-
Carthamus tinctorius L. (safflower, 2n = 24) is the only cultivated species of the Carthamus genus, which belongs to the Carduoideae subfamily of the Asteraceae family[1]. Safflower is a thistle-like annual herbaceous plant with many branches and stems. It grows from 30 to 150 cm in height. At the end of each stem, there is a globular flower capitulum packaged by bracts. As the capitula develops, each capitulum contains 15−30 or more achenes. The four-sided achenes are typically white, and smooth with a thick pericarp[2]. The flower colors among the different safflower genotypes vary from red, orange, yellow and white (Fig. 1).

Figure 1.
Morphological development map of safflower. (a) The whole plant of safflower at the stage of blooming. (b) Main flower colors, red, yellow and white, emerging in different safflower varieties. (c) Main flower developmental stage, including closed buds at stage 1, a few small florets occurring at the center of the capitulum at stage 2, large florets occurr at the center of the capitulum and the ovules are enlarged at stage 3, and florets begin to wilt at stage 4. (d) Seed morphology among the different safflower varieties.
Safflower is a medicinal plant that possibly originated in the Fertile Crescent region more than 4,000 years ago[3,4]. It has been grown for various purposes such as medicine, fabric dye, food coloring, ornamental, biofuel and animal feeding. Safflower is known for its ability to resist drought, salinity, extreme temperatures and hail storms, and can adapt to diverse growth conditions worldwide. Presently, it is cultivated in over 60 countries, including India, Mexico, America, Kazakhstan, Russia, Argentina, China, Turkey, Kyrgyzstan, Australia, Egypt, France, Japan and Pakistan, mainly in arid and semi-arid regions[2]. The geographical distribution of safflower cultivation and production distribution of safflower seeds have been depicted (Fig. 2), which is shown by GBIF[5] and FAOSTAT[6], respectively. Despite the worldwide cultivation, the major production areas for safflower seeds are Asia and America, with a proportion of 90% cumulative production (Fig. 2).
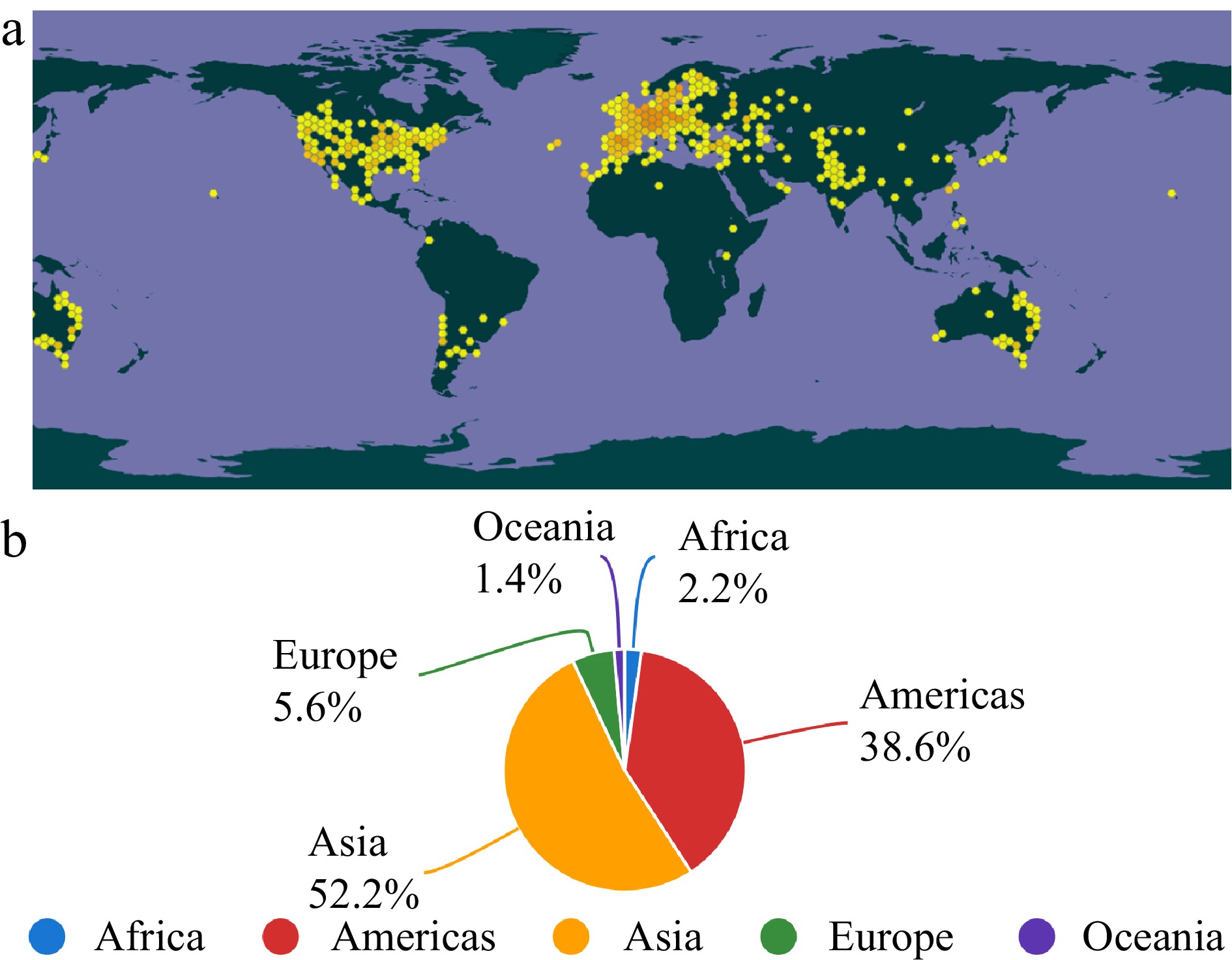
Figure 2.
The geographical distribution of safflower cultivation and production distribution of safflower seeds. (a) Recorded geographical distribution of worldwide cultivated safflower (Carthamus tinctorius L.) from 1999 until 2023. Note: Yellow dots indicate georeferenced occurrences. (b) The production distribution of safflower seeds from 1994 until 2021 recorded in FAOSTAT.
Safflower is known as 'honghua' or red flower in China, 'kusum' in India and Pakistan, 'golrang' in Iran and 'alazor' in Spain. In China, safflower was introduced by Zhang Qian through the Silk Road more than 2000 years ago. Now, it is mainly cultivated in these provinces of China, Xinjiang (Xinhonghua, 新红花), Yunnan (Yunhonghua, 云红花), Sichuan (Chuanhonghua, 川红花), Zhejiang (Duhonghua, 杜红花), Henan (Weihonghua, 卫红花), Anhui, among others[7].
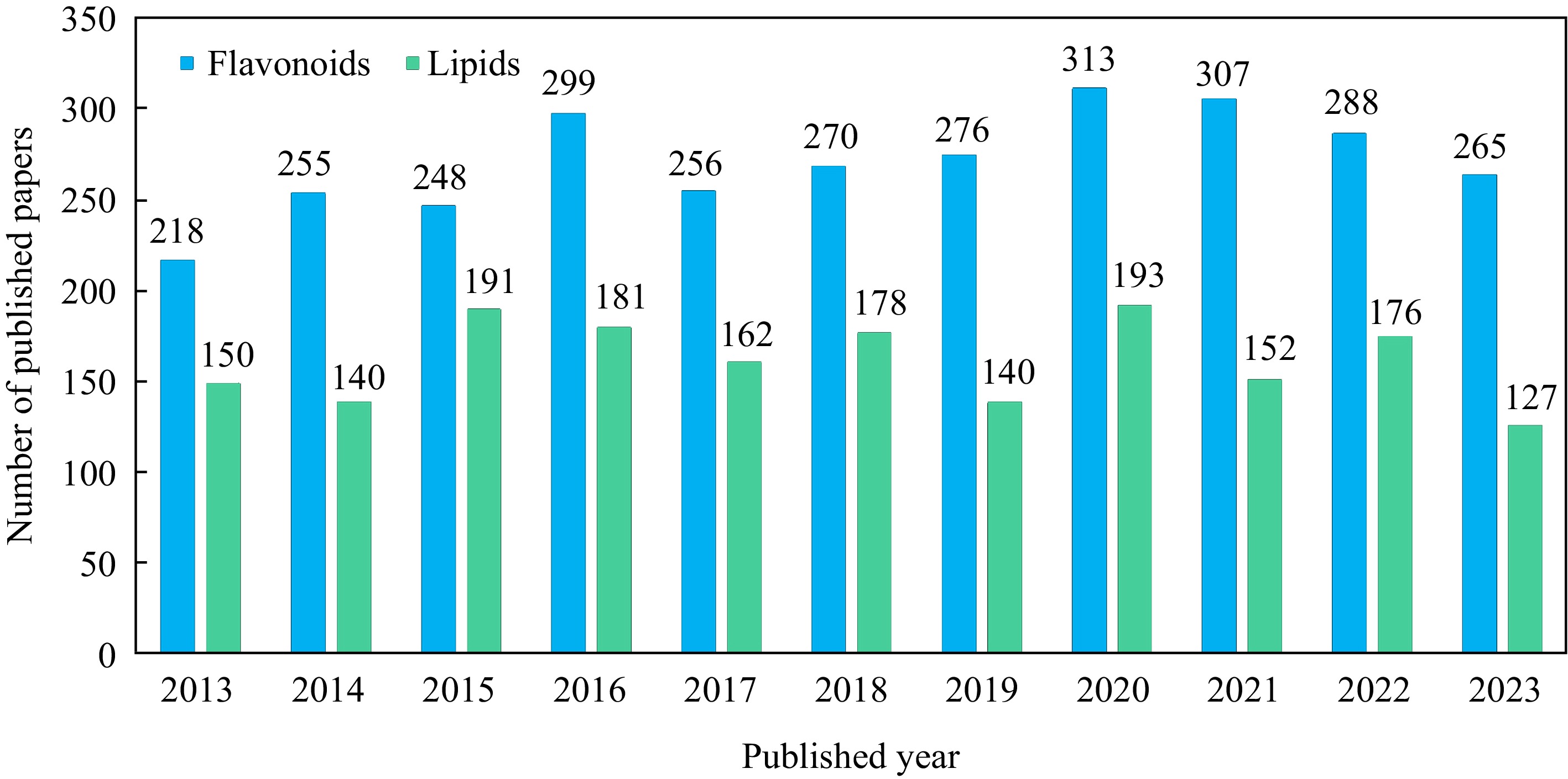
The glycosylated flavonoids of flowers and unsaturated fatty acids of seeds have become two main hotspots in the study of safflower's medicinal and edible values. By the detailed search of CNKI, ScienceDirect, PubMed and Web of Science databases using the combined keywords of 'Carthamus tinctorius', 'safflower', 'flavonoids', 'chalcones', 'HSYA', 'pigments', 'lipids', 'oil', and 'fatty acids', over 100 papers for respective safflower flavonoids and lipids have been published each year (Fig. 3). During the past 10 years, it seems that research on safflower flavonoids has received more attention compared to safflower lipids. 2020 and 2021 were the two years with the highest number of flavonoid research papers, reaching 313 and 307, respectively. Most reviews mainly emphasise the medicinal value of safflower and the pharmacological mechanism of safflower flavonoids in animals[8,9]. Therefore, we focus on review and summary of the application and regulation of safflower flavonoid biosynthesis in this study. This review will provide new insights into the further development and utilisation of safflower flavonoids for human health.
-
Safflower flowers produce two kinds of pigments, safflower red (SR) and safflower yellow (SY) pigments, which are C-glucosylquinochalcones and their derivatives consisting of diverse flavonoids[10]. Natural safflower pigments are still popular in dyes of food products due to their non-carcinogenic and non-allergic characteristics. As one of the main flavonoids from the SR pigment, carthamin can be used for red fiber and cosmetics dyes, while the SY pigment has been increasingly used in pharmacy, food coloring and beverages. Presently in Japan, the SR pigment is usually used for colored chocolate, and the SY pigment is used in colored candy, jelly and juice. In China, both SY and SR pigments are also used in food additives, such as fruit juice, fruit syrup, fruit powder, soda water, and soybean and meat products[2]. As for cosmetics, the SY pigment is used in perfume, shampoo and hair cream, while the SR pigment, known as 'beni' in Japan, is mainly rouge and lipstick bath soap[11].
Natural dye application
-
Safflower has been used as a traditional dye for a long time. Archaeological studies show that safflower in ancient Egypt was mainly used to dye clothes red or yellow[12,13]. In Japan, the safflower SR pigment is used for traditionally dyeing Japanese kimonos[11]. In Turkey, it is also used for coloring of silk, colored cotton and carpet-weaving. New wedding clothes, red flower quilts, red ribbon and rouge have been dyed with the SR pigment in Zhejiang, China for over 400 years. However, the natural dye manufacturing of safflower has almost ceased due to the complex dye processes, which are only prepared on a small scale for national culture, religious occasions and local customs.
Medicine application
-
The flavonoids from safflower flowers are considered to play important roles in dispersing blood stasis and relieving, as well as the activation, of blood circulation and meridians, due to the main active quinochalcone flavonoid (hydroxysafflor yellow A, HSYA) of SY pigment. Therefore, HSYA is considered one of the quality standards of safflower in the Chinese Pharmacopoeia besides flavonol kaempferol[14]. There are more than 20 types of formulae in Chinese Pharmacopoeia 2020, such as decoctions of Xuefu Zhuyu and Taohong Siwu, where safflower is the main ingredient. These safflower-containing formulae are mainly used for prevention and treatment of cardio-cerebrovascular diseases[15,16]. In addition, some curative medicines have also been developed. For example, safflower injection, a flower aqueous extract of safflower, has been widely applied in the protection from cardiovascular and cerebrovascular diseases[17]. Safflower is also used to treat different gynecological diseases. Kangen-karyu, one Chinese prescription containing safflower, is applied for the treatment of symptoms related to blood circulation deficiency[18]. Safflower has also become an indispensable element of Iranian folkore medicine due to its laxative effects in Iran[19]. Safflower is also typically used for mastalgia, scabies, and arthritis in Indian traditional medicine.
-
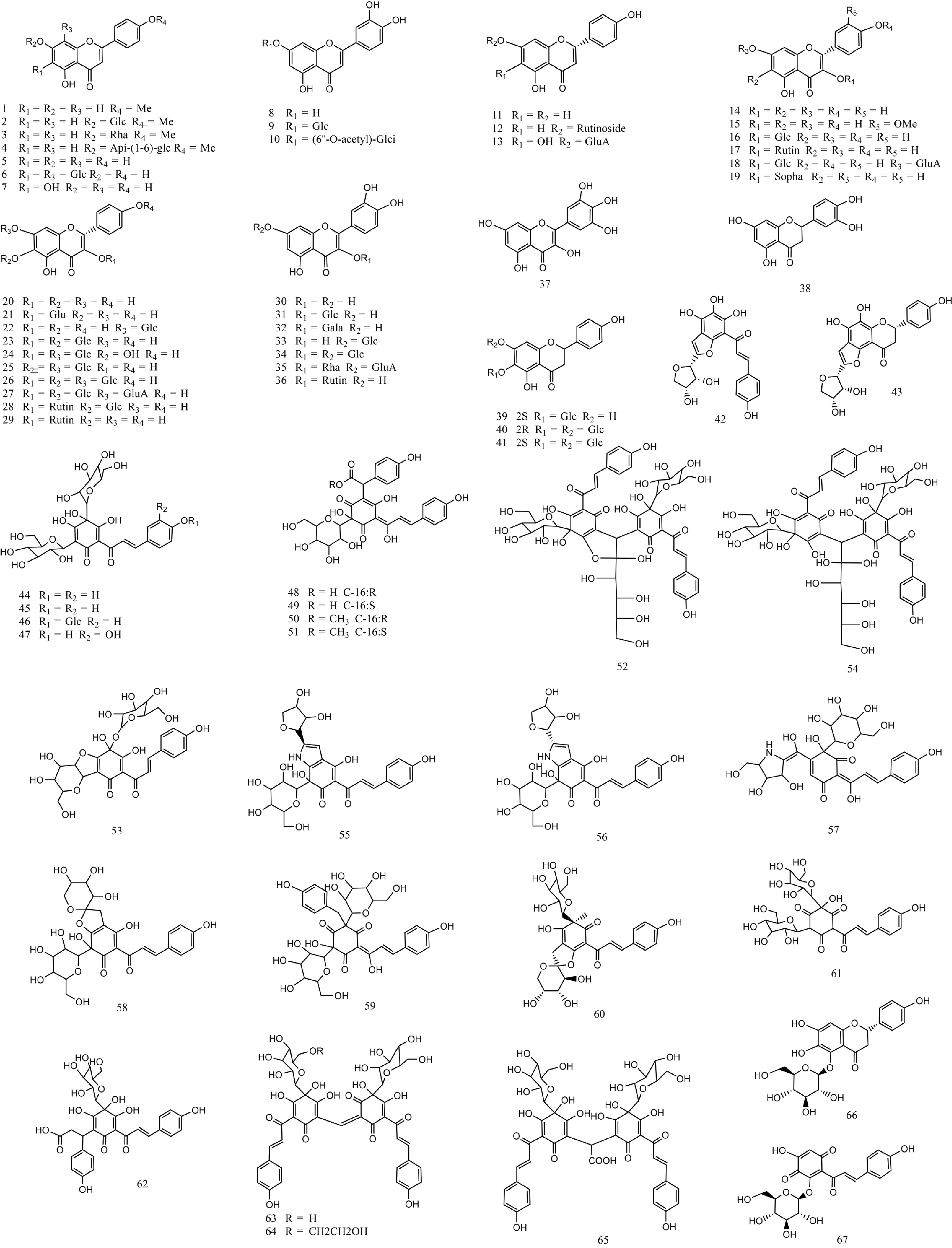
Due to their well-known roles in cardio-cerebrovascular health, safflower flavonoids are considered the main active product among the safflower metabolites. So far, over 60 kinds of flavonoids have been identified and characterized in safflower (Fig. 4)[20]. According to the differential flavonoid components compared with other plants, safflower flavonoids can be divided into two categories: unique and common. The unique flavonoids are only identified in safflower, containing most quinochalcones, such as SR-derived carthamin, YR-derived safflor yellow A, HSYA, safflor yellow B, and anhydrosafflor yellow B (AHSYB). The unique flavonoids with significant pharmacological activity make safflower a popular herbal medicine for treating cardio-cerebrovascular diseases. Most of these unique flavonoids belong to C-glycosides. Therefore, the unique pathway of flavonoid biosynthesis in safflower has long been the focus of biological and medical scientists. In contrast, safflower common flavonoids were also identified in many other species and mainly consist of naringenin, kaempferol, hyperoside, luteolin and quercetin, glycosylation products of which belong to O-glycosides[21].
The main active constituent of medicine in safflower is water-soluble SY pigment (such as HSYA and AHSYB), but the alcoholic extracts from safflower are also used in some preparations. The different cultivars of safflower with red, yellow and white head-like inflorescences own differential flavonoids[22]. The major flavonoid glycosides of the safflower cultivars with three different colors were C-glucosylquinochalcones of HSYA and carthamin for red or orange flowers, flavonols of kaempferol 3-rutionside and 6-hydroxy kaempferol in yellow flowers, flavonols of kaempferol 3-rutionside and kaempferol 3-β-D glucoside in white flowers, respectively.
The main bioactive component-hydroxysafflor yellow A (HSYA)
-
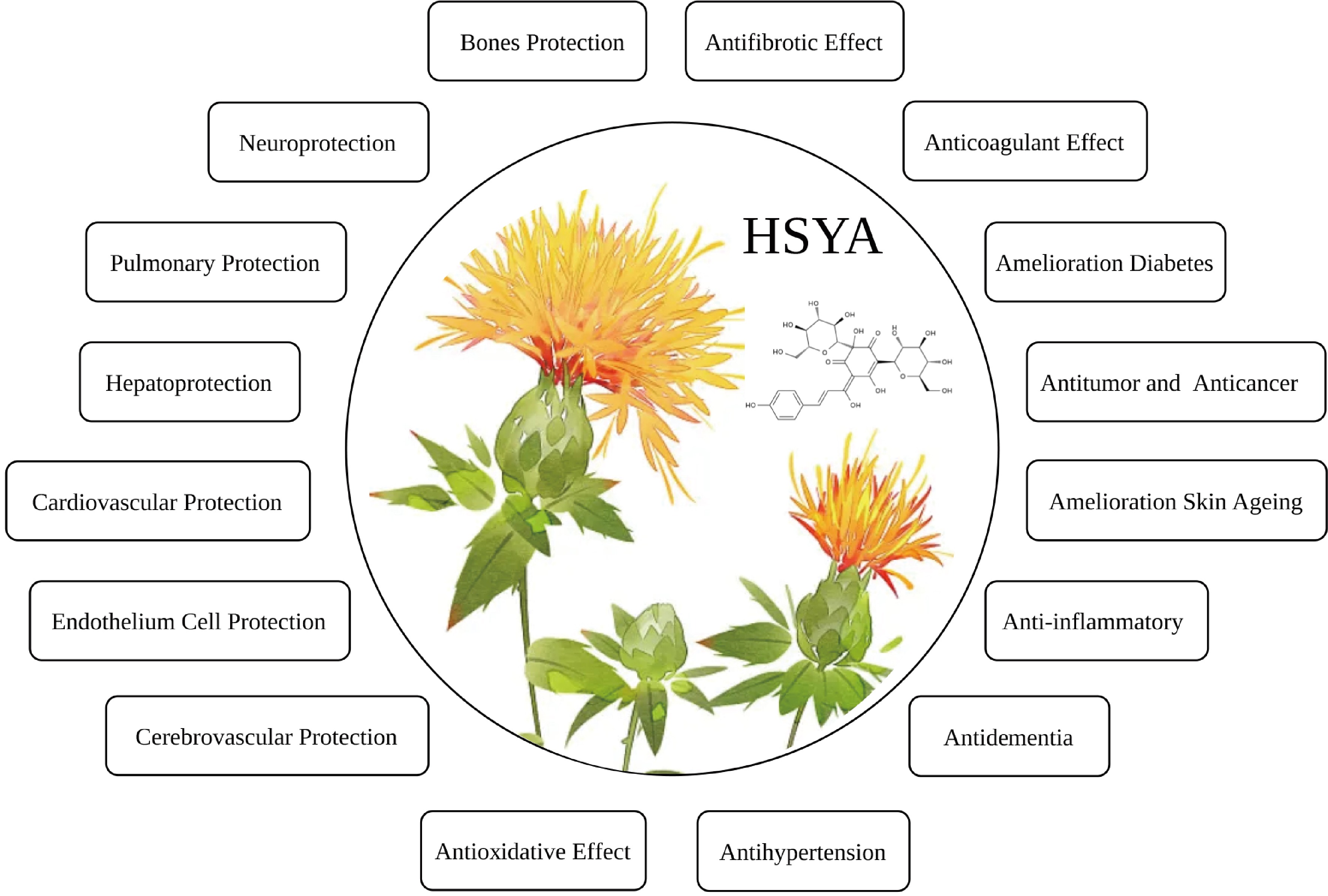
Hydroxysafflor yellow A (HSYA) has a molecular formula of C27H32O16. HSYA will be a promising drug for the treatment of broad-spectrum diseases, including glycolipid disorder, oxidative stress, liver injury, nerve injury, inflammation, cancer, apoptosis, anti-allergy and blood pressure (Fig. 5).
The HSYA content has become the main factor affecting the quality of safflower and an important basis for screening excellent varieties of safflower. At present, HSYA is mainly obtained from safflower accounting for about 1% to 3%. The HSYA content in safflower is affected by various factors, such as genotype, geographical origins, planting environment, color, harvest time and climate. During the flowering period of safflower, the HSYA content shows a trend of increasing first and then decreasing. Usually, HSYA in Chinese cultivars are higher than that in India, Kenya and Turkey. From the perspective of color, the red flower has the highest HSYA content than the orange flower followed by the yellow flower, while the white flower does not contain HSYA[23]. To obtain a high content of HSYA, the morning of the third or fourth day after flowering is considered as the most appropriate time to pick safflower. Currently, the HSYA amount is not enough for clinical applications. In vitro biosynthesis and genetic improvement of safflower varieties are two promising ways to improve the HSYA amount.
-
The main branch of the flavonoid biosynthesis pathway is well-studied and conserved in the plant kingdom[24−26]. At the beginning of the process, phenylalanine ammonia lyase (PAL) catalyzes the first committed step in the general phenylpropanoid pathway, followed by cinnamic acid 4-hydroxylase (C4H), and 4-coumarate: CoA ligase (4CL), resulting in the formation of trans-cinnamic acid, coumaric acid and coumaroyl-CoA in turn. Subsequently, the polyketide synthase (chalcone synthase, CHS) first produces chalcones from three molecules of malonyl-CoA and one molecule of coumaroyl-CoA. Then, chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), dihydroflavonol 4-reductase (DFR) and anthocyanidin synthase (ANS) catalyze the downstream flavonoid biosynthesis, including the (2S)-flavanones, dihydroflavonols, anthocyanidins, respectively. The anthocyanidins are generally glycosylated by UDP-glycosyltransferase (UGT) using UDP-glucose. Besides these enzymes in the conserved main pathway, there are other enzymes involved in the branches of the flavonoid pathway for different species, such as flavone synthase (FNS), isoflavone synthase (IFS), flavanone 3',5'-hydroxylase (F3'5'H), flavonol synthase (FLS), flavanone 4-reductase (FNR), leucoanthocyanidin reductase (LAR) and anthocyanidin reductase (NAR)[26]. In safflower, the unique flavonoids are major C-glucosylquinochalcones (such as HSYA and carthamin), which are derivatives from naringin chalcone further catalyzed by the members of UGT or cytochrome P450 (P450) gene families (Fig. 6).
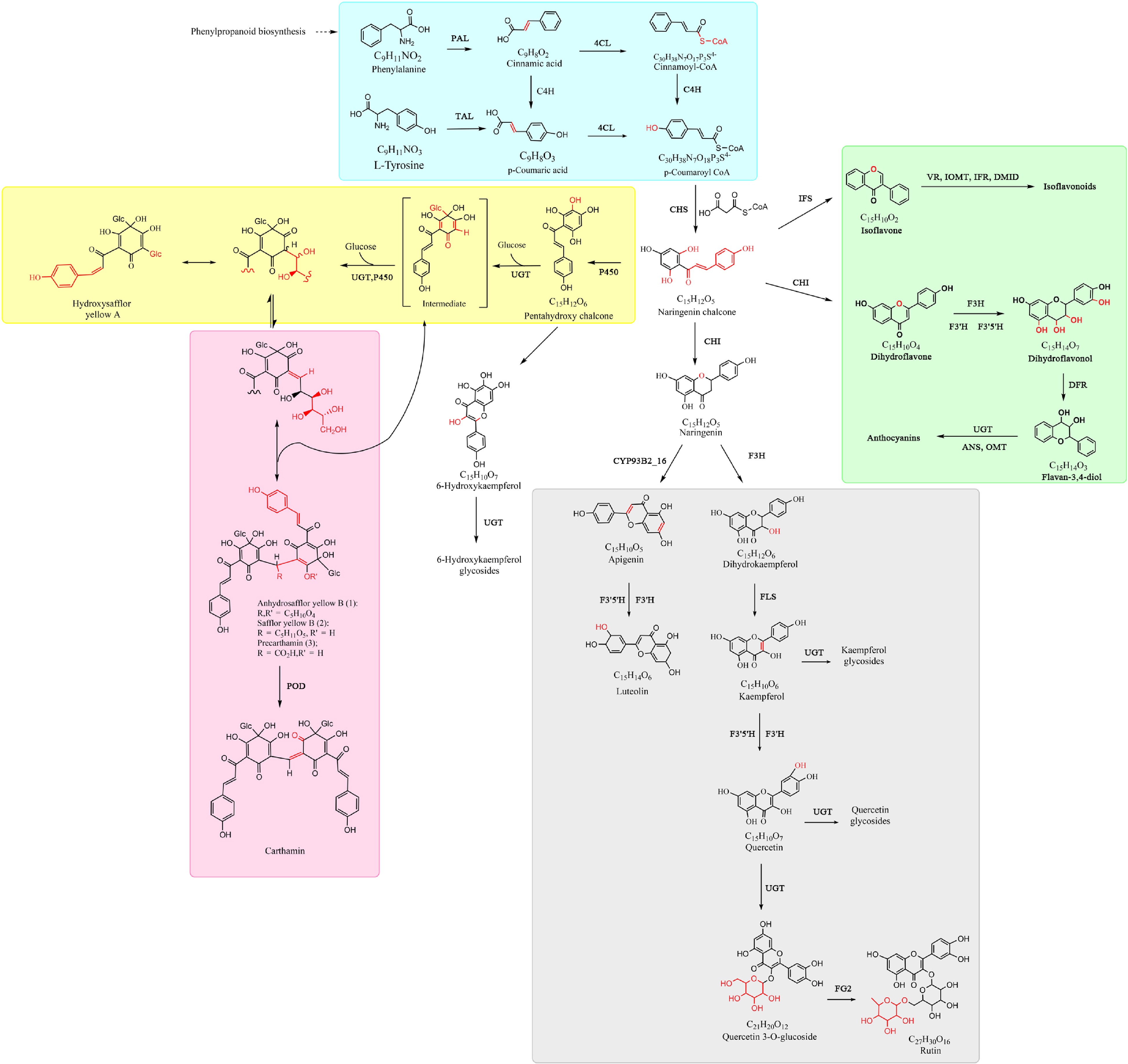
Figure 6.
Proposed pathway of flavonoid biosynthesis in safflower. The yellow-shaded box and red-shaded box indicate the unique flavonoids of safflower yellow and red pigments, respectively. Other color-shaded boxes represent the detected common flavonoids in safflowers and other plants. Note: The enzyme names and flavonoid compounds are abbreviated as follows: PAL, phenylalanine ammonia lyase; TAL, tyrosine ammonia lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, 4-coumarate: CoA ligase; CHS, chalcone synthase; UGT, UDP-glycosyltransferase; P450, cytochrome P450; POD, peroxidase; CYP93B2_16, one P450 member; CHI, chalcone isomerase; IFS, isoflavone synthase; F3H, flavanone 3-hydroxylase; F3'H, flavanone 3'-hydroxylase; F3'5'H, flavanone 3',5'-hydroxylase; FLS, flavonol synthase; VR, vestitone reductase; IOMT, isoflavanone O-methyl transferase; OMT, O-methyl transferases; DMID, 7,2'-dihydroxy-4'-methoxyisoflavanol dehydratase; IFR, isoflavone reductase; ANS, anthocyanidin synthase; DFR, dihydroflavonol 4-reductase; FG2, flavonol-3-O-glucoside L-rhamnosyltransferase.
Synthetic biology has greatly promoted the biosynthesis of natural plant products. As a multi-step enzymatic process, biosynthesis is continuous, effective, low-carbon and environmentally-friendly. In future, the main source of HSYA will depend on biosynthesis. Therefore, clarifying the regulatory mechanisms of the HSYA biosynthetic pathway in safflower is vital, which will mine key functional genes for special flavonoid biosynthesis. Now, genetics and molecular biological technologies, including DNA sequencing, transient expression and genetic transformation, have been applied in the research of safflower flavonoid biosynthesis.
Genetic regulation of safflower flavonoid biosynthesis
-
Association analysis of molecular markers with traits is a powerful tool to identify the important candidate gene loci. Previously, the associated markers of amplified fragment length polymorphisms (AFLP) and cDNA-AFLP in the safflower population showed that HSYA content was regulated by one nuclear gene with two alleles (HSya and hsya). HSya is completely dominant over hsya[27−29]. In addition, two other genes were identified as negatively correlated to HSYA biosynthesis by the cDNA-AFLP approach. One gene was annotated as a small heat shock protein (CTL-hsyapr) in safflower[30], and another was CT-wpr (TDF-11), likely a non-coding RNA[31]. Therefore, both coding and non-coding genes involve in HSYA biosynthesis, regulation of which is more complex than expected. Complete genetic information, containing the genomes of nuclear, chloroplast and mitochondrion, is vital for understanding the genetic mechanism of special traits in a plant species. Wu et al. have established a complete genomic map of safflower via genetic linkage map and genomic sequencing[32−34]. These genomic resources will lay a foundation for the characterization of HSYA-associated functional genes, as well as the development of HSYA-associated high-quality molecular markers.
Regulation of structural genes on safflower flavonoid biosynthesis
-
The biosynthesis of HSYA in SY pigment and carthamin in SR pigment are the main topics in the safflower flavonoid field. The function and evolution of rate-limiting enzyme, CHS in safflower flavonoid biosynthesis have received more attention[33,35−37]. Compared genomic analysis showed that the tandem duplication event and functional divergence of CHS gene family may contribute to HSYA accumulation in safflower[33]. Also, overexpression of CtCHI1 and CtF3H helped to accumulate HSYA, indicating their positive role in HSYA biosynthesis in safflower[38]. To explore the potential regulatory mechanisms of safflower flavonoids, various experiments of transcriptomic and metabolomics have also been performed for flower samples, including different colors[23,39,40], color-transition stages[41], flower development stages[42,43]. Metabolomics analysis showed differential kinds and contents of flavonoids in safflower flowers with different colors. For example, the content of HSYA was higher in red flowers than in orange and yellow flowers, but it was not detected in white flowers. The proposed genes encoding CHS, P450, and UGT are possibly responsible for this difference by integrated multi-omics analysis[23]. As for carthamin, carthamin synthase (CarS), the peroxidase homologs of CtPOD1~3, were reported to be responsible for the carthamin biosynthesis from pre-carthamin[44]. Another report showed that UGT family members were also the potential flavonol glycosyltransferases presenting different functional characterization of flavonoid biosynthesis in yellow and white flowers[45]. Therefore, UGT enzyme is a universal modification for flavonoid glycosylation in safflower, while the speculated P450 and POD enzymes function as oxidases in further addition of oxygen groups in HSYA or carthamin. However, the biosynthetic relationship between HSYA and carthamin in the flavonoid pathway is still unclear.
Regulation of transcription factors on safflower flavonoid biosynthesis
-
Flavonoid biosynthesis in plant-specific tissues and developmental stages is finely regulated by transcription factors (TFs). The MYB and bHLH family TFs are well known for regulating the structural genes in the flavonoid biosynthesis pathway[46,47]. In safflower, the genomic features of MYB and bHLH family members have been identified at the genome-wide level, some of which may be associated with the biosynthesis of safflower flavonoids, such as anthocyanin[48,49]. The MBW protein complexes consist of MYB, bHLH and WDR TFs, and take part in various biological processes, including the initiation of the flavonoid biosynthesis pathway, cell fate determination, vacuolar acidification adjustment, as well as response to environmental factors[50,51]. The identified safflower MBW complexes (CtbHLH41-CtMYB63-CtWD40-6) positively regulate HSYA biosynthesis[52]. Besides the well-known TFs, other regulators like bZIP, WRKY, C2C2 Zing finger, AP2/ERF and HD-zip, have also been implicated in the regulation of flavonoid biosynthesis directly or indirectly[53,54]. Therefore, more kinds of TFs and different members of the same TF should be investigated in safflower flavonoid biosynthesis in future.
Effect of phytohormones on safflower flavonoid biosynthesis
-
Phytohormones play important roles in regulating plant growth, development, and adaptation through the flavonoid biosynthesis of chalcones, flavone, flavonol, anthocyanin, and other flavonoids[55]. In safflower, external Methyl jasmonate (MeJA) was reported to participate in the accumulation of quinochalcones and flavonols (such as carthamin, HSYA, kaempferol-3-O-β-rutinoside, kaempferol-3-O-β-D-glucoside, luteolin, and rutin) via the upregulation of flavanone 3-hydroxylase gene (F3H)[56]. Further MeJA promotes the biosynthesis of quinone chalcones, such as HSYA, possibly by differentially regulating the expression of upstream genes (such as HCTs, CHIs and CHSs) and downstream genes (such as ANRs, ANSs and F3Ms) in the flavonoid biosynthesis pathway, respectively[57]. Safflower F3'5'H may be mediating MeJA-induced flavonoid biosynthesis in Arabidopsis transgenic plants via enhanced anthocyanin accumulation[58]. Overexpression of MeJA-induced CtCYP82G24 in Arabidopsis showed increased expression levels of other key flavonoid biosynthetic genes such as AtDFR, AtANS and AtFLS, and higher levels of flavonoid and anthocyanin accumulation[59]. In addition, CtACO1, encoding one of 1-aminocyclopropane carboxylic acid oxidase family and involving ethylene biosynthesis, also regulates the flavonoid profile in safflower. The overexpression of ACO promotes the biosynthesis of rutin, quercetin and quercetin 3-β-D-glucoside, but inhibits the accumulation of quinochalcones carthamin and HSYA in safflower[60]. However, another number, CtACO3, significantly improved the content of carthamin and HSYA[61]. The diverse functions of ACO homologs indicated the complex regulatory mechanisms of phytohormones in safflower HSYA biosynthesis.
Effect of environmental factors on safflower flavonoid biosynthesis
-
As one kind of secondary metabolite, flavonoid biosynthesis is also widely regulated by various environmental factors, such as temperature[62,63], light[64,65], and nutritional deficiency stress[66], which contribute to the adaptation of plants to complex terrestrial environments. In safflower, there were several reported environmental factors involved in the biosynthesis of both common and unique flavonoids, possibly mediated by regulation on structural genes. Different light intensities appear not to influence the level of total safflower flavonoids, but they significantly regulate the contents of certain flavonoids, including common kaempferol and unique HSYA[67]. For example, high light intensity significantly decreased the HSYA content compared to middle or low light intensity. It was reported that the elongated hypocotyl 5 (CtHY5) transcription factor regulated the biosynthesis of common flavonoids in safflower, such as common flavonoids of rutin and kaempferol, by binding to the promoter of CtCHS1[68]. However, the regulatory mechanism of unique HSYA biosynthesis by light intensity remains unclear. Drought stress also induced the accumulation of total flavonoids in safflower. For example, CtCYP81E8 gene encoding Isoflavone 2'-Hydroxylase positively regulated expression of CHS, F3'H, and DFR genes in flavonoid pathway, and overexpression of CtCYP81E8 in Arabidopsis showed it can promoted the total amount of safflower flavonoids, including kaempferol, naringin, and chalcones[69]. However, the unique flavonoids, such as HSYA, have not been detected in CtCYP81E8-overexpressed Arabidopsis. In addition, CtC4H1 in safflower, encoding one cinnamate 4-hydroxylase, was significantly upregulated in Arabidopsis under drought stress. The overexpressed-CtC4H1 in Arabidopsis improves flavonoid accumulation, accompanied by the enhancement of antioxidant defense capabilities[70]. However, the molecular regulatory mechanism of safflower flavonoids, especially for unique flavonoids, mediated by environmental factors remains unclear. It is well known that safflower has very strong adaptability to drought and saline-alkali stress. Whether the unique flavonoids (such as HSYA) were the adaptive response of safflower to abiotic stress is well worth exploring.
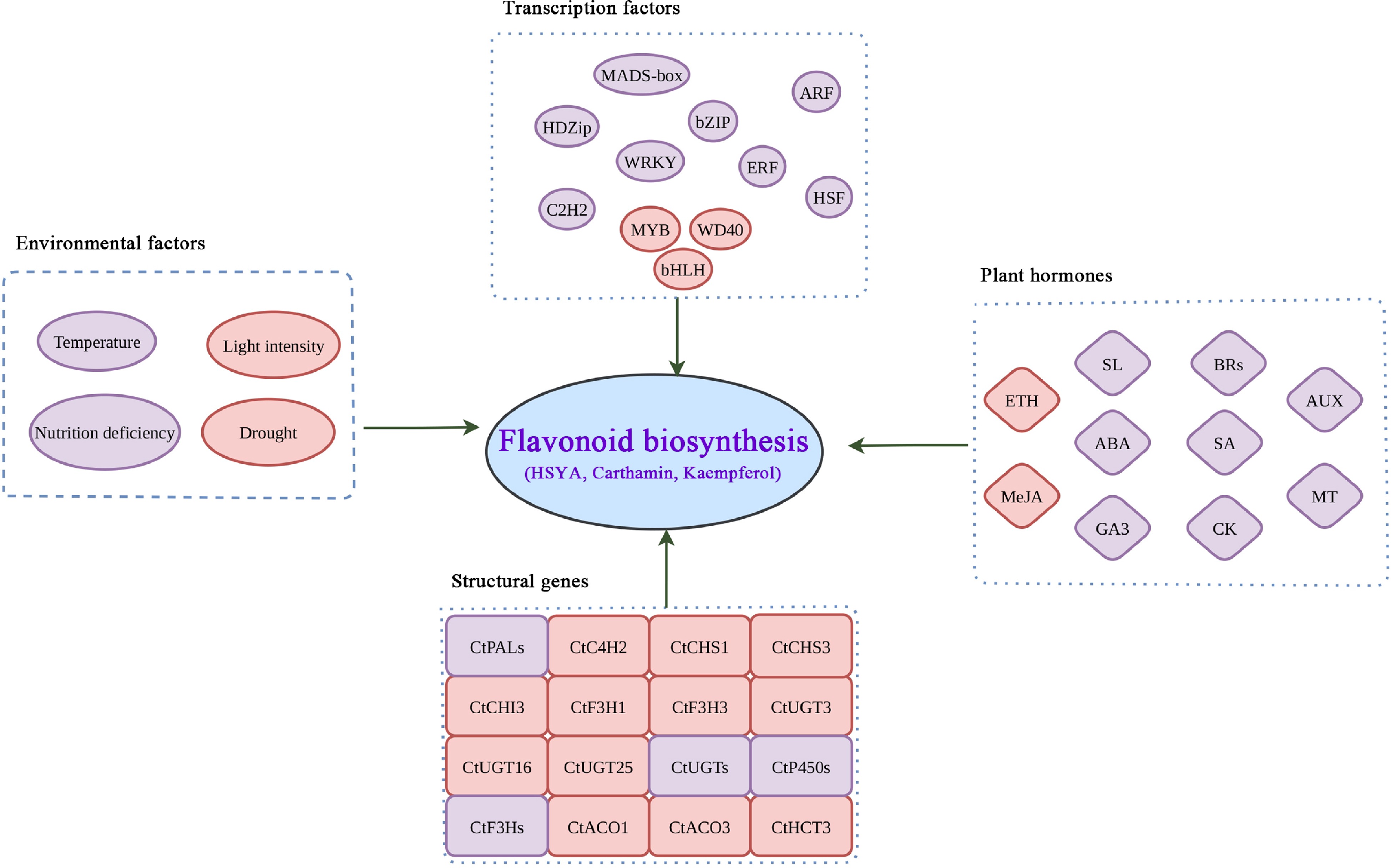
In summary, environmental factors and phytohormones may influence the process via the expression of structural genes or transcription factors, coordinately (Fig. 7). A complete gene regulatory network integrating endogenous and exogenous environmental signals will be helpful to understand the flavonoid biosynthesis in safflower. In future, the roles of phytohormones and environmental factors, as well as their crosstalk should be investigated to reveal their regulatory mechanisms in safflower flavonoid biosynthesis.
-
Safflower has been used for multiple purposes for over 2000 years in China due to its high value in medicine and food. Here, we make a detailed systematic review of the research progress on the application and regulation of flavonoid biosynthesis. Safflower flavonoid biosynthesis is regulated at multiple levels, including genetic information, developmental processes, and environmental factors. How to improve the content of unique flavonoids is a hot topic in safflower improvement. Combining the potential regulation of environmental factors and phytohormones with the advance of molecular biotechnologies, we propose that the following possible strategies, including molecular breeding, synthetic biology and cultivation optimization, which would be helpful to achieve this goal.
Based on genomic sequences and germplasm resources, the development of HSYA-associated molecular markers will provide a foundation for marker assisted selection (MAS) of high-content HSYA varieties. Together with the advance of other omics (such as phenomics, transcriptomics, proteomics, metabolomics and functional genomics), population genomics is becoming a powerful discipline for identifying masses of genes and excellent genotypes in important economic traits. Despite the existence of safflower abundant germplasm resources, safflower genetic improvement has been impeded for a long time due to the lack of genomic information. Along with the recent release of complete genetic information of safflower[32−34], the high-throughput molecular marker of single nucleotide polymorphisms would accelerate construction of core germplasm resources and high-resolution genetic mapping, as well as application of MAS in safflower breeding of high-content flavonoids. In addition, some statistical methods, such as GWAS (genome-wide association study) and TWAS (transcriptome-wide association analyses)[71], are efficient ways to mine candidate genes in crops, and can also be used in safflower for mining flavonoid-biosynthesis genes. Once, the functional genes involved in safflower HSYA biosynthesis are confirmed by molecular technologies, the HSYA-associated markers can be further developed and applied in the breeding of high-content HSYA varieties. Also, the gene-editing tool of CRISPR[72] is highly-efficient for germplasm innovation. Therefore, combination of gene-editing and MAS technologies would greatly prompt the genetic engineering of safflower germplasms for high-content valuable flavonoids in the future.
Mining structural genes responsible for unique flavonoid biosynthesis in safflower will lay a foundation for their highly efficient biosynthesis. For example, a new glycosyltransferase (UGT73AE1) from safflower showed the first reported trifunctional plant glycosyltransferase, demonstrating the functional versatility of the glycosylation for natural bioactive products[73]. Synthetic biology is a promising interdisciplinary field, which combines systems biology with genetic engineering technology to construct artificial biocatalytic systems[74]. It can solve our demands in food and energy production, medicine, and manufacturing[75]. Nowadays, plant natural product synthesis is mainly carried out in either a microbial (such as yeast) or plant chassis (such as suspension cells). For the biosynthesis of plant natural products, the plant chassis usually is more advantageous than microbial in that plants have more similar genetic information and offer a more finely compartmentalized cellular structure[76]. A stable tissue culture system of safflower has also been established[77]. Along with the genetic dissection of the unique flavonoid-biosynthesis pathway, synthetic biology can be applied in the improvement of specific flavonoids in safflower by integration of structural genes into the genomes of microbes and other engineered cells.
In addition, optimizing the proportions of phytohormones and environmental factors during safflower cultivation will contribute to the accumulation of specific safflower flavonoids. Jasmonic Acid (JA) is widely involved in the regulation of plant secondary metabolites and responses to biotic and abiotic stresses. Ethylene can positively affect the anthocyanin biosynthesis in the fruits from Rosaceae plants, such as apple[78], but negatively regulates anthocyanin biosynthesis in some species, such as red Chinese pear fruits[79]. Currently, MeJA and ethylene have been reported to be positively and negatively involved in safflower flavonoid biosynthesis, respectively. Other phytohormones should also be investigated in regulating the contents of unique flavonoids in safflower. For example, exogenous application of Gibberellin, Melatonin and Abscisic Acid (ABA) significantly influences the content of the flavonoid in grape fruit[80,81]. By integrating the light signaling pathway, ABA can regulate colorless flavonol biosynthesis, and vice versa, namely flavonol can also regulate the ABA signaling network[82,83]. Therefore, integrated regulation under multiple phytohormones and environmental factors is important for flavonoid biosynthesis in specific plants. In future, construction and comparison of hierarchically regulatory networks will not only provide insight into the regulatory mechanisms of safflower flavonoid biosynthesis under the interaction of phytohormones and environmental factors, but also lay a basis for improving the contents of safflower unique flavonoids under the application of phytohormones and environmental factors.
In conclusion, although significant progress on safflower flavonoid application and research has been made, there are still important challenges, which limit further exploration and utilization of safflower. Due to the abundance of secondary metabolites such as safflower flavonoids, there are certain difficulties in constructing a stable genetic transformation system. The establishment of a genetic transformation system for safflower will not only help to reveal gene functions in safflower flavonoid biosynthesis, but also provide a reference for other medical plants rich in flavonoids. In addition, the identification of potential enzymes (such as UGT and P450) and their catalytic mechanisms responsible for HSYA biosynthesis have not yet been elucidated in detail. By overcoming these challenges, it would be helpful to efficiently realize biosynthesis of HSYA using microorganisms or safflower as the chassis in future.
-
The authors confirm their contribution to the paper as follows: study conception and design: Wu Z; safflower material provision: Hu X, Hu Z, Jiao P, Zhai J; data collection: Wu Z, Li R, Sun M, Xiao M, Zhang J; analysis and interpretation of results: Wu Z, Li R, Sun M, Pu S; draft manuscript preparation: Wu Z, Li R. All authors approved the final version of the manuscript.
-
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
This work was supported by the Zhejiang Provincial Natural Science Foundation of China (No. LY24C020002), and the National Natural Science Foundation of China (No. 32160355).
-
The authors declare that they have no conflict of interest.
-
# Authors contributed equally: Zhihua Wu, Ruting Li
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Wu Z, Li R, Sun M, Hu X, Xiao M, et al. 2024. Current advances of Carthamus tinctorius L.: a review of its application and molecular regulation of flavonoid biosynthesis. Medicinal Plant Biology 3: e004 doi: 10.48130/mpb-0024-0005
Current advances of Carthamus tinctorius L.: a review of its application and molecular regulation of flavonoid biosynthesis
- Received: 19 September 2023
- Revised: 28 January 2024
- Accepted: 31 January 2024
- Published online: 04 March 2024
Abstract: Safflower is a valuable medicinal plant due to its diverse flavonoids, known as safflower yellow (SY) and safflower red (SR) pigments, which are popular in the fields of medicine, food and natural dyes. The main pharmacological ingredient in safflower is SY pigment, which is a kind of water-soluble flavonoid, such as hydroxysafflor yellow A (HSYA). In contrast, SR pigment mainly contains carthamin, which is sparingly soluble in water and has a historic reputation as a red colorant. These flavonoids from both SY and SR pigments belong to C-glucosylquinochalcones and their derivatives, which only exist in safflower. Over the past few decades, genetics, molecular biology and multi-omics strategies have been used to study flavonoid biosynthesis in safflower. However, there has not been a comprehensive summary of the application and molecular progress of safflower flavonoid biosynthesis. In this review, we summarize the various applications of safflower in medicine, natural dyes, and food, as well as the molecular regulation of flavonoid biosynthesis. We also propose future research directions on safflower flavonoids. This work aims to provide an objective national and international review of literature on the application and biosynthetic regulation of safflower flavonoids.
-
Key words:
- Safflower /
- Application /
- Flavonoid biosynthesis /
- HSYA /
- Molecular regulation.