-
Astragalus root, the roots of Astragalus membranaceus, is widely used as herbal medicine and dietary supplements in many countries[1,2]. It is rich in pterocarpan glycosides which were considered as its bioactive components. Pterocarpan glycosides showed hypoglycemic and antioxidant activities, with the glucosyl group playing a significant role[3]. Moreover, pterocarpan glycosides can be absorbed and metabolized by the intestine to yield free pterocarpans[4], exhibiting anti-cancer[5], antimicrobial[6], and antimyotoxic[7] bioactivities.
Glycosylation is common in the biosynthesis of plant secondary metabolites. After forming the basic framework of pterocarpans, they are often modified by uridine diphosphate (UDP)-glycosyltransferases (GTs) and exist in plants as glycosides[8]. Though flavonoid GTs have been intensively studied[9−11], glycosyltransferases for pterocarpans have been scarcely reported. Only a few promiscuous GTs, including GuGT10[9], GuGT14[9], GgCGT[10], and UGT73AE1[11], could utilize pterocarpan as one of the substrates. However, none of these GTs showed remarkable selectivity to pterocarpans, and the conversion rates for the former three GTs were relatively low. Consequently, they were not further investigated for their application on pterocarpans, and the pterocarpan glycosylation tool for whole-cell catalysis is still lacking.
In this study, two pterocarpan GTs, AmGT28 and AmGT44, from A. membranaceus were identified. They exhibited high catalytic efficiency and high selectivity towards pterocarpans. Moreover, a whole-cell catalytic system was successfully established for the glycosylation of pterocarpan compounds.
-
Compounds 1−3, 11−12, 1a, and 2a were purchased from MedChemExpress (Shanghai, China). Substrate 4 was purchased from Adamas-beta (Shanghai, China). Substrates 6 and 13 were purchased from Aladdin Biotechnology (Shanghai, China). Substrates 5, 7−10, 14, 15 and sugar donors were purchased from YuanYe Biotechnology Co., Ltd. (Shanghai, China). Products 7a, 9a, 10a, and 10b were purchased from Must Bio-technology Co., Ltd. (Chengdu, China). Methanol, acetonitrile, and formic acid were HPLC grade (Fisher Scientific, USA). All other chemicals were purchased from Beijing Chemical Corporation (Beijing, China) and Solarbio (Beijing, China).
Plant sample collection and transcriptome data analysis
-
The seeds of A. membranaceus were purchased from Anguo of Hebei province in China, and were cultivated in the authors’ laboratory. They were grown in an incubator under the following conditions: 25 °C, 80% relative humidity, 12 h photoperiod. The 40-day-old roots were harvested and used for RNA extraction. The transcriptome data (SRR923811) was utilized for Basic Local Alignment Search Tool (BLAST) searches to identify candidate pterocarpan glycosyltransferases. LaUGT1 and LaUGT2 (Genbank No. MT427396 and MT427397), legume isoflavonoid glycosyltransferases, were used as template genes.
RNA extraction, molecular cloning, heterologous expression and protein purification
-
The fresh roots of 40-day-old A. membranaceus plants were frozen and ground. The RNA was extracted using TransZol method according to the manufacturer's instructions (Transgen Biotech, Beijing, China). cDNA was synthesized from the total RNA using the ABScript II cDNA First-Strand Synthesis Kit (ABclonal Technology, Beijing, China). PCR was carried out using 0.3 µL of cDNA as a template, AmGT28F/R and AmGT44F/R as primers (Supplemental Table S1) under the following conditions: 95 °C for 5 min, followed by 36 cycles of 95 °C for 20 s, 55 °C for 30 s, 72 °C for 80 s, with a final extension at 72 °C for 10 min. The target gene fragments were cloned into the expression vector pET-28a(+) (Invitrogen, California, USA) through seamless splicing. After sequencing, the recombinant plasmid was transformed into Escherichia coli BL21 (DE3) (TransGen Biotech, China) for expression.
A single E. coli BL21 (DE3) colony was inoculated with a plasmid containing the required expression fragment in Luria-Bertani (LB) medium containing 50 µg/mL kanamycin. The E. coli cells were cultured at 37 °C until OD600 = 0.6−0.8. Subsequently, the culture was cooled to 18 °C for 30 min, and 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was added to induce the expression of the recombinant protein at 18 °C. Following incubation with shaking for 18−20 h, E. coli cells were harvested by centrifugation at 7,600 rpm. The cells were then disrupted using ultrasound to collect the supernatant. Protein purification was performed using nickel affinity columns (ProteinIso Ni-NTA Resin, TransGen Biotech, Beijing, China). Foreign proteins were eluted with 30 mM imidazole, while the target proteins were eluted with 300 mM imidazole. The purified protein solution was stored in a storage buffer (20 mM Tris, 500 mM NaCl, 20% glycerol, pH = 7.5). After rapid freezing with liquid nitrogen, the protein was stored at −80 °C.
In vitro enzyme catalytic activity assays
-
To identify the functions of the enzymes, assays were performed in a reaction solution composed of 0.1 mM substrate, 0.5 mM UDP-Glc and 10 μg of purified enzymes in a reaction buffer (50 mM Na2HPO4-NaH2PO4 buffer, pH 7.0). The reaction mixtures were then incubated at 37 °C for 4 hours. The reaction was terminated by adding 100 μL methanol. The resultant mixture was subjected to vacuum drying. The residue was dissolved in 150 μL of methanol and centrifuged at 15,000 rpm for 30 min before UHPLC analysis. The conversion rates were calculated using the following equation: Aproduct / (Asubstrate + Aproduct) × 100%, where A represents the peak area from the UHPLC/UV chromatogram.
To explore the sugar donor selectivity of AmGT28 and AmGT44, various sugar donors including UDP-Glc, UDP-xylose (UDP-Xyl), UDP-galactose (UDP-Gal), and UDP-N-acetylglucosamine (UDP-GlcNAc) were evaluated using maackiain (2) as acceptor. The sample processing method remained consistent with the last paragraph.
To determine the kinetic parameters of AmGT44, enzymatic reactions were carried out in a final volume of 100 μL 50 mM Na2HPO4-NaH2PO4 buffer (pH 7.0), containing 0.1 mM of UDP-glucose and 0.1 ng purified protein. When the substrate was maackiain (2) or 7-hydroxyisoflavone (4), the final concentration range of the substrate was 0.1−1,000 μM or 0.1−4,000 μM, respectively. The reactions were conducted at 37 °C for 30 min and then stopped by adding 100 µL of ice-cold methanol. The resulting mixture was dried under vacuum. The residue was dissolved in 150 μL of methanol and centrifuged at 15,000 rpm for 30 min for UHPLC analysis. The data obtained were analyzed by Origin 2018 (OriginLab, USA).
Analytical instruments and methods
-
Enzymatic products were analyzed by a Vanquish ultra-high-performance liquid chromatography (UHPLC) instrument coupled with a Q-Exactive Orbitrap mass spectrometer through a heated ESI source (Thermo Scientific, CA, USA). ESI source parameters: positive ion polarity mode; sheath gas (N2), 45 arb; auxiliary gas (N2), 10 arb; spray voltage, 3.5 kV; capillary temperature, 350 °C; collision energy, 10 V. The samples were separated on a Waters T3 column (2.1 mm × 100 mm, 1.8 μm, USA). The column temperature was 50 °C. Samples were eluted using the program as in Supplemental Tables S2 & S3.
Preparative-scale reactions and purification of the products
-
To purify the glycosylated product 5a, the experimental system described above was scaled up proportionally to approximately 100 mL, with 10 mg of substrate 5 being utilized. The reaction mixtures were incubated at 37 °C for 12 h, combined and mixed with 100 mL of methanol. Subsequently, the mixtures were centrifuged at 15,000 rpm for 30 min, and the supernatants were concentrated and dissolved in 4 mL of methanol. The glycosylated product 5a was then separated using reversed-phase semi-preparative HPLC on an Agilent 1200 instrument (Germany) equipped with a Zorbax SB-C18 column (9.4 × 250 mm, 5 μm, Agilent). Mobile phase A consisted of water containing 0.03% trifluoroacetic acid, and the mobile phase B was acetonitrile. The elution program was as follows: 0−9 min, 10% B; 9−10 min, 10%−65% B; 10−29 min, 65% B; 29−30 min, 65%−100% B; 30−40 min, 100% B. The flow rate was 2.0 mL/min.
Nuclear magnetic resonance (NMR) spectra of 5a were recorded on a Bruker AVANCE III-400 instrument at 400 MHz for 1H NMR and 100 MHz for 13C NMR, in DMSO-d6. Chemical shifts (δ) are reported in parts per million (ppm) and coupling constants (J) are reported in Hertz (Hz) (Dataset 1).
Whole-cell catalytic activity assay
-
The optimization of the whole-cell catalytic reaction condition was performed in a 500-μL system using maackiain (2) as the substrate. The E. coli BL21 (DE3) strain harboring the AmGT28/AmGT44 gene was cultured at 37 °C until the OD600 reached 5.0−5.5, and then shifted to 18 °C with the addition of IPTG (0.1 mM). The method was optimized regarding the following factors: timing of substrate addition, culture temperature, and culture duration. In methods 1/2/3, the substrates were added simultaneously with IPTG and the cultures were maintained at 18 °C for 15/20/25 h, respectively. In methods 4/5/6, the strains were cultured at 37 °C for 10 h, followed by the addition of IPTG, and then further incubated for an additional 5/10/15 h. In methods 7/8/9, the strains were cultured at 18 °C for 10 h, then IPTG was added, and the cultures were further incubated for an additional 5/10/15 h. The products were extracted with 600 μL of ethyl acetate. The organic phase was dried under vacuum, and the residue was dissolved in 150 μL of methanol. Subsequently, the solution was centrifuged at 15,000 rpm for 30 min for UHPLC analysis.
-
The transcriptome data of A. membranaceus was screened using the reported legume isoflavonoid GTs LaUGT1 and LaUGT2 as templates (e < 10−100)[12]. Through the BLAST search, three candidate genes were found, and only two of them (AmGT28 and AmGT44, Genbank No. PP597340 and PP597341) contained complete open reading frames. These genes were cloned into the pET-28a(+) vector and expressed in an E. coli BL21 (DE3) strain. The purified recombinant proteins were obtained using Ni-NTA affinity chromatography (Supplemental Fig. S1), and their catalytic activities were characterized through enzyme catalysis reactions (containing 10 μg of purified enzymes, 0.1 mM substrate, 0.5 mM UDP-Glc, 50 mM Na2HPO4-NaH2PO4 buffer, pH 7.0). Subsequently, AmGT28 and AmGT44 was found to catalyze (−)-astrapterocarpan (1) to yield a new product 1a with an almost 100% conversion rate (Fig. 1a, b). In LC/MS analysis, the product displayed an [M+H]+ ion at m/z 463.2, which was 162 Da greater than that of (−)-astrapterocarpan (m/z 301.1). The MS/MS spectrum of the [M+H]+ ion generated an [M-162+H]+ ion at m/z 301.1 (Fig. 1c). Finally, product 1a was confirmed to be (−)-astrapterocarpan 3-O-glucoside by comparing with a reference standard. Therefore, AmGT28 and 44 were identified as pterocarpan 3-O-glycosyltransferases.
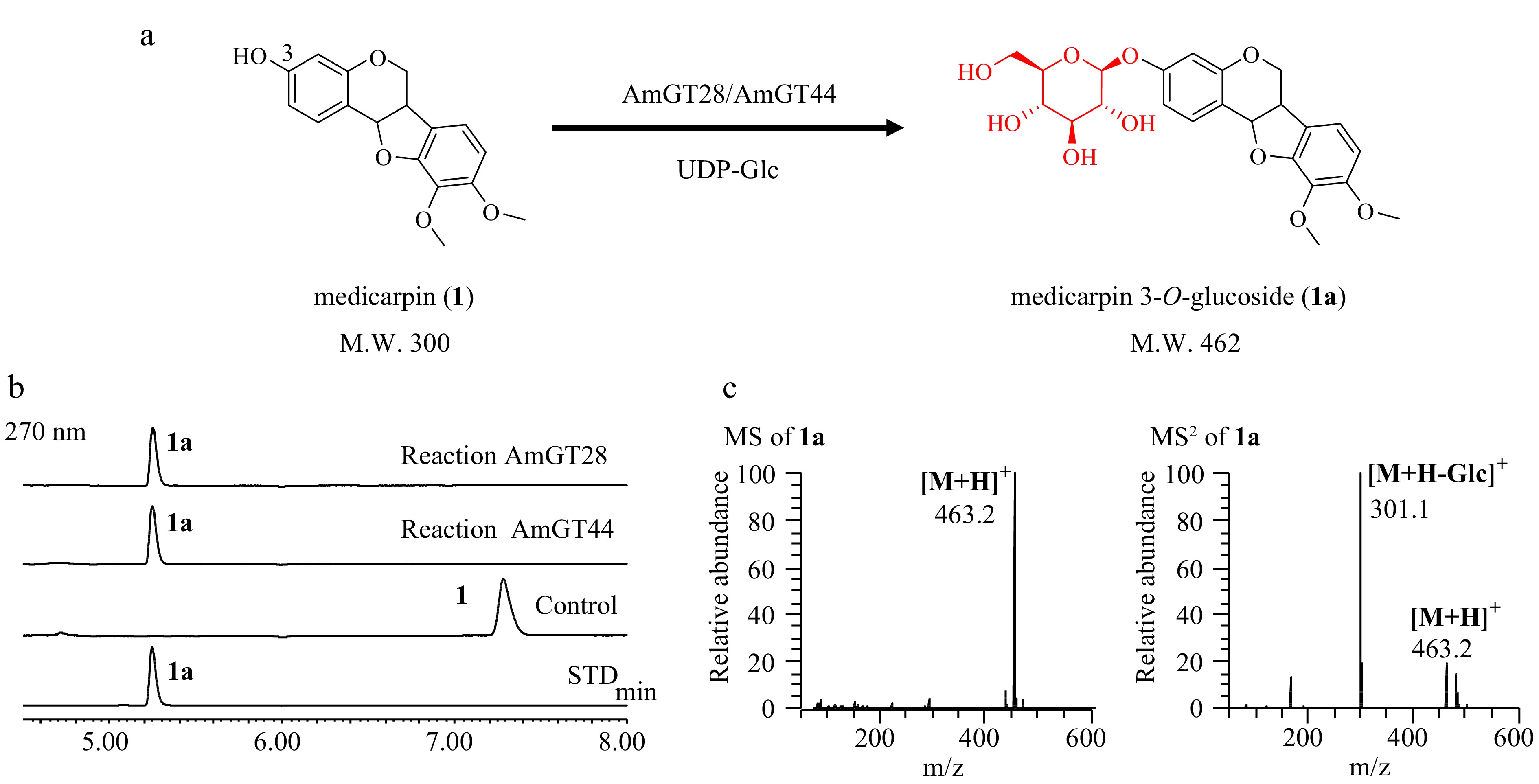
Figure 1.
Functional characterization of AmGT28 and AmGT44. (a) Glycosylation of 1 by recombinant AmGT28/AmGT44 to produce 1a using UDP-Glc as the sugar donor. (b) UHPLC/UV chromatograms of the enzymatic reaction mixture. Control, reactions conducted using boiled protein; STD, reference standard. (c) MS and MS/MS spectra of 1a in the positive ion mode from the reaction catalyzed by AmGT28.
Substrate and sugar donor promiscuity of AmGT28 and AmGT44
-
The substrate promiscuity of AmGT28 and AmGT44 was explored by utilizing aglycones from Astragalus root or their simple derivatives[1]. These substrates include pterocarpans (1−3), isoflavonoids (4−13), and flavonoids (14, 15). The reactivity of AmGT28 and AmGT44 towards these compounds was assessed using UDP-Glc as the sugar donor (Fig. 2; Supplemental Figs S2−S15). In total seven products (1a, 2a, 5a, 7a, 9a, 10a, 10b) were identified by comparing with reference standards.
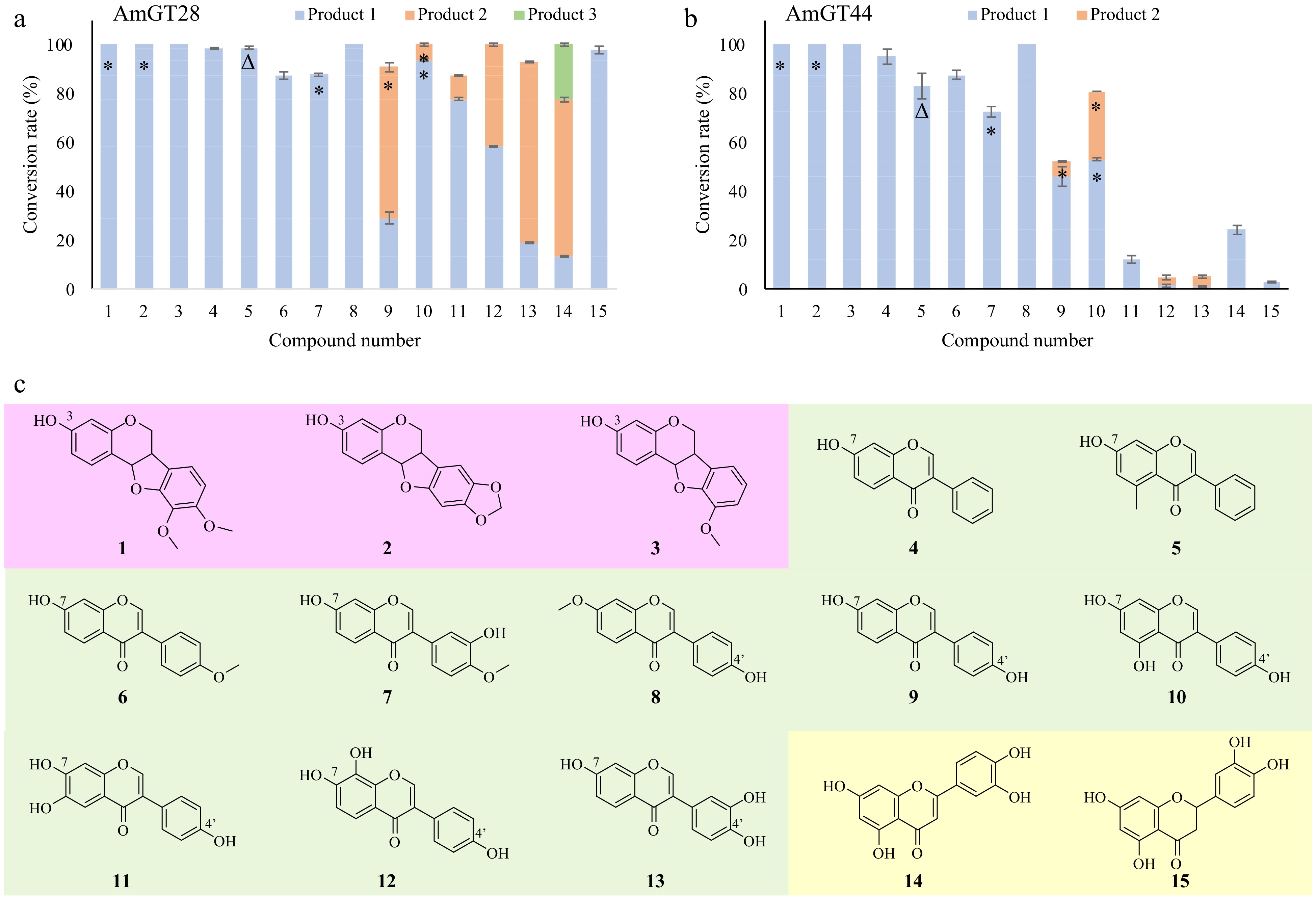
Figure 2.
Substrate specificity of AmGT28 and AmGT44. (a), (b) Conversion rates (%) of glycosylated products for substrates 1−15, using UDP-Glc as the sugar donor (n = 3). *, Products identified by comparing with reference standards; Δ, product purified from scaled-up reactions and characterized by NMR. (c) Structures of substrates 1−15.
For all three pterocarpans, both AmGT28 and AmGT44 exhibited a 100% conversion rate. Since these compounds have only one hydroxyl group at C-3, the glucosylation site was further confirmed by comparing products 1a and 2a with reference standards. In the case of isoflavonoids, AmGT28 and AmGT44 exhibited relatively high conversion rates for the majority of the substrates. Compounds 4−8 were consumed to produce individual products 4a−8a, respectively. Product 5a was synthesized through a scaled-up reaction and was characterized by NMR spectroscopy (Supplemental Figs S16−S20). The glucosyl group was attached to C7-OH, as evidenced by the HMBC correlation between C-7 (δC 160.1) and H-1′′′ (δH 5.08). The structure of 5a had not been previously reported. The identity of 7a was confirmed by comparing it with a reference standard as a 7-O-glucoside. The substitution position was assigned as C7-OH for 4a and 6a, and as C4′-OH for 8a, as the corresponding substrates contain only one hydroxyl group available. For isoflavonoids containing both C7-OH and C4′-OH (9−13), multiple glucosylated products were observed. Interestingly, AmGT28 and AmGT44 utilized 9 to produce 9a (7-O-Glc) and 9b (4-O-Glc), respectively. In the case of compound 10, both enzymes predominantly produced 10b (4-O-Glc). These findings suggested that the regioselectivity for AmGT28 and AmGT44 was limited, particularly when working with isoflavonoids possessing similar structures on ring A and ring B as substrates. Moreover, AmGT28 exhibited relatively high conversion rates for substrates 10−13, whereas AmGT44 showed lower conversion rates. This discrepancy may be attributed to the presence of multiple hydroxyl groups on the (iso)flavonoid backbone, potentially influencing hydrogen bonding and the binding mode of the substrate, thereby leading to varying regioselectivity.
To investigate the sugar donor preference of AmGT28 and AmGT44, three alternative sugar donors, namely UDP-Xyl, UDP-Gal, and UDP-GlcNAc were examined alongside UDP-Glc. When maackiain (2) was used as the substrate, AmGT44 exhibited the capacity to utilize UDP-Glc, UDP-Xyl and UDP-GlcNAc, resulting in O-glycosides with conversion rates of 100%, 100%, and 60%, respectively. The products were identified using LC/MS (Supplemental Fig. S21). On the other hand, AmGT28 could utilize three additional donors with a conversion rate of lower than 20%.
Kinetic parameters of AmGT44
-
Since AmGT28 and AmGT44 could glucosylate both pterocarpan and isoflavonoid at similar positions (3-OH for pterocarpan and 7-OH for isoflavonoid, respectively), their preference for these substrates were compared. Maackiain (2) and 7-hydroxyisoflavone (4) were chosen as substrates because of their high commercial availability. To explore the optimal reaction conditions, AmGT28 and AmGT44 were tested using 2 as the substrate (Supplemental Fig. S22, S23). Both enzymes exhibited maximal activity at pH 7.0 (50 mM Na2HPO4-NaH2PO4 buffer) and a temperature of 37 °C. The presence of divalent metal ions did not significantly impact the catalytic activity of the enzymes.
The optimal condition was used to measure the catalytic parameters of AmGT44. When 7-hydroxyisoflavone (4) was used as the substrate, the Km value of AmGT44 was 449.83 μM, and the Kcat/Km was 1.44 × 10−7 μM−1·s−1 (Fig. 3b). When maackiain (2) was used as the substrate, the Km value of AmGT44 was 69.28 μM, and the Kcat/Km was 5.05 × 10−5 μM−1·s−1 (Fig. 3a), which was 350 times higher than the value with 4. For AmGT28, although the conversion rates were both 100% for 2 and 4 at 100 μg/mL protein concentration (Fig. 2a), the conversion rate of 2 was higher than that of 4 at lower protein concentrations (40, 10, and 4 μg·mL−1) (Supplemental Fig. S24). These results indicated that AmGT44 and AmGT28 preferred pterocarpan 2 over isoflavonoid 4, supporting their primary function as pterocarpan 3-O-glucosyltransferases.
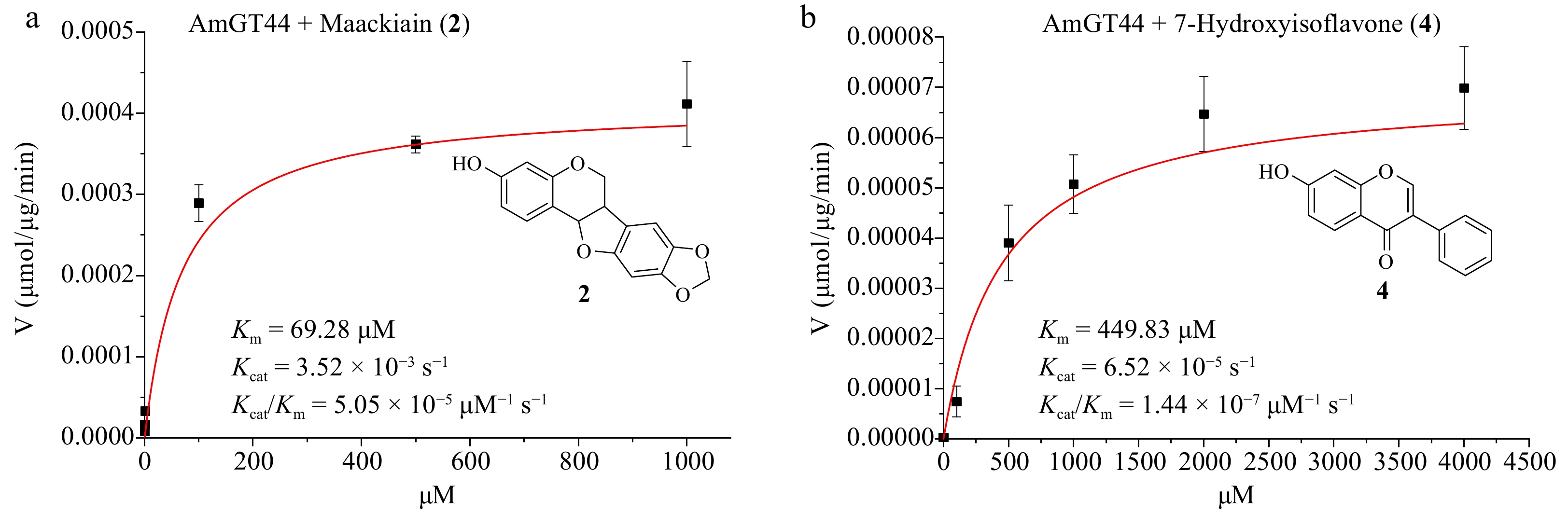
Figure 3.
Substrate preference of AmGT44. (a) Kinetic parameters of AmGT44 using maackiain (2) as the substrate and UDP-Glc as the sugar donor. (b) Kinetic parameters of AmGT44 using 7-hydroxyisoflavone (4) as the substrate and UDP-Glc as the sugar donor.
Whole-cell catalytic reactions
-
Since the purification of the glycosyltransferases is time-consuming, a whole-cell biocatalytic system was established, utilizing E. coli cells harboring the AmGT28 or AmGT44 gene. Given that wild-type E. coli cells could produce UDP-Glc in a considerable amount[13], glycosylation can be achieved without the addition of sugar donors. The optimization of the whole-cell catalytic reaction system involved the culture temperature, culture duration, and the timing of substrate addition (Fig. 4a). Maackiain (2, 0.1 mM) was added into the system as the substrate. As a result, method 6 was identified as the most suitable condition: after inducing expression of the recombinant protein at 18 °C for 10 h, the substrate was added and co-incubated at 37 °C for 15 h. Under these conditions, AmGT28 and AmGT44 demonstrated conversion rates of 76% and 100% for 2, respectively (Fig. 4b). The culture conditions were then applied to all the substrates in Fig. 4c and d (Supplemental Fig. S25). Interestingly, all three pterocarpans exhibited high conversion rates in whole-cell catalytic reactions. The conversion rates of 1-3 were 100% for AmGT44 and 58%−72% for AmGT28. To determine the titer, calibration curves for 1a and 2a were established (Supplemental Fig. S26), and the semi-quantitation of 3a was conducted using the curve of 1a. In the 500-μL whole-cell catalysis system of AmGT44, the highest titers of 1a−3a achieved 30.95, 38.62, and 33.71 μg·mL−1, respectively. For further scaled-up reactions of AmGT44, substrate concentrations of 0.2, 0.4, and 1.0 mM were tested using 2 as the substrate, and a conversion rate of > 90% could be achieved at 0.2 mM (Supplemental Fig. S27). The 0.2 mM substrate was then applied to a 10-mL system, where the conversion rate was calculated to be 95%, and the titer was determined to be 78.66 μg/mL (Supplemental Fig. S28).
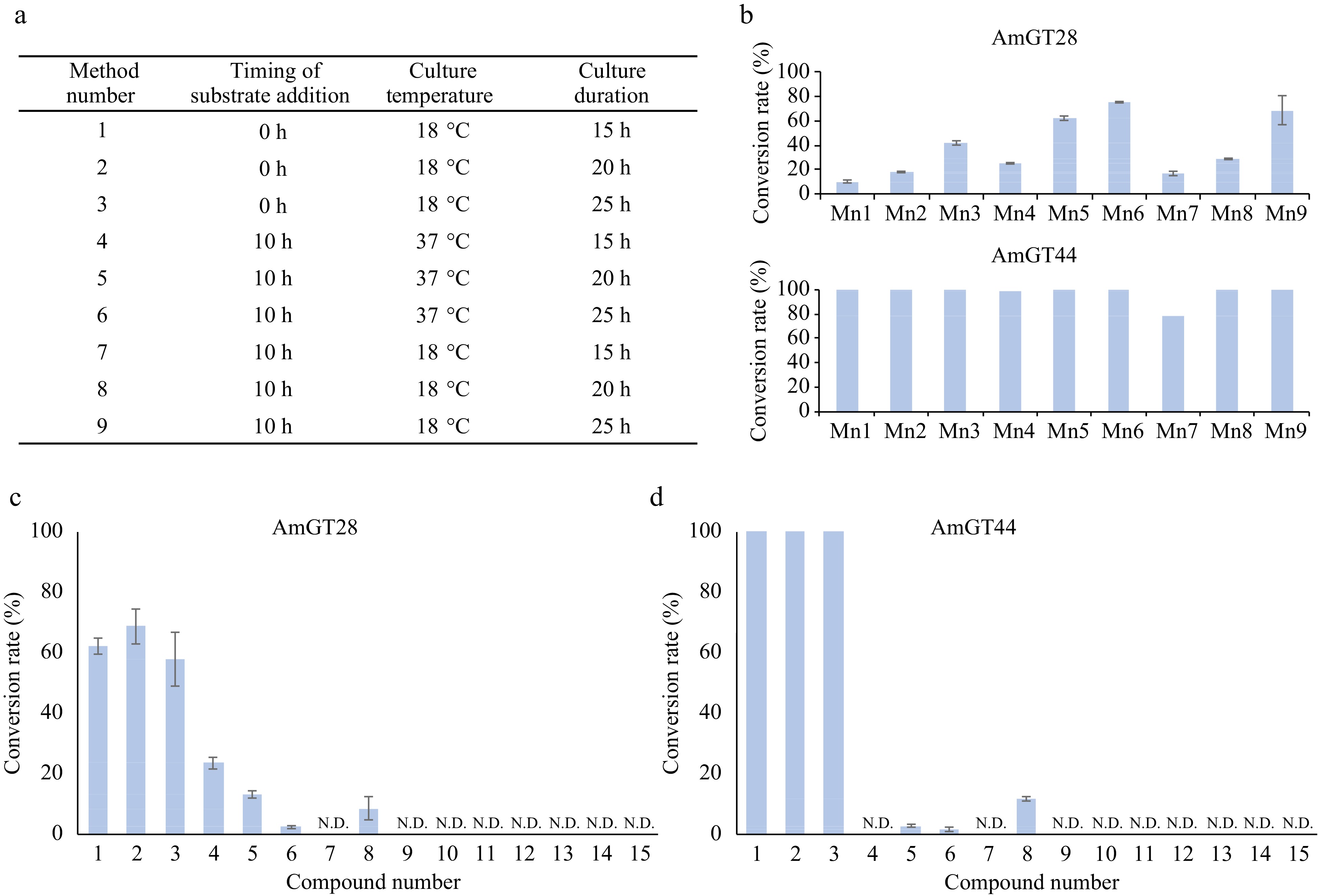
Figure 4.
Whole cell catalytic reactions for AmGT28 and AmGT44. (a) Optimization of reaction conditions using methods 1−9. (b) Conversion rates of compound 2 using AmGT28 and AmGT44 under different conditions 1−9. Mn: Method number. (c), (d) Conversion rates of glycosylated products for substrates 1−15. For all experiments, n = 3.
For other compounds, only isoflavonoids 4-6 and 8 could be used in whole-cell catalytic reactions with a low conversion rate (Supplemental Fig. S25). This observation could be attributed to the lipophilicity of the substrates, as evidenced by the partition coefficient (log P). Specifically, the log P value was 2.35 to 3.01 for 1−6 and 8, while they were in the range of 1.24 to 2.13 for compounds 7 and 9−15 (Supplemental Table S4). Enhancing the conversion rates of polar compounds would require further investigation.
-
In conclusion, we cloned and characterized two pterocarpan 3-O-glycosyltransferases, AmGT28 and AmGT44, from A. membranaceus. AmGT28 and AmGT44 demonstrate efficient catalysis in the O-glycosylation of at least ten pterocarpans and isoflavonoids, achieving high conversion rates exceeding 70%. They utilize UDP-Glc as the sugar donor and exhibit a preference for pterocarpans over isoflavonoids in their glycosylation reactions. Subsequently, we established a whole-cell biocatalytic system utilizing AmGT44, which enabled the glucosylation of pterocarpans 1−3 with a conversion rate of 100% within 25 h. The highest titer of maackiain 3-O-glucoside (2a) reached 78.66 μg·mL−1. This study has provided efficient catalytic tools for the biosynthesis of pterocarpan glycosides.
-
The authors confirm contribution to the paper as follows: study conception and design: Qiao X, Ye M; analysis and interpretation of results: Gao BH, Zhang M, Chen K, Li HY, Xiong M, Huang YX, Zhang Y; draft manuscript preparation and revision: Gao BH, Wang LL, Yao MJ, Liu QL, Guo J, Qiao X. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analyzed during this study are included in this published article and its supplementary information files.
This work was supported by National Natural Science Foundation of China (82122073, 81973448), National Key Research and Development Program of China (No. 2023YFA0914100), and Key project at central government level: The ability establishment of sustainable use for valuable Chinese medicine resources (2060302). We are thankful for the support from the State Key Laboratory of Natural and Biomimetic Drugs.
-
The authors declare that they have no conflict of interest.
- Supplemental Table S1 PCR primers used in this study.
- Supplemental Table S2 UHPLC elution program for the enzymatic properties and the sugar donor preference experiments.
- Supplemental Table S3 UHPLC elution program for the substrate preference and whole-cell catalysis experiments.
- Supplemental Table S4 Partition coefficient (Log P) of substrates.
- Supplemental Dataset 1 NMR data compound 5a.
- Supplemental Fig. S1 SDS-PAGE of His-tagged AmGT28/AmGT44 purified by Ni-NTA affinity chromatography.
- Supplemental Fig. S2 UHPLC/UV and UHPLC/MS analysis of AmGT28/AmGT44 catalytic reaction mixtures for compound 2 with UDP-Glc.
- Supplemental Fig. S3 UHPLC/UV and UHPLC/MS analysis of AmGT28/AmGT44 catalytic reaction mixtures for compound 3 with UDP-Glc.
- Supplemental Fig. S4 UHPLC/UV and UHPLC/MS analysis of AmGT28/AmGT44 catalytic reaction mixtures for compound 4 with UDP-Glc.
- Supplemental Fig. S5 UHPLC/UV and UHPLC/MS analysis of AmGT28/AmGT44 catalytic reaction mixtures for compound 5 with UDP-Glc.
- Supplemental Fig. S6 UHPLC/UV and UHPLC/MS analysis of AmGT28/AmGT44 catalytic reaction mixtures for compound 6 with UDP-Glc.
- Supplemental Fig. S7 UHPLC/UV and UHPLC/MS analysis of AmGT28/AmGT44 catalytic reaction mixtures for compound 7 with UDP-Glc.
- Supplemental Fig. S8 UHPLC/UV and UHPLC/MS analysis of AmGT28/AmGT44 catalytic reaction mixtures for compound 8 with UDP-Glc.
- Supplemental Fig. S9 UHPLC/UV and UHPLC/MS analysis of AmGT28/AmGT44 catalytic reaction mixtures for compound 9 with UDP-Glc.
- Supplemental Fig. S10 UHPLC/UV and UHPLC/MS analysis of AmGT28/AmGT44 catalytic reaction mixtures for compound 10 with UDP-Glc.
- Supplemental Fig. S11 UHPLC/UV and UHPLC/MS analysis of AmGT28/AmGT44 catalytic reaction mixtures for compound 11 with UDP-Glc.
- Supplemental Fig. S12 UHPLC/UV and UHPLC/MS analysis of AmGT28/AmGT44 catalytic reaction mixtures for compound 12 with UDP-Glc.
- Supplemental Fig. S13 UHPLC/UV and UHPLC/MS analysis of AmGT28/AmGT44 catalytic reaction mixtures for compound 13 with UDP-Glc.
- Supplemental Fig. S14 UHPLC/UV and UHPLC/MS analysis of AmGT28/AmGT44 catalytic reaction mixtures for compound 14 with UDP-Glc.
- Supplemental Fig. S15 UHPLC/UV and UHPLC/MS analysis of AmGT28/AmGT44 catalytic reaction mixtures for compound 15 with UDP-Glc.
- Supplemental Fig. S16 1HNMR spectrum of compound 5a in DMSO-d6 (400 MHz).
- Supplemental Fig. S17 13CNMR spectrum of compound 5a in DMSO-d6 (100 MHz).
- Supplemental Fig. S18 HMBC spectrum of compound 5a in DMSO-d6 (400 MHz).
- Supplemental Fig. S19 HSQC spectrum of compound 5a in DMSO-d6 (400 MHz).
- Supplemental Fig. S20 HR-MS (positive) spectrum of compound 5a.
- Supplemental Fig. S21 UHPLC/UV and UHPLC/MS analysis of AmGT28/AmGT44 catalytic reaction mixtures for compound 2 with UDP-Glc, UDP-Xyl, UDP-Gal and UDPGlcNAc.
- Supplemental Fig. S22 Effects of reaction buffer (a), reaction temperature (b), and divalent metal ions (c) on the activities of AmGT28. Maackiain (2) was used as the acceptor and UDP-Glc was used as the sugar donor. An optimized reaction time of 4 hour was used. AmGT28 exhibited its maximum activity at pH 7.0 (50 mM Na2HPO4-NaH2PO4) and 37 °C.
- Supplemental Fig. S23 Effects of reaction buffer (a), reaction temperature (b), and divalent metal ions (c) on the activities of AmGT44. Maackiain (2) was used as the acceptor and UDP-Glc was used as the sugar donor. An optimized reaction time of 4 hour was used. AmGT44 exhibited its maximum activity at pH 7.0 (50 mM Na2HPO4-NaH2PO4) and 37 °C.
- Supplemental Fig. S24 UHPLC/UV chromatograms of enzyme catalytic prodcuts demonstrating substrate preference of AmGT44 and AmGT28.
- Supplemental Fig. S25 UHPLC/UV chromatograms of the extracts from whole-cell biocatalysis by E. coli cells harboring AmGT28/AmGT44 gene. Control, reactions conducted using boiled protein; STD, reference standard. The structures and numbers of substrates are given in Fig. 2c.
- Supplemental Fig. S26 Calibration curves for 1a and 2a.
- Supplemental Fig. S27 UHPLC/UV chromatograms of the whole-cell catalytic reaction mixtures with different substrate (2) concentration (0.2 mM, 0.4 mM, 1.0 mM, 500 μL). STD, reference standard.
- Supplemental Fig. S28 UHPLC/UV chromatograms of the whole-cell catalytic reaction mixture with a substrate (2) concentration of 0.2 mM and a volume of 10 mL. STD, reference standard.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Gao BH, Zhang M, Chen K, Wang LL, Yao MJ, et al. 2024. Characterization of two pterocarpan glycosyltransferases in Astragalus membranaceus and their application in whole-cell biocatalysis. Medicinal Plant Biology 3: e010 doi: 10.48130/mpb-0024-0013
Characterization of two pterocarpan glycosyltransferases in Astragalus membranaceus and their application in whole-cell biocatalysis
- Received: 15 December 2023
- Revised: 10 April 2024
- Accepted: 07 May 2024
- Published online: 05 June 2024
Abstract: Pterocarpan glucosides are biologically active natural products, in which the glucosyl group plays an important role. However, specific pterocarpan glycosyltransferases have rarely been reported. In this study, two highly efficient glycosyltransferases, AmGT28 and AmGT44, from Astragalus membranaceus, catalyzing the conversion of (−)-astrapterocarpan (1) to (−)-astrapterocarpan 3-O-glucoside (1a) were identified. They could also use other pterocarpans and isoflavonoids as substrates, showing a preference for pterocarpans over isoflavonoids. A new isoflavone 7-O-glucoside 5a was prepared through a scaled-up enzymatic reaction. Furthermore, AmGT44 was expressed in E. coli to establish a whole-cell biocatalytic system (500 μL with 0.1 mM substrate), which could glucosylate pterocarpans 1−3 with a conversion rate of up to 100% and a titer of 30−38 μg/mL within 25 h. The titer of maackiain 3-O-glucoside (2a) could reach 78.66 μg/mL in a 10-mL system. The study provides efficient catalytic tools for the biosynthesis of pterocarpan glycosides.













