-
This study examined the dried aerial parts of Schizonepeta tenuifolia Briq. (Lamiaceae), a medicinal plant which serves as the primary botanical source for the traditional Chinese medicine Jingjie (Schizonepetae Herba). Renowned for its marked efficacy in dispelling wind and releasing the exterior, Jingjie is included in the pharmacopoeias of multiple countries[1]. Pharmacological investigations have revealed that the p-menthane monoterpenes present in Jingjie (constituting > 90% of its volatile oil) exhibit broad-spectrum biological activities, including anti-inflammatory, antibacterial, and antiviral effects, as well as inhibition of tumor cell proliferation[2−4]. Notably, these characteristic chemical constituents not only underpin the clinical application of Jingjie for indications such as the common cold and fever, but have also earned the plant the distinct designation 'Japanese catnip' in international botanical circles due to its unique metabolic profile[5,6].
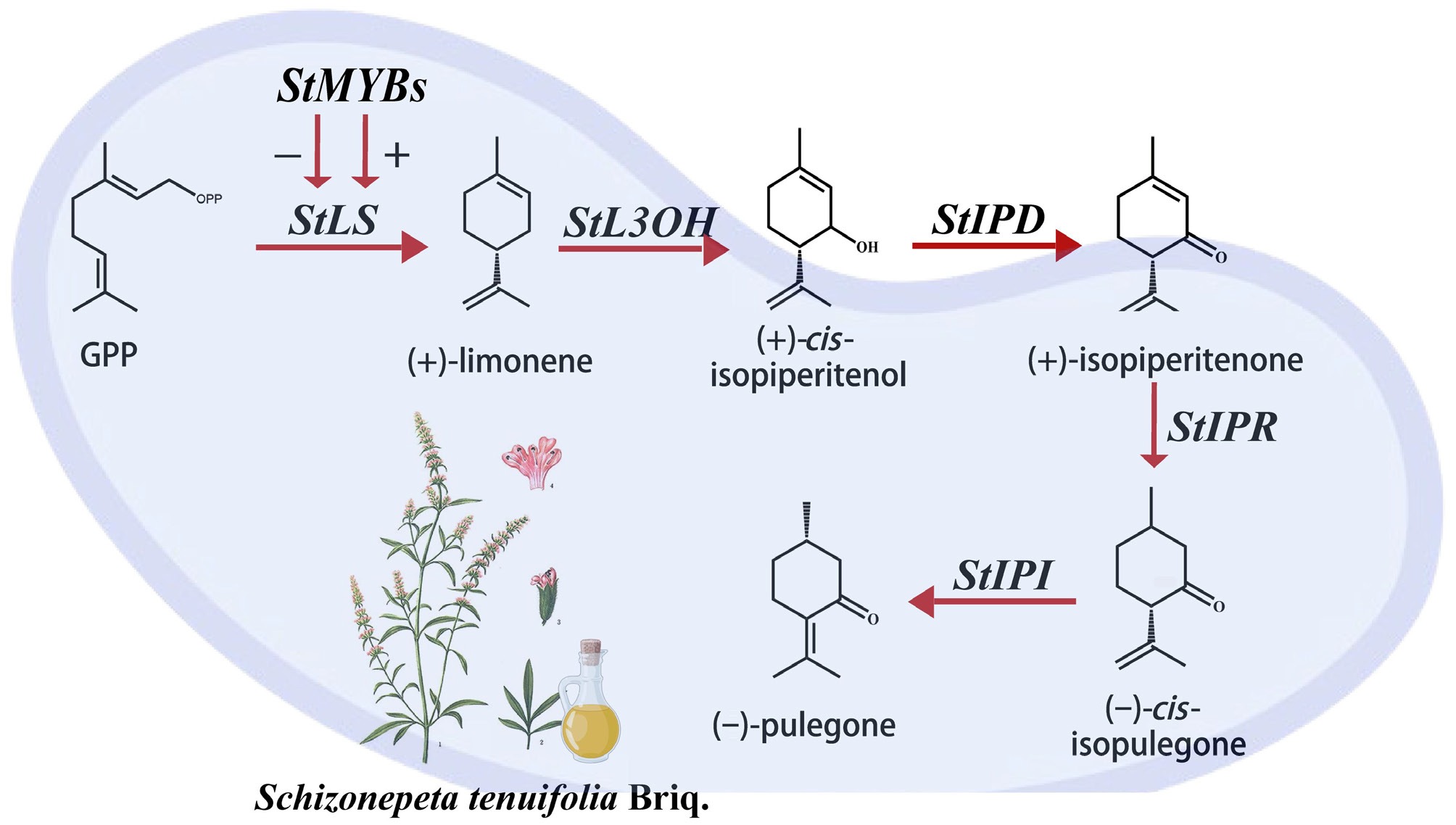
Given the pharmacological significance of these monoterpenes, elucidating their biosynthetic pathways is crucial for understanding and potentially enhancing the medicinal properties of S. tenuifolia. Recent research has made significant progress in understanding monoterpene biosynthesis, particularly in characterizing the metabolic pathways within the Lamiaceae family. Studies have established that the methylerythritol phosphate (MEP) pathway serves as the primary route for monoterpene production, with limonene synthase (LS) playing a pivotal role by catalyzing the cyclization of geranyl pyrophosphate (GPP) into limonene[7,8]. Leveraging multi-omics technologies, metabolic networks in Lamiaceae species have been mapped. Employing comparative genomics and transcriptomics, our team, in collaboration with international partners, identified the first gene set responsible for the biosynthesis of (−)-pulegone in S. tenuifolia, involving a four-step enzymatic process[9]. The StLS gene (Sch000026966) was found to be specifically expressed in glandular trichomes. Subsequent enzymatic assays confirmed that the encoded StLS enzyme catalyzes the formation of limonene[3]. Furthermore, metabolic flux analysis demonstrated that the StL3OH hydroxylase cooperates with the StIPR isopiperitenone reductase to efficiently convert limonene into (−)-pulegone and (+)-menthone, thereby enhancing the production of these monoterpene end-products.
As a core catalytic enzyme in monoterpene biosynthesis, the molecular properties and regulatory mechanisms of limonene synthase (StLS) have been progressively elucidated. Key aspects are outlined below:
(1) Enzymatic properties and catalytic mechanism: Limonene synthase belongs to the terpene synthase superfamily. Its tertiary structure features an α-helical fold and contains conserved metal ion-binding domains (e.g., DDxxD motifs). The enzyme utilizes Mg2+ or Mn2+ to stabilize its substrate, geranyl pyrophosphate (GPP), during the cyclization reaction that forms limonene[10−12]. Following post-translational glycosylation modification in the endoplasmic reticulum, StLS is translocated to plastids to facilitate substrate conversion. The resulting monoterpenes are subsequently transported and stored in specialized compartments[7,13]. The specific subcellular localization of StLS provides critical insights into the spatial organization of monoterpene biosynthesis.
(2) Tissue-specific regulation: StLS expression is regulated by Myb-binding motifs and other cis-acting elements within the -797 to -598 bp region of its promoter. Its expression is predominantly high in glandular trichomes, consistent with their role as storage sites for terpenoids[14,15]. Silencing the StLS gene using virus-induced gene silencing (VIGS) resulted in a significant decrease in limonene content within the leaf volatile oil, underscoring its essential role in monoterpene production[9].
(3) Regulatory network of environmental factors: External environmental stimuli modulate StLS activity through epigenetic modifications and signal transduction pathways. Light intensity positively correlates with StLS transcript levels, with high light intensity elevating its mRNA abundance and consequently enhancing monoterpene accumulation[16]. Moreover, hormones such as methyl jasmonate (MeJA) indirectly influence StLS expression by activating the MYB-WRKY transcriptional module, illustrating the intricate interplay between environmental stress signaling and secondary metabolic regulation[7].
The biosynthesis of secondary metabolites in medicinal plants is governed by a complex interplay of transcription factors (TFs) and epigenetic mechanisms, forming a dynamic hierarchical regulatory network. TFs directly activate or repress the transcription of key biosynthetic genes (e.g., in terpenoid and flavonoid pathways) by binding to specific cis-acting elements (e.g., MYB binding motifs) within the promoters of their target genes[17]. This process often involves chromatin remodeling complexes. For instance, MYB TFs can recruit histone acetyltransferases (HAT) to promote chromatin accessibility or interact with polycomb repressive complexes (PRC2) to deposit repressive histone marks (e.g., H3K27me3), thereby establishing persistent epigenetic memory[18].
Despite the established regulatory roles of MYB transcription factors in various medicinal plants, the species-specific regulatory mechanisms governing MYB TFs in S. tenuifolia remain largely unexplored. In this study, the role of R2R3-MYB transcription factors in regulating monoterpene biosynthesis in S. tenuifolia were systematically investigated. Focusing on the promoter region (2,000 bp upstream) of StLS—the rate-limiting enzyme for (−)-pulegone biosynthesis—and integrating transcriptome data with cis-acting element analysis, candidate TFs potentially binding to the MYBHV1 motif were screened. This led to the identification of StMYB76 and StMYB13. Yeast one-hybrid assays confirmed the direct binding of both TFs to the StLS promoter and their ability to activate its transcriptional activity. Critically, transient overexpression of StMYB76 and StMYB13 in plants significantly upregulated the expression of StLS, as well as the downstream enzymes StL3OH and StIPR, resulting in substantially elevated levels of (−)-pulegone compared to control plants.
Collectively, the present findings represent a significant breakthrough in understanding the molecular regulation of monoterpene biosynthesis in S. tenuifolia, which is highlighted by the following three key aspects:
(1) This study reveals, for the first time, the central regulatory role of MYBHV1 cis-elements in monoterpene biosynthesis.
(2) The specific mechanism whereby R2R3-MYB transcription factors coordinate multi-enzyme gene expression through a cascade amplification effect were determined.
(3) A functional StMYBs-StLS molecular regulatory module was constructed, providing precise, editable targets for genetic manipulation aimed at the targeted optimization of volatile oil components in S. tenuifolia.
These findings not only expand the theoretical framework of plant secondary metabolism regulatory networks but also provide novel regulatory elements for medicinal plant synthetic biology research, specifically addressing the knowledge gap regarding species-specific regulatory mechanisms in S. tenuifolia.
-
Mature seeds of Schizonepeta tenuifolia Briq were used as experimental materials, and the species' authenticity was verified with Prof. Wu Qinan of Nanjing University of Traditional Chinese Medicine. Seeds were cultivated in a light incubator at 25 °C, with a photoperiod of 16 h of light, and 8 h of darkness, 65% humidity, and a light intensity of 1,000 μmol·m−2·s−1. On the 20th day of cultivation, the apical 3–4 completely grown leaves were harvested from the seedlings, promptly snap-frozen in liquid nitrogen, and preserved in an ultra-low-temperature refrigerator at −80 °C. Three biological replicates were established for each sample, which were subsequently preserved at −80 °C in an ultra-low temperature freezer. Three biological replicates for each sample for subsequent tests were established.
Analysis of cis-elements in the StLS promoter
-
Utilizing the attributes of the core promoter region in plants (TATA-box localization and transcription start site distribution pattern)[19], the regulatory region 2,000 bp upstream of the ATG start codon of the StLS gene (Sch000026966) was delineated, and cis-regulatory elements were meticulously annotated via the PlantCARE database (Release 2023). Regulatory elements have been comprehensively annotated[20]. The analytical parameters were set as follows: identification of core promoter elements requires a threshold score of ≥ 5 (confidence level > 95%), while cis-acting element matching necessitates an E-value of ≤ 1 × 10−5 (Bonferroni's multiple testing correction), emphasizing the screening for transcription factors associated with terpene metabolism, including MYB, bHLH, and other binding motifs. Calculations of component distribution density were executed using TBtools v2.032 to produce heatmaps, and graphical standardization was accomplished with Adobe Photoshop CC 2023 (v24.7). This methodological framework has been effectively utilized for the functional investigation of drought-responsive promoters in transgenic rice[21], with its parameter configurations and visualization procedure validated through third-party replication.
Prediction of transcription factor binding sites and construction of regulatory networks
-
Utilizing the fully annotated protein sequences of S. tenuifolia, a homology comparison was conducted via PlantTFDB (v5.0), and the binding motif prediction module of TBtools was employed to align the conserved binding motifs (ATMACHR, MYBHv1, etc.) of the Arabidopsis thaliana R2R3-MYB family with the S. tenuifolia transcription factor library, thereby finalizing the construction of the transcription factor network. The conserved binding motifs of the R2R3-MYB family in Arabidopsis thaliana (ATMACHR, MYBHv1, etc.) were aligned with the transcription factor library of S. tenuifolia to finalize the motif enrichment analysis. Subsequently, the 2,000 bp sequence of the StLS gene promoter region was analyzed using the FIMO tool (v5.5.2) from the MEME Suite to identify significant binding sites (threshold setting: p < 1 × 10−5, Bonferroni correction). To elucidate the regulatory hierarchy, the transcription factor-target gene interaction network was constructed using Cytoscape (v3.9.1), with topological parameters computed via the Network Analyzer plug-in: node degree (degree ≥ 8) indicated connection strength, while betweenness centrality (≥ 0.15) identified pivotal nodes, ultimately delineating the core set of regulatory factors.
Physicochemical identification characteristics of StMYBs transcription factors
-
Based on S. tenuifolia. transcriptome data, the amino acid sequences of StMYB76 (Gene ID: Sch000018969) and StMYB13 (Gene ID: Sch000028205) were extracted. Sequence translation was performed by the Batch Translate CDS to Protein module of TBtools v2.032. Protein physicochemical properties were resolved using the ExPASy ProtParam tool system.
Examination of conserved structural domains and motifs of StMYB transcription factors
-
Motif prediction was conducted using MEME Suite (v5.5.0) with parameters configured for a maximum of 10 motifs, a width range of 6–50 amino acids, and an E-value cutoff of ≤ 1e-5. The NCBI CD-Search tool (Conserved Domain Database v3.19) was employed for structural domain validation, with screening criteria of predicted coverage ≥ 70% and similarity E-value ≤ 1e-3. The final map integration was executed using the Motif View module of TBtools v2.032, in conjunction with Adobe Photoshop CC 2023.
Phylogenetic examination of StMYBs
-
Homologous protein sequences of R2R3-MYB from Labiatae species were acquired from the NCBI RefSeq database, with screening criteria of BLASTP comparison E-value < 1 × 10−20, coverage ≥ 80%, and consistency > 60%. Multiple sequence comparison was conducted using Clustal Omega v1.2.4, and the optimal evolutionary model, as indicated by ModelTest-NG assessment, was LG + G + I based on the BIC criterion. Neighbor-joining trees were generated using MEGA11 (gamma correction α = 1.2), with node support (> 70% retention) evaluated using 1,000 bootstrap tests. Final optimization was visualized using iTOL v6.7.1.
Construction of yeast one-hybrid (YIH) vectors
-
Total RNA was isolated from S. tenuifolia leaves utilizing TRIzol® Reagent (Invitrogen, 15596-026). The A260/A280 ratio, measured by NanoDrop One (Thermo Scientific), was 1.9 ± 0.1 (n = 3), and the RIN value was ≥ 8.0 (Agilent 2100 Bioanalyzer). The initial strand of cDNA was synthesized using PrimeScript™ RT Master Mix (Takara, RR036A). According to the distribution features of cis-elements in the StLS promoter region, the 2,000 bp sequence was categorized into three functional modules: P1 region (−671 to 0 bp), which encompasses the TATA-box core promoter and MYB binding site; P2 region (−1,364 to −916 bp), enriched with light-responsive elements (G-box, ACE) and MYBHV1 motifs; P3 region (−2,000 to −1,365 bp), which contains one ABA response element and one W-box. Each fragment was amplified using the high-fidelity enzyme PrimeSTAR® Max (Takara, R045A) (see Supplementary Table S1 for the primers), and the fragments were directionally cloned into the pGADT7 (ADA) vector utilizing the Nimble Cloning Kit (Sangon Biotech, C601). We used the Nimble Cloning Kit (Sangon Biotech, C601) to insert the fragments into both the pGADT7 (AD) vector and the pAbAi bait vector. The recombinant plasmids were confirmed using Sanger sequencing (BGI), exhibiting 100% sequence identity (BLASTn E-value < 1e-50).
Self-activation assay and AbA threshold assessment
-
Ten μg of recombinant pAbAi plasmid was subjected to digestion with BstBI (NEB, R0515S) at 37 °C, and linearization was verified as complete using a 1% agarose gel (120 V, 30 min). Y1H Gold receptor cells (Clontech, 630489) were transformed using the LiAc/SS Carrier DNA technique and subsequently plated on SD/-Ura agar at 30 °C for 72 h. Five individual colonies were randomly selected for colony PCR validation using primers AbAi-F/R, achieving a 100% positive rate. The inhibitory concentration was established using gradient AbA screening (0–600 ng/mL). We seeded positive strains in SD/-Ura liquid medium (OD600 = 0.8) with serial AbA concentrations and grew them at 30 °C with shaking for 48–72 h.
Verification of transcription factor-promoter relationships
-
pGADT7-StMYB76/StMYB13 was co-converted with pAbAi-StLS-P1/P2/P3 into Y1H Gold, spliced, and transformed by the PEG/LiAc technique, then plated on SD/-Leu/-Ura solid medium at 30 °C for 48 h. Following the confirmation of positive clones via colony PCR, a logarithmic phase bacterial solution (OD600 = 0.8 ± 0.05) was subjected to a 10-fold gradient dilution (0−103), and subsequently spotted onto AbA-containing screening medium (SD/-Leu/-Ura + 600 ng/mL AbA). The vacant vector, pGADT7, served as a negative control, whereas pGADT7-StMYB76/StMYB13 functioned as a positive control.
Functional validation of StMYB transcription factors
-
Utilizing the combined approach of virus-induced gene silencing (VIGS), and transient overexpression, the function of StMYB76/StMYB13 in the control of (−)-pulegone biosynthesis was systematically elucidated. The experiments utilized the transient transformation system of S. tenuifolia, optimized in the laboratory[16]: specific silencing fragments (300–350 bp, Tm value 55 ± 2 °C, GC content 45–55%) were designed for the R2R3-MYB structural domains of StMYB76 (Sch000018969) and StMYB13 (Sch000028205), and these silencing fragments were expressed via Gateway. Cloned into the pTRV2 vector via Gateway® technology (Thermo Fisher) at 55%, the ORF was fully driven by the CaMV 35S promoter, while the overexpression vector pHREAC, which includes the GFP reporter gene, was created using the Golden Gate assembly technique (NEB) (see Supplementary Table S2 for the primers). The recombinant plasmid was electrotransformed into Agrobacterium tumefaciens GV3101 and grown in YEP medium (50 μg/mL rifampicin + kanamycin) at 28 °C until an OD600 of 0.8 ± 0.05 was achieved. The bacterial solution was resuspended using a permeabilization buffer (10 mM/L MES, 10 mM/L MgCl2, 200 µM/L AS) and subsequently infiltrated into S. tenuifolia leaves with a needle-free syringe.
Analysis of gene expression profiles
-
Utilizing the S. tenuifolia endogenous reference gene Actin as a baseline, customized primers were developed via Primer-BLAST (NCBI) to target key enzyme genes involved in the (−)-pulegone production pathway (StLS, StL3OH, StIPR, StIPD, StPR) and the regulatory factors StMYB76/StMYB13. The specifications for primer design include a product length of 80–150 bp, a Tm value of 60 ± 1 °C, and an amplification efficiency of 90%–110% (R2 > 0.99 as confirmed by the standard curve); detailed sequences can be found in the attached Supplementary Table S3. Total RNA (RIN ≥ 8.5) was obtained via the FastPure® Plant Total RNA Isolation Kit (Vazyme, RC401). cDNA was synthesized using HiScript III Reverse Transcriptase (Vazyme, R323-01). qRT-PCR was conducted utilizing ChamQ Blue Universal SYBR Master Mix (Vazyme, Q711), with the reaction protocol outlined in Supplementary Table S4. Relative expression was quantified using the 2−ΔΔCᴛ technique, with three biological and technical replicates (n = 9) established.
Monoterpene metabolite analysis
-
Leaf tissue (0.1 g) was precisely weighed and added to a 2 mL centrifuge tube containing 1.5 mL of camphor internal standard (1 μg·mL−1, Sigma, C0407). The cells were disrupted in a grinder (60 Hz, 60 s), and the supernatant was collected after centrifugation at 12,000 × g for 5 min. Subsequently, the supernatant underwent dehydration with anhydrous Na2SO4 and was filtered through a 0.22 μm filter membrane. Each sample was replicated three times. An Agilent 8860-GC/MS system (with an HP-5MS capillary column: 30 m × 0.25 mm × 0.25 μm) was employed, and the chromatographic separation and data processing techniques were based on established methodologies[22].
Data integration and analysis
-
Statistical significance was determined using paired comparison plots in Origin 2021 software (OriginLab, USA), with thresholds set at p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***). Gene expression and metabolite data were analyzed by two-tailed Student's t-tests (α = 0.05) with Benjamini-Hochberg correction for multiple testing. Data are presented as mean ± SD (n = 9; three biological replicates × three technical replicates). Promoter element distributions and phylogenetic analysis were evaluated using descriptive statistics.
-
Essential transcription factors governing StLS from the S. tenuifolia genome were identified using the cross-species motif matching technique. Utilizing the PlantTFDB binding motif prediction module within TBtools, the binding motifs of A. thaliana transcription factors (TFs) were correlated with those of S. tenuifolia, resulting in the identification of S. tenuifolia-specific binding motifs for its transcription factors. The StLS promoter region was analyzed using the FIMO tool to identify significant binding sites (p < 1 × 10−4) based on this motif library. A total of 1,192 notable binding sites were discovered, relating to transcription factors from the MYB, WRKY, AP2, and other families (see Supplementary Table S5 for details). The regulatory network of StLS and transcription factors was established using Cytoscape software, with core regulators identified using degree and betweenness centrality analysis, and the interactions were visualized (Fig. 1a).
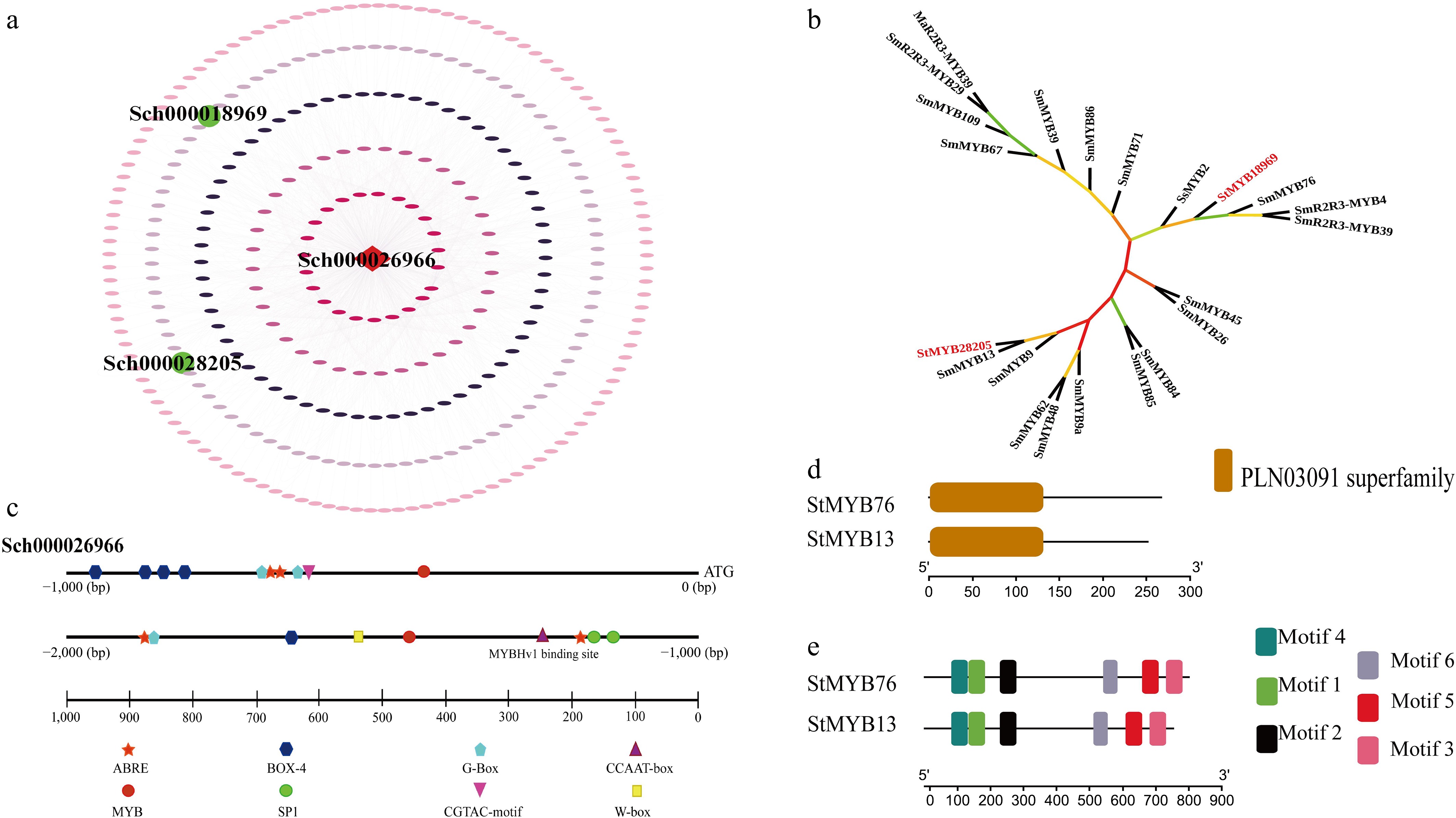
Figure 1.
Screening and identification of StMYB transcription factors. (a) The regulatory network schematic illustrates the functional core of StLS-interacting transcription factors. The regulatory network was established utilizing 1,192 binding sites identified by the FIMO tool, with green nodes denoting StMYB transcription factors and square nodes signifying the target gene StLS. (b) Phylogenetic analysis of StMYBs. The maximum likelihood method (bootstrap = 1,000) demonstrated that StMYB76/StMYB13 constitutes a distinct evolutionary branch in conjunction with Salvia miltiorrhiza SmMYB76/StMYB13, thus affirming the conserved terpenoid regulatory function within the Labiatae family. Evolutionary distances suggest that both display a strong affinity for MYBs linked to mint monoterpene synthesis (branch length < 0.2). (c) Distribution map of cis-elements in the StLS promoter. Three categories of functional modules were identified within 2,000 bp of the promoter region: metabolic regulatory elements (MYB-Hv1, G-box), stress-responsive elements (ABRE, MBS), and cross-species conserved motifs exhibiting greater than 75% homology to barley MYB-Hv1. (d), (e) Structural domain and motif composition of StMYBs. Analysis of conserved structural domains indicated that both possess a characteristic R2R3-MYB structural domain at the N-terminus (PLN03091 superfamily, E-value < 1 × 10−15). Six conserved motifs were identified using MEME, with motif 1 (N-terminal α-helix), and motif 4 (C-terminal transcriptional activation domain) forming the functional core.
The subsequent functional analysis of the StLS promoter identified essential regulatory modules (Fig. 1c and Supplementary Table S6). Bioinformatic analysis found important elements related to metabolism, like MYB-Hv1 (CAACGG), and G-box (CACGTT), as well as elements that respond to stress (ABRE, MBS) and light (G-box) in the promoter region. However, their biological functions need to be tested through experiments. Nonetheless, their biological activities necessitate experimental verification. Homology analysis indicated that StMYB76 and StMYB13 exhibit sequence conservation with barley (Hordeum vulgare) MYB-Hv1 (Gene ID: 123400161). The structural analysis of StMYB genes revealed (Fig. 1d–e) that StMYB76 (801 bp CDS) and StMYB13 (753 bp CDS) encode 267 and 251 amino acids, with isoelectric points of 5.10 and 5.11, and molecular weights of 65.53 and 59.91 kDa, respectively. Meme motif analysis indicated that the N-terminal region of the two possessed the PLN03091 distinctive R2R3 structural domain (motif 1/4, E-value < 1 × 10−15). Phylogenetic analysis indicated that both formed a distinct evolutionary branch (bootstrap = 1,000) with Salvia miltiorrhiza SmMYB76/SmMYB13, demonstrating a significant level of conservation in terpenoid regulatory activities within the Labiatae family (Fig. 1b).
Promoter areas of StMYBs interacting with StLS
-
This study established a yeast one-hybrid validation system to elucidate the regulatory mechanism of StMYBs on the StLS promoter, based on the distribution characteristics of cis-acting regions (Fig. 2a). The StLS promoter segments P1–P3 were inserted into the pAbAi bait vector by Nimble Cloning, and self-activation was confirmed to be absent at a dose of 600 ng/mL AbA (Fig. 2b and Supplementary Fig. S1). Following this, pGADT7-StMYBs effector vectors were co-transformed with each bait vector into the Y1HGold strain and subsequently introduced into the AbA-containing screening system after initial screening in SD/-Ura/-Leu medium. The results indicated that the StMYB76 and StMYB13 recombinant strains demonstrated notable proliferation features in the P2 zone media (Fig. 2c–e); however, no growth signals were observed in the empty vector control. The P2 region contains the conserved MYBHV1 cis-element (CCAAT-box), suggesting that this transcription factor activates reporter gene expression by specifically recognizing this regulatory motif, thereby demonstrating its direct binding properties to the StLS promoter.
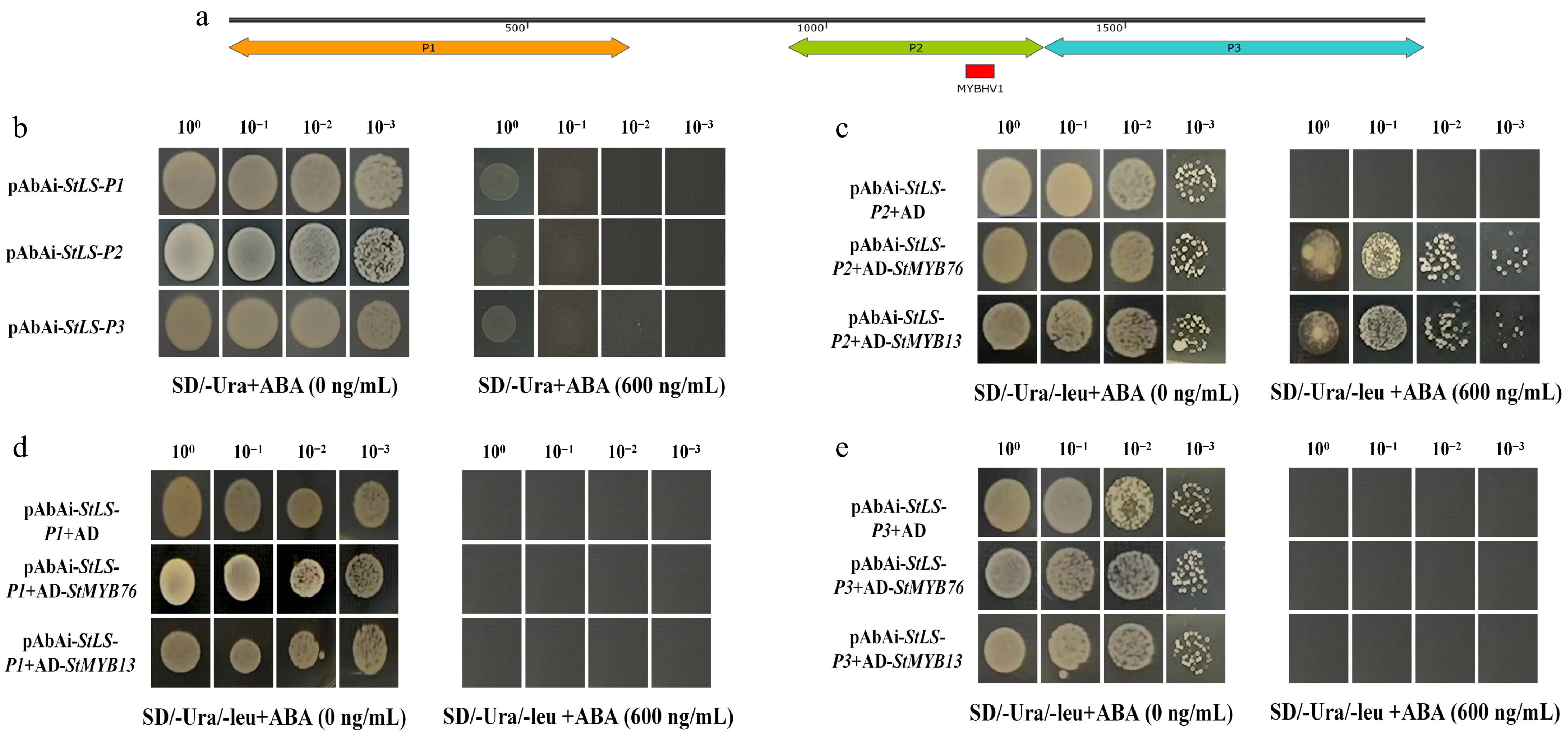
Figure 2.
Using a yeast one-hybrid system, transcription factor-promoter interactions are validated. (a) Diagrammatic representation of the StLS gene's shortened 2,000 bp promoter region upstream of the ATG start site, which includes the P1–P3 functional segments. (b) The self-activating effect of the decoy vector pAbAi-StLS-P1/P2/P3 was detected; at a concentration of 600 ng/mL AbA, no non-specific activation was seen. (c)–(e) StMYB transcription factors' binding to various promoter segments are characterized. StMYB76 and StMYB13 specifically activate the reporter system that contains the P2 fragment, while the negative control shows no growth signal.
Molecular mechanism of StMYBs in regulating the monoterpene synthesis pathway
-
This study employed virus-induced gene silencing (VIGS) technology to develop the pTRV-TFs recombinant vector to elucidate the biological activities of StMYBs, and confirmed the efficacy of gene silencing by RT-qPCR. As illustrated in Fig. 3a and b, the transcript levels of StMYB13/76 in the experimental group were markedly diminished in comparison to the TRV empty vector control (CK) (p < 0.05), signifying the successful development of VIGS. Quantitative analysis demonstrated no significant difference (p = 0.32, Student's t-test) in baseline expression levels between StMYB13 (1.00 ± 0.15) and StMYB76 (0.92 ± 0.11) in wild-type plants (Fig. 3a, CK group), thereby affirming their functional equivalency. Subsequent investigation revealed that the suppression of StMYBs expression resulted in a concurrent reduction in the mRNA levels of the downstream target gene StLS (Fig. 3b), demonstrating its positive regulatory characteristics. The expression of crucial genes in the monoterpene synthesis route, StL3OH, StIPD, and StPR, was considerably down-regulated, while StIPR remained unaffected, indicating that StMYBs preferentially control the pathway node genes to alter metabolic processes (Fig. 3b). GC-MS analysis (Fig. 3c) revealed that the (−)-pulegone concentration did not differ statistically between the experimental group (CK-StMYBs) and the control group, even though the silencing of StMYBs markedly reduced the expression of essential enzyme genes. This finding indicates that StMYBs can influence monoterpene biosynthesis by modulating the expression of genes at critical points in the process; nevertheless, their silencing did not result in substantial alterations in end-product content, highlighting the intricacy of metabolic homeostasis regulation.
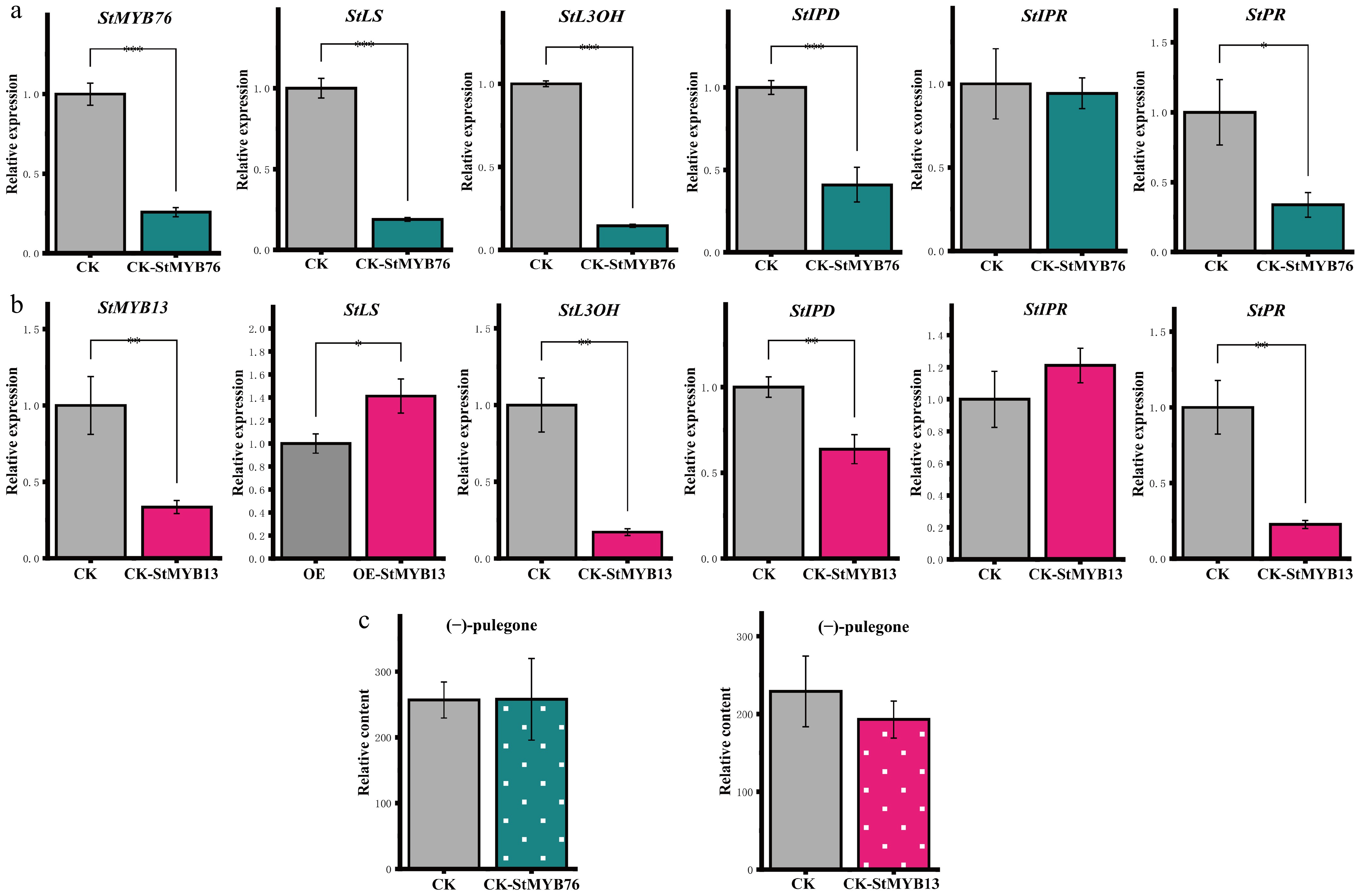
Figure 3.
Impact of StMYBs gene silencing on the (−)-pulegone biosynthetic pathway. (a), (b) The silencing of the StMYB76/StMYB13 gene leads to changes in the expression levels of critical genes in the (−)-pulegone biosynthesis pathway. (c) Changes in the concentration of (−)-pulegone monoterpene in S. tenuifolia following the silencing of StMYBs. * p < 0.05, ** p < 0.01, *** p < 0.001, 'ns' denotes not significant.
StMYBs positively regulate the biosynthetic pathway of (−)-pulegone.
-
This work further confirmed the actions of StMYBs through an investigation of the gene silencing system, and a transient overexpression system. The RT-qPCR results indicated (Fig. 4a, b) that the overexpression of StMYBs in S. tenuifolia leaves greatly enhanced the transcription of the StLS gene (p < 0.001) and upregulated the expression of essential genes in the monoterpene biosynthesis pathway. This phenomenon contrasted with the silencing experiments, demonstrating its positive regulatory function. GC-MS examination revealed (Fig. 4c) that the concentration of (−)-pulegone in the overexpressing plants was markedly elevated compared to the wild type (CK) (p < 0.001), establishing an expression-level-dependent pattern association with alterations in gene expression patterns. The results demonstrated that StMYBs facilitate the buildup of (−)-pulegone by activating StLS expression and upregulating essential enzyme-encoding genes in the menthane monoterpene biosynthesis pathway. The consistency of this regulation pattern at the transcriptional level offers fresh insights into elucidating the transcriptional regulatory network of terpenoid secondary metabolism, although the VIGS results showed some discrepancies between gene expression and metabolite data.
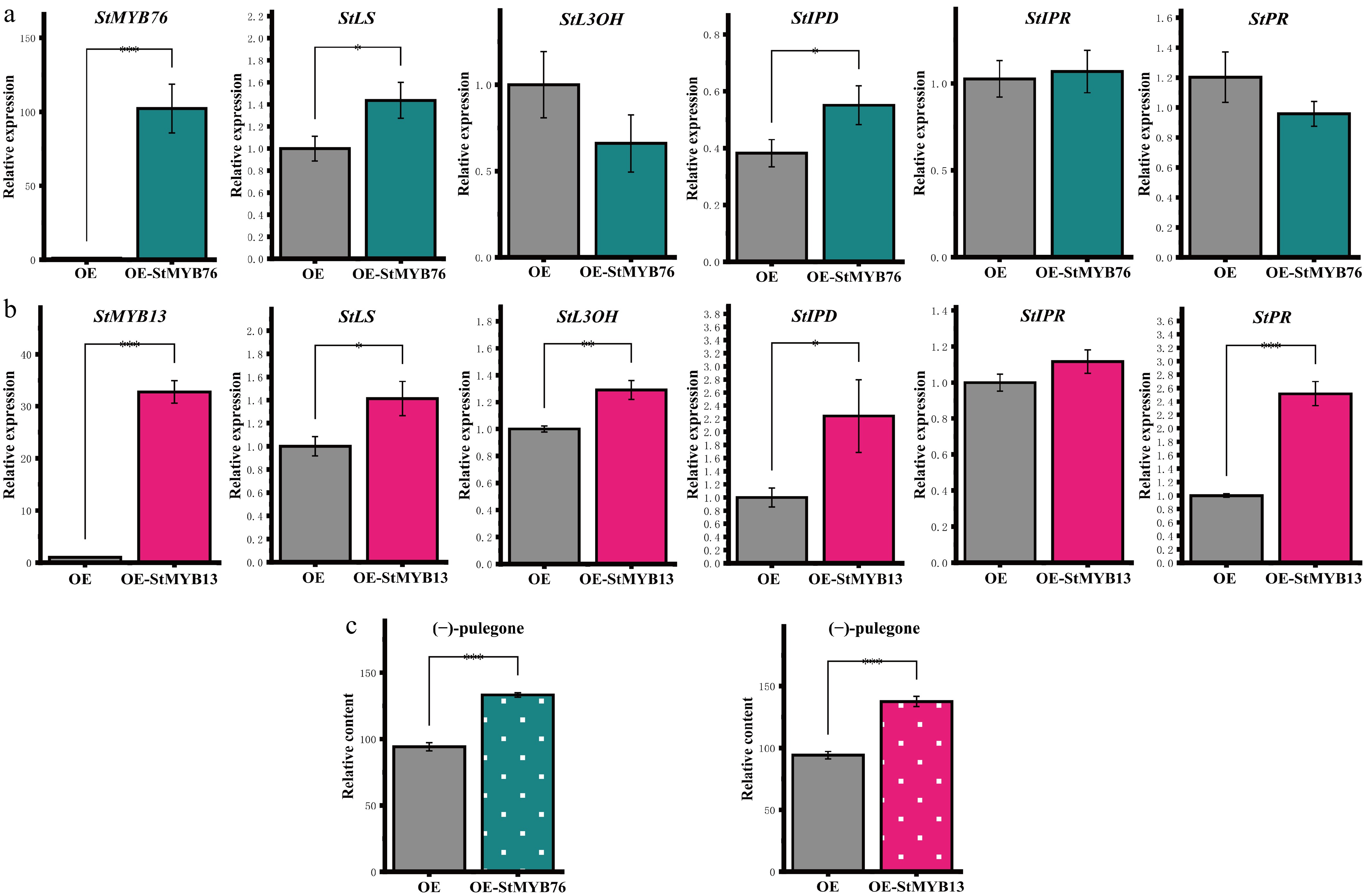
Figure 4.
Regulatory effects of StMYBs overexpression on the (−)-pulegone biosynthesis pathway. (a), (b) The figure illustrates the changes in the expression levels of StMYB76/StMYB13 and critical genes within the (−)-pulegone biosynthesis pathway. (c) The overexpression of StMYBs leads to changes in the concentration of (−)-pulegone monoterpene in S. tenuifolia. * p < 0.05, ** p < 0.01, *** p < 0.001, 'ns' denotes not significant.
-
Terpenoids are crucial in Labiatae species as key elements of plant primary and secondary metabolic pathways, and their oxygenated monoterpene derivative (−)-pulegone holds significant biological relevance. This compound constitutes the primary active ingredient (30%–50%) in the volatile oils of S. tenuifolia, Mentha haplocalyx, and Mosla spp. It is also recognized in drug discovery for its notable anti-inflammatory, antiviral, and neuroleptic properties[23]. This molecule has gained prominence in pharmacological development owing to its exceptional anti-inflammatory, antiviral, and neuroleptic characteristics[24,25]. Recent studies indicate that examining the biosynthetic pathway of (−)-pulegone and its transcriptional regulation can elucidate the molecular foundations of secondary metabolism in Labiatae, while also offering theoretical support for the targeted enhancement of essential oil constituents and the optimization of industrial production systems.
Transcription factors, as the fundamental regulatory components of gene expression, dynamically modulate the transcriptional activity of target genes by specifically recognizing cis-acting regions. In plant secondary metabolism, such regulatory proteins function as molecular switches for metabolite synthesis by precisely controlling genes that encode essential enzymes of metabolic pathways[26]. Representative instances comprise strawberry FaMYB63, which meticulously modulates the biosynthesis of eugenol by bi-directionally influencing the expression dynamics of secondary metabolic gene clusters (FaEGS1/2, FaCAD1)[27], and the SmMYC2-SmMYB36 regulatory axis in Salvia miltiorrhiza facilitates the simultaneous co-accumulation of phenolic acids and tanshinones through the cascade activation of SmGGPPS1 expression[28]. Despite the conserved roles of R2R3-MYB family members in regulating terpene metabolism, substantial knowledge gaps persist regarding the upstream regulatory network of S. tenuifolia StLS genes. The mechanisms of their transcriptional activation, and the regulatory hierarchy remain inadequately defined, presenting a crucial opportunity to delineate the molecular regulatory framework of monoterpene synthesis in Labiatae.
R2R3-MYB transcription factors regulate target genes specifically through the synergistic interplay of their N-terminal conserved DNA-binding domain (R2R3-MYB repeat), and the C-terminal variable functional domain[29]. This modular structure imparts functional versatility to the members of this family, making them functionally diverse in the regulation of plant development and metabolism. It is significant that, despite the variations in regulatory targets of R2R3-MYB proteins among different species, their DNA-binding motifs remain highly conserved throughout these species. Using Pistacia chinensis as a case study, PcMYB113 markedly increases anthocyanin accumulation in the seed coat and leaves of Arabidopsis via heterologous expression through specific interaction with PcF3H promoter elements[30]; Erigeron breviscapus EbMYBP1 positively modulates flavonoid biosynthesis genes by binding to cis-regulatory elements of flavonoid synthesis genes, including EbFLS2 and EbCHS, thereby enhancing the flavonol metabolic pathway[31]. These instances demonstrate that the molecular mechanisms of the R2R3-MYB family are conserved across species in the regulation of secondary metabolic pathways, thereby offering a theoretical framework for predicting the metabolic networks of medicinal plants through the use of regulatory elements derived from model organisms, and establishing an evolutionary basis for the development of artificial regulatory modules in synthetic biology.
It is demonstrated that StMYB13 and StMYB76 cooperatively maintain (−)-pulegone metabolic homeostasis through functional redundancy, evidenced by three interconnected tiers of evidence: constitutive expression equivalence, synchronized perturbation responses, and metabolite homeostasis preservation. Specifically:
(1) Their basal expression showed no significant difference in wild-type plants (Fig. 3a)
(2) VIGS-mediated silencing induced synergistic downregulation (p < 0.001), concomitantly suppressing downstream genes StLS, StL3OH, and StIPD—while overexpression coordinately enhanced their transcription (Figs 3b, 4b)
3. Despite significant gene suppression under silencing, (−)-pulegone content remained stable (Fig. 3c); conversely, overexpression elevated its accumulation by 2.3-fold (p < 0.001; Fig. 4c)
Mechanistically, both factors specifically bound the MYB-Hv1 element (CAACGG) within the StLS promoter P2 region (Fig. 2c–e), and selectively regulated downstream genes (StL3OH, StIPD) via conserved CCAAT-box motifs in their promoters (Supplementary Table S7). Notably, StIPR—lacking this motif—remained unaffected (Fig. 3b), revealing hierarchical regulation:
StMYB13/76 act as master regulators directly activating StLS, subsequently coordinating pathway progression through homologous cis-element recognition and metabolite-mediated signaling cascades (Fig. 5).
This compensatory mechanism mirrors PnMYB1/PnMYB4-modulated saponin biosynthesis in Panax ginseng[32], providing novel targets for terpenoid metabolic engineering in Lamiaceae.
MYB family transcription factors serve a pivotal function in plant secondary metabolic pathways by dynamically constructing modular regulatory networks. This functional variety demonstrates considerable environmental adaptation and molecular plasticity across several species. Using Fragaria × ananassa flavonoid synthesis as an illustration, FaMYB5 functions not only as a monomeric activator for anthocyanin and proanthocyanidin synthesis but also assembles an MBW ternary complex with the bHLH-type protein FaEGL3 and the WD40-repeat protein FaLWD1. This complex enhances the chromatin accessibility of target genes (e.g., FaANS, FaDFR) through the cooperative recruitment of histone acetylases, resulting in a transcriptional activation efficiency that is increased by 3–5 times compared to the monomeric model[33,34]. This complex-mediated synergistic regulation mechanism elucidates the molecular foundation for the functional diversification of MYB proteins via conformational alterations. Conversely, investigations into the volatile monoterpene metabolism of Antirrhinum majus revealed the dynamic reconfiguration of the MYB regulatory network in response to environmental stimuli: in the basal condition, AmMYB24 forms an inhibitory complex with AmMYC2, thereby diminishing the transcriptional activity of the ocimene synthase gene (AmOCS) through competitive binding; however, upon blue light induction, AmMYB24 undergoes phosphorylation of its C-terminal structural domain and subsequently associates with the MYBCOREATCYCB1 motif of the photoreceptor AmCRY24, resulting in the formation of a light-responsive activation complex that amplifies the expression of AmOCS by eightfold, and triggers the rapid release of ocimene[35]. Collectively, these cases demonstrate that MYB transcription factors facilitate exact metabolic regulation across species and pathways via mechanisms including conformational plasticity, post-translational modification, and heterologous protein interactions, offering a novel perspective for examining the molecular architecture of plant environmental adaptive evolution.
MYB family transcription factors serve as essential regulatory components of the plant environmental stress response network, governing secondary metabolism and stress tolerance through the integration of many signaling pathways. Research indicates that this protein family can selectively identify cis-acting regions in stress-responsive gene promoters (e.g., MBS, CCAAT-box) and dynamically modulate the expression of downstream targets[36]. Salt stress promotes the accumulation of the Salix matsudana SmMYB1R1-L protein, which markedly improves plant salt tolerance by activating the SmEXPA13 gene[37].
In the realm of secondary metabolic regulation, the MYB interaction network in Artemisia annua exemplifies functional complexity and provides a framework to interpret environmental integration in S. tenuifolia. Specifically, AaMYB108—a core activator of artemisinin biosynthesis—promotes pathway assembly by forming a complex with the gland-specific WRKY factor AaGSW1[38,39]. This process is dynamically constrained by: (1) AaCOP1-mediated ubiquitination and degradation of AaMYB108 under darkness, blocking AaGSW1 interaction; (2) AaJAZ8 inhibition through competitive binding to AaMYB108, disrupting complex formation[40]. This light-hormone cross-regulatory paradigm, revealing spatiotemporal precision in MYB control, parallels our identification of light/hormone-responsive elements (G-box, ABRE) in the StLS promoter (Fig. 1c), suggesting conserved environmental sensing mechanisms. Unlike AaMYB108's dual regulation, however, StMYBs may exhibit distinct stress-responsive partnerships (e.g., bZIPs), warranting future exploration (Fig. 5).
-
This study demonstrates that StMYB13 and StMYB76 are essential in regulating the biosynthesis of (−)-pulegone in S. tenuifolia. Tests in yeast were used a method to turn off genes with viruses, and did short-term experiments to show that these transcription factors directly connect with the StLS promoter and control the activity of important genes needed to produce (−)-pulegone. Our findings improve our understanding of how plants control the production of monoterpenes and provide a scientific basis for possibly enhancing (−)-pulegone production through genetic engineering.
Finding that StMYB13 and StMYB76 are important controllers of (−)-pulegone production highlights the complicated and overlapping ways that regulation works. The similar functions of StMYB13 and StMYB76 suggest that different regulatory pathways might help keep the (−)-pulegone production process stable. This insight has important implications for developing strategies to enhance the production of this valuable monoterpene in S. tenuifolia, and potentially other plant species.
Future research will focus on understanding how StMYB13, StMYB76, and other transcription factors interact, as well as looking into how light and hormones affect the StLS promoter. We will also look at how the flow of substances and the activity of enzymes change due to StMYB regulation to fully understand how (−)-pulegone is made. These studies will provide a foundation for creating planned methods to improve the production of (−)-pulegone and other terpenoids in medicinal plants.
This work was supported by the National Natural Science Foundation of China (Grant No. 82373978), the Natural Science Foundation of Jiangsu Province (Grant No. BK20231307), the First-Class Discipline 'Leadership Plan' Scientific Research Special Project of Nanjing University of Chinese Medicine (Grant No. ZYXYL2024-002), and the Nanjing University of Chinese Medicine School's Signature Talent Cultivation Program (Grant No. RC202411).
-
The authors confirm contributions to the paper as follows: study conception and design: Wu Q, Liu C; data collection: Wang X, Dang J; analysis and interpretation of results: Li D, Pan M; draft manuscript preparation: Wu Q, Liu C, Yang F. All authors have read and agreed to the final published version of the manuscript.
-
All data analyzed during this study are available from the corresponding author upon reasonable request.
-
The authors declare that they have no conflict of interest.
- Supplementary Table S1 Yeast single hybrid primer sequence.
- Supplementary Table S2 Primer sequences for the StMYBs silencing fragment and overexpression.
- Supplementary Table S3 RT-qPCR specific primer sequences of genes.
- Supplementary Table S4 Reaction procedure for RT-qPCR.
- Supplementary Table S5 Prediction of inter-gene regulatory relationship based on transcription factor binding StLS Motif.
- Supplementary Table S6 Distribution of cis-acting elements of key genes in the monoterpene biosynthetic pathway.
- Supplementary Table S7 The distribution of StMYBs promoter cis-regulatory element.
- Supplementary Fig. S1 Validation of the self-activation of pAbAi-StLS-P1/P2/P3 in yeast. Yeast cells were diluted with distilled water (10−1 to 10−3) and grown on SD/−Ura medium containing 100, 200, 400 ng/mL aureobasidin A (AbA).
- Copyright: © 2025 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Wang X, Dang J, Li D, Pan M, Yang F, et al. 2025. Screening and first validation of MYB transcription factors influencing the biosynthesis of (−)-pulegone monoterpene constituents in Schizonepeta tenuifolia Briq.. Medicinal Plant Biology 4: e038 doi: 10.48130/mpb-0025-0035
Screening and first validation of MYB transcription factors influencing the biosynthesis of (−)-pulegone monoterpene constituents in Schizonepeta tenuifolia Briq.
- Received: 14 May 2025
- Revised: 26 July 2025
- Accepted: 25 August 2025
- Published online: 01 December 2025
Abstract: This study investigates the secondary metabolism regulatory mechanism of the Labiatae medicinal plant Schizonepeta tenuifolia Briq. The monoterpene synthase StLS catalyzes the manufacture of (−)-pulegone, a distinctive monoterpene active component of this species, and its expression level directly impacts secondary metabolite accumulation. Due to the limited knowledge of the StLS transcriptional regulatory network, this study employed a multi-omics combined analysis to investigate the role of the R2R3-MYB transcription factor in plant metabolic regulation. It was discovered that StMYB13 and StMYB76 are regulatory elements after a deep analysis of the Arabidopsis thaliana transcriptome database using bioinformatics screening. The yeast one-hybrid system (Y1H) showed that StMYB13/StMYB76 could specifically bind CAACGG in the StLS promoter region from −1,234 to −1,274 bp. Virus-induced gene silencing (VIGS) and transient overexpression were used to unveil the StMYB transcription factor's monoterpene metabolic pathway regulatory network. The transcription factor controlled the monoterpene synthase StLS's transcriptional activity and downstream biosynthetic enzyme-encoding gene expression. The silencing of StMYB genes led to a dramatic reduction (p < 0.001) in the expression of the pivotal gene StIPD, whereas the overexpression system caused a substantial elevation in its mRNA levels. Further analysis demonstrated that this regulatory process exhibits a distinct expression-level-dependent pattern: (−)-pulegone content was significantly elevated in overexpression lines, whereas no statistically significant difference in (−)-pulegone accumulation was observed in silenced groups. These findings revealed StMYBs' transcriptional regulation mechanism and provided molecular evidence for the analysis of S. tenuifolia's volatile oil synthesis nodes. This discovery deepened the monoterpene biosynthesis pathway and laid the theoretical groundwork for metabolic engineering and molecular-assisted breeding systems of S. tenuifolia.
-
Key words:
- Schizonepeta tenuifolia Briq. /
- StMYBs /
- StLS /
- (−)-pulegone monoterpene