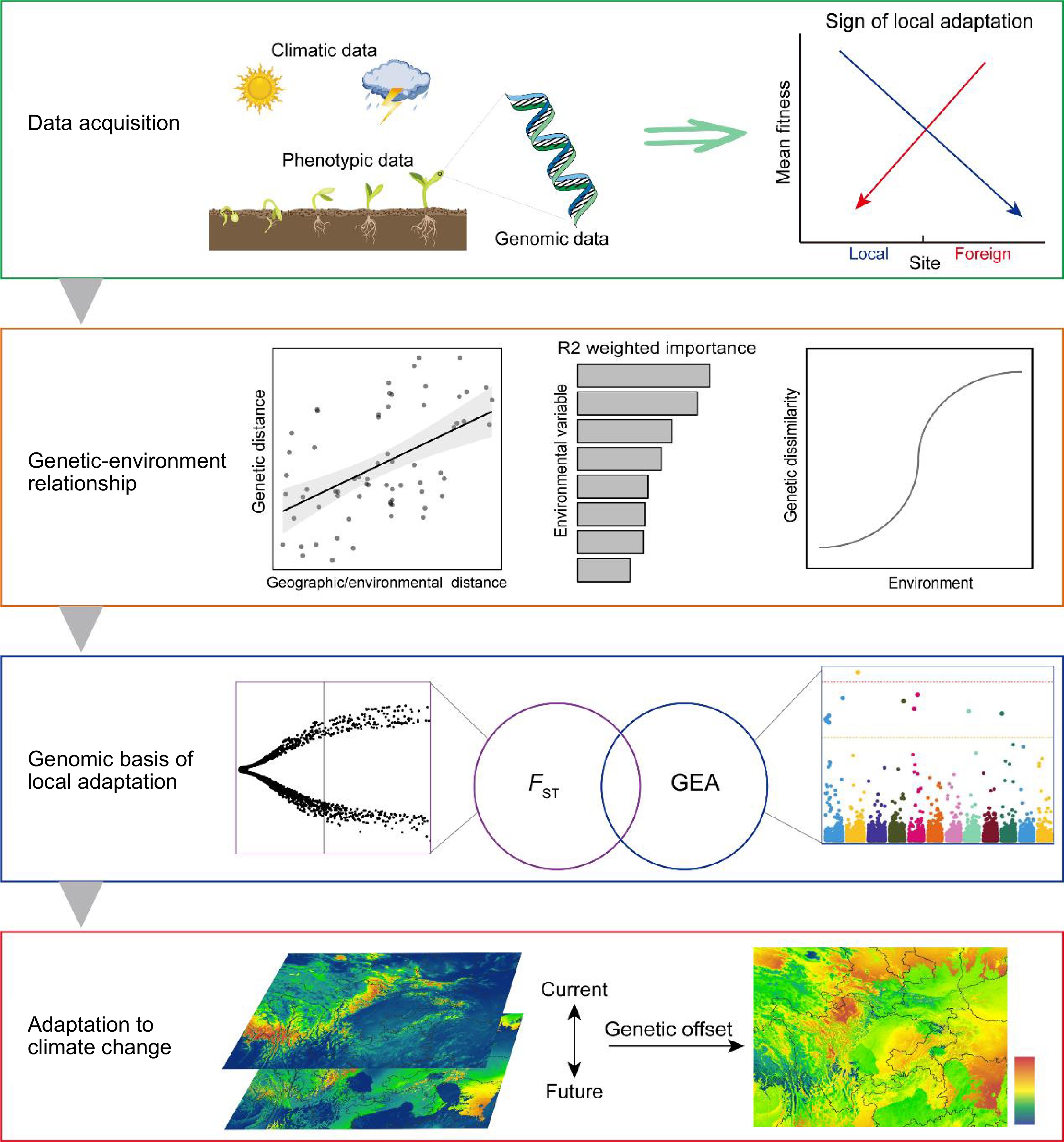
Figure 1.
A summary of performing local adaptation studies. Four steps are provided, including: 1) acquisition of climatic, phenotypic, and genomic data to demonstrate the sign of local adaptation; 2) revealing the association between genomic divergence and environmental variables using Mantel tests, GF modelling and GDM; 3) identifying genomic basis of local adaptation by FST-based and GEA outlier detection methods; and 4) prediction of genomic offset to measure adaptive potential to climate change based on the current genotype–environment relationships. FST is the fixation index quantifying genetic differentiation among subpopulations. GF, gradient forest. GDM, generalized dissimilarity modelling, GEA, genotype-environment association.
-
Local adaptation occurs when species exhibit higher fitness in their local environment compared to individuals from other regions[13]. This is due to natural selection favoring certain traits based on the specific conditions of the environment, leading to ecological divergence among populations and ultimately resulting in speciation. Examples of local adaptation in plants include variations in leaf morphology, root architecture, seed dormancy and germination, flowering time, and allocation of resources to growth and reproduction[4]. Investigating the pattern of local adaptation and revealing the genetic basis of local adaptation are critical steps in addressing fundamental questions related to natural selection and adaptive speciation[7].
Classical studies demonstrating local adaptation have established the use of common garden experiments and reciprocal transplant as effective tools for elucidating the role of local conditions in driving adaptive divergence[14]. Reciprocal transplant and common garden experiments have demonstrated that local adaptation is prevalent, in which observed phenotypic variation among populations from different habitats are heritable or potentially adaptive rather than being plastic in nature, as seen in model plant Arabidopsis thaliana and Mediterranean oaks[15−17]. While these field experiments are effective in identifying traits associated with local adaptation and ecological factors that drive selection, but they require space, time, and resources which may not be feasible for long-lived fruit tree species. Additionally, these field experiments have limitations in providing direct insights into molecular-level evolutionary processes due to their reflection of selection over relatively short periods of time that may not represent their real historical conditions.
The advent of next-generation sequencing technologies has catalyzed a proliferation of population genomics research, thereby substantially augmenting the investigative potential for elucidating the genetic underpinnings of adaptation[4]. Population genomics facilitates the detection of genetic variation such as single nucleotide polymorphisms (SNPs), insertions, deletions and structural variations, that are associated with specific ecological factors or adaptive traits[18]. The availability of genomic data from multiple individuals within a population provides unprecedented opportunities to address fundamental questions in species adaptation and conservation[19]. For example, Zhang et al. revealed the genomic and epigenetic footprint of local adaptation to variable climates in kiwifruit based on the methods of landscape genomics and predicted species vulnerability to climatic change, which has provided appropriate breeding and conservation strategies for kiwifruit[12] .
-
Landscape genomics is a rapidly advancing field that integrates techniques from landscape ecology, population genetics, and community ecology to identify the environmental factors shaping adaptive genetic variation distribution and the underlying genetic materials responsible for local adaptation[20]. This field focuses how landscape elements influence the spatial distribution of genetic variation and correlates environmental variables with genomic regions to understand the genetic basis of local adaptation[21−23]. In the context of climate change, landscape genomics has the potential to not only deepen our understanding of how environmental factors shape genetic diversity and adaptation, but also to assess the magnitude of disruption in genotype-environment relationships that may be triggered by climate change, i.e., genomic offsets or vulnerability[24−27].
Landscape genomic methods enable the examination of correlations between climate variables and genotype frequencies across landscapes, as well as the characterization of distribution patterns of adaptive genetic variation based on the existing or novel genomic resources. As such, it represents a powerful tool for understanding how climate has influenced the geographic genetic structure of populations in the past and for predicting the populations that may be vulnerable to future climate change[28−30]. Landscape genomics allows researchers to address three core questions in evolutionary biology.
Which environmental/spatial variables shape the distribution of adaptive genetic variation?
-
Climate and landscape shifts can have significant impacts on the spatial pattern of adaptive genetic variation in populations[31]. Changes in temperature, precipitation, and other factors (e.g. soil factors, species interactions and competition, etc.) can alter the distribution and abundance of species and their habitats, leading to changes in the selection pressures that shape genetic variation. This, in turn, can affect the spatial distribution of adaptive genetic variants and the genetic diversity of populations.
Population genetics provided an opportunity to unravel how is adaptive genetic variation distributed across time and space by calculating indices of genetic diversity and the degree of genetic isolation, i.e. genetic structure. However, the question of how environmental variables influence patterns of genetic variation across geographic space remains unanswered. Landscape genomic approaches are useful for assessing the relative contributions of geographic distance and environmental variables in shaping population structure, specifically to determine whether spatial patterns of genetic variation are primarily driven by isolation by environment (IBE) or isolation by distance (IBD)[32]. Both the patterns of IBE and IBD suggest that divergent selection is responsible for maintaining population differentiation in the face of possible gene flow, indicating local genetic adaptation. (Partial) Mantel tests have been widely used to reveal the signatures of IBD/IBE on the influence of genetic differentiation[33]. In addition, to enhance the statistical power for quantitative comparison of the extent to which genetic divergence is influenced by IBD vs IBE, both structural equation modeling (SEM) and multiple matrix regression with randomization (MMRR) have been widely employed[34,35].
In landscape genomic researches, multivariate statistical models are more appropriate because they enable the simultaneous analysis of multiple environmental variables and their effects on genetic variation, while accounting for potential correlations among the variables[36]. Redundancy analysis (RDA) and canonical correlation analysis (CCA) are commonly used to quantify the relative importance of environmental and spatial variables[36]. Both methods are based on the concept of linear regression, in which one variable (the dependent variable) is predicted from one or more other variables (the independent or explanatory variables). In addition, recently developed gradient forest (GF)[37], a regression tree-based method and generalized dissimilarity modelling (GDM)[38], a distance-based modelling method, have promising applications in quantifying the role of spatial and environmental variables in structuring genetic variations[20]. These multivariate models can identify patterns of genetic variation that are not easily detected by univariate analyses, which may help to uncover previously unknown ecological drivers of population genetic structure. For example, in an annual alpine herb Circaeaster agrestis (Circaeasteraceae), both GF and RDA results suggested that temperature variations were important drivers of population divergence[29], while in the conifer tree Pinus densata, wet-day frequency (WET) and annual mean UV-B (UVB1) were most strong factors association with genomic variation[39].
What are the genetic bases underlying local adaptation to environmental conditions?
-
One of the principal objectives of landscape genomics is to discern the genome-wide and locus-specific influences on the distribution of genetic variation across landscapes[21]. Two types of genome-scan approaches for detecting molecular underpinnings of adaptation have been widely used: one that detects loci of exceptionally high genetic differentiation among populations (FST-based outlier detection, mining for outliers with high genetic differentiation) and one that searches for regressions between allele frequencies and climatic variables (genotype-environment association methods). FST-based outlier detection takes advantage of differences in allelic frequency differentiation between populations to identify loci that show higher levels of differentiation than expected under neutral evolutionary processes[40]. In contrast, genotype-environment associations (GEA) relate environmental gradients to genome-wide distributions of allele frequencies, searching for genetic signatures of selection as a result of landscape heterogeneity[41].
Commonly used GEA methods include: 1) Statistical approaches based on mixed effect models, such as Bayesian models (BayEnv[41], BayScEnv[42] and BayPass[43]) and latent factor mixed models (LFMM[44]). These methods identify adaptive genetic variations by establishing correlations between population allele frequencies (response variable) and environmental variables (fixed factors), while accounting for population structure (random factor)[45]; 2) Methods based on constrained ordinations, such as RDA and partial RDA (pRDA), which models linear relationships among environmental factors and genetic variation, identifying covarying allele frequencies associated with the multivariate environmental variables[46]; 3) GF modelling, a machine-learning method that can be used to predict the response of individual loci to environmental gradients, accounting for both linear and non-linear relationships[20]. Most studies use more than the above two methods to identify the adaptive loci in order to reduce the false-positive rate[45].
What are the impacts of future climate/landscape shifts on adaptive potential of populations?
-
Once the association between candidate adaptive loci or genomic regions and climatic variables have been identified, these predicted rasters can then be used to model how the frequency of alleles might change across the landscape as temperatures and other environmental factors shift in response to climate change. This allows inferring spatial gradients of adaptation or maladaptation, or the extent to which a given population or species is optimally adapted to its local environment[2]. By predicting how these gradients might shift over time, one can gain insights into the temporal dynamics of adaptation to climate change, which is important for understanding how plant populations and ecosystems might respond to future environmental change and can help inform conservation and management strategies[27].
To predict genomic vulnerability under climate change, GF algorithms and GDM are applied to develop non-linear models that correlate candidate loci with environmental variables. GF algorithms predict how changes in environmental variables will affect the frequency of associated genetic variants. GDM, on the other hand, models how genetic distance between populations changes along environmental gradients. By applying these models, one can estimate the expected genetic distance between present populations and populations that will face different environmental conditions in the future, helping to quantify the extent of genomic vulnerability or risk of maladaptation in different populations[20,24]. For example, using the established genotype-environment relationships and climate-associated loci, GF analysis predicted a highly spatiotemporal shift of Populus koreana in response to future climate change and identified the most vulnerable populations that need conservation priority[26].
-
Studying the local adaptation of fruit tree using landscape genomics comes with several challenges, some of which include:
1. Limited sampling: The first challenge is to ensure that the sampling design is comprehensive and well-planned. Landscape genomics requires large-scale sampling and appropriate statistical power to detect genomic regions or genes associated with local adaptation[45]. This can be difficult as horticultural species often have a limited range and may be subject to human-induced selection pressures. Therefore, it is crucial to collect data from a diverse range of populations so that genetic variation related to adaptation to different environmental conditions can be detected.
2. Gene flow and population structure: Gene flow between populations can limit the evolution of local adaptations[8], and population structure can lead to high levels of genetic differentiation between populations[7]. Both of these factors can complicate the identification of genomic signature involved in local adaptation. In fruit tree, gene flow can occur naturally through pollination or artificial hybridization through human-mediated breeding programs[47]. This means that the population structure and relationship between genetic variation and environmental factors may be more complex than in natural systems. Consequently, modeling approaches should account for the complex and interactive effects of multiple environmental factors as well as potential gene flow.
3. Complex genetic architecture: The adaptive traits involved in local adaptation is often controlled by multiple loci (polygenic adaptation), which can interact with each other, epistatically or additively, and have complex genetic architecture[48]. This complexity can make it challenging to identify the specific genes or genomic regions responsible for local adaptation. Advanced statistical and computational models that integrate information from multiple loci are required to identify candidate loci associated with environmental variables.
4. Gene functional validation: Finally, once candidate genomic regions have been identified, functional validation in the laboratory is essential to confirm their role in adaptation. Despite the increasing popularity of genome-editing as a tool for improving fruit crops, there are still several challenges that need to be addressed in order to ensure its sustainable application[49].
-
In summary, local adaptation of fruit trees to climate is an important yet understudied area of research. With the emergence of landscape genomics, there is now an opportunity to identify candidate adaptive loci and genetic offsets to infer spatial adaptive gradients and their temporal shifts. This information can help to better understand the genomic and molecular underpinnings of local adaptation in fruit trees and to forecast possible maladaptation under rapid climate change. Additionally, these studies can inform breeding programs aimed at developing climate-resilient cultivars and conservation efforts aimed at preserving locally adapted populations. Further research can investigate the specific genes and molecular mechanisms underlying the adaptation to different climatic conditions. This can integrate genome-wide association studies (GWAS), transcriptomics, and epigenetics analyses[50−52]. Moreover, studies can focus on the physiological and morphological traits that contribute to plant adaptation, such as drought tolerance, heat tolerance, and flowering time. Overall, a better understanding of local adaptation of fruit trees can help ensure food security and biodiversity conservation in the context of climate change.
-
The authors confirm contribution to the paper as follows: study conception and design: Yao X, Wang H; draft manuscript preparation: Zhang X; manuscript revision: Jiang Q, Shen Y. All authors reviewed the results and approved the final version of the manuscript.
-
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
This study was funded by the National Natural Science Foundation of China (Grant No. 32070377) and the Key Projects of the Joint Fund of the National Natural Science Foundation of China (Grant No. U1802232).
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Zhang X, Jiang Q, Shen Y, Wang H, Yao X. 2024. Using landscape genomics to assess local adaptation of fruit trees to current and future climatic conditions. Fruit Research 4: e003 doi: 10.48130/frures-0023-0039
Using landscape genomics to assess local adaptation of fruit trees to current and future climatic conditions
- Received: 30 May 2023
- Revised: 22 August 2023
- Accepted: 16 October 2023
- Published online: 02 January 2024
Abstract: Local adaptation has been proven to be common in plants and extensively studied, from increasing plant yields to predicting species responses to the future changing climate. Compared to main crops and forest trees, however, investigations into the local adaptation of fruit trees across current and future climatic landscapes are still lacking. With the explosion of large-scale genomic data, landscape genomics has emerged as a new approach to identify candidate loci that are related to environmental variations (i.e., genotype-environment associations or GEA), while allowing for downstream analyses such as the calculation of adaptive indices and genetic offsets, which can be used to predict spatial-temporal shifts of populations in response to future environmental change. Here, by summarizing the cutting-edge methods for investigating species' local adaptation as well as evaluating the genetic offsets based on the current genotype-environment association, we call for more efforts on elucidating genomic and molecular underpinnings of local adaptation of fruit trees and forecasting the possible maladaptation under rapid climate change. In summary, the study of local adaptation in fruit trees is important for ensuring long-term sustainability and productivity. The emergence of landscape genomics has great potential to advance our understanding of the genomic and molecular mechanisms underlying local adaptation and to predict responses to environmental change.
-
Key words:
- Landscape genomics /
- Local adaptation /
- Climate change /
- Fruit trees.












