-
During domestication and breeding, bread wheat (Triticum aestivum) underwent extensive artificial selection. The bread wheat genome is the result of two polyploidy events. Tetraploid wheat (AABB, 2n = 4x = 28) is the result of an ancient polyploidy event between Triticum urartu (AA, 2n = 2x = 14) and Aegilops speltoides (BB, 2n = 2x = 14). The second polyploidy event occurred approximately 10,000 years ago in the Fertile Crescent between presumably cultivated tetraploid wheat and wild Aegilops tauschii (DD, 2n = 2x = 14)[1], and the resulting hybrid (AABBDD, 2n = 6x = 42) is the bread wheat used today. Archaeobotanical evidence indicates that both the transitions from diploid wild einkorn (Triticum monococcum ssp aegilopoides; AmAm) to domesticated diploid forms (T. monococcum ssp monococcum) and from wild tetraploid emmer wheat (Triticum turgidum ssp dicoccoides; BBAA) to domesticated tetraploid cultivars (T. turgidum ssp dicoccum) are associated with a trend toward larger grains. For hexaploid wheat, grain size continues to be an important selection trait. Today, wheat grains provide one-fifth of the calories and a substantial proportion of protein consumed by humans[2]. The main attributes of grain size, including grain width, grain length, and length/width ratio, are greatly different among the hexaploid wheat varieties[3]. Compared to the early common wheat landraces, such as T. aestivum ssp. spelta and ssp. macha, grain width was dramatically increased and grain length was decreased in the early stages of the bread wheat breeding process[3]. Therefore, the wheat grain shape has changed from long and thin to shorter and wider in modern bread wheat varieties[3]. In addition, as wheat is a staple commercial crop, its market requirements also distinctly influence the selection of grain size and shape. Important attributes, such as grain density, uniformity, end-use quality, protein content, and trace elements are also associated with grain size, and directly influence grain yield and flour quality, which determines the market value of wheat grains[4].
Fertilization initiates grain development. Generally, the development process of wheat grains is divided into three stages: the cell division and expansion stage that occurs 1−14 d after anthesis (DAA), during which the basic structure of wheat grains are formed; the grain filling stage 14−28 DAA, during which the accumulation of starch and protein occurs and the grain dry weight increases by approximately twofold[5]; and grain maturation and desiccation that occurs from 28 DAA until complete maturation[6]. During the last stage, the grain filling slows and is complete by approximately 35 DAA. The fresh weight subsequently decreases rapidly until ~42 DAA due to desiccation[6].
During wheat grain development, gene expression, and translation profiles change dynamically[6−9]. A substantial change in the transcriptome occurs during grain development[6]. Moreover, different subgenomes of wheat are unbalanced at both the transcriptional and translational levels[7, 8]. Dynamic chromatin landscape changes during embryogenesis is correlated with biased gene expression among homeolog gene triads and divergent expression among three subgenomes[9].
Although grain size is a key yield component in wheat[10], the large polyploid wheat genome substantially impedes linkage mapping of genes regulating grain size and identification of molecular functions. More information on wheat grain size control has been obtained by population genetic studies, in which putative regulatory genes or QTLs are found by genetic mapping or genome-wide association studies (GWASs). With the use of molecular markers, many QTLs related to grain size or grain weight have been identified on all 21 wheat chromosomes in recent decades, and utilized in breeding[11−19]. Recently, with the rapid development of wheat genome sequencing, genetic mapping has been utilized to identify causal genes for grain traits[20, 21]. A subset of these causal genes was independently confirmed by gene knockout and/or overexpression methods. There are excellent recent reviews centered on the molecular regulation pathways of grain size in rice and wheat[22−26]. In this review, we instead focused on the source-flow-sink system to summarize the genes involved in photosynthesis, and carbohydrate transportation, as well as their roles in grain size determination. We also investigated genes directly affect grain size and grain filling, with particular attention given to the regulation of starch and seed storage protein (SSP) biosynthesis (Fig. 1). We also discussed difficulties and feasible strategies for grain size improvement.
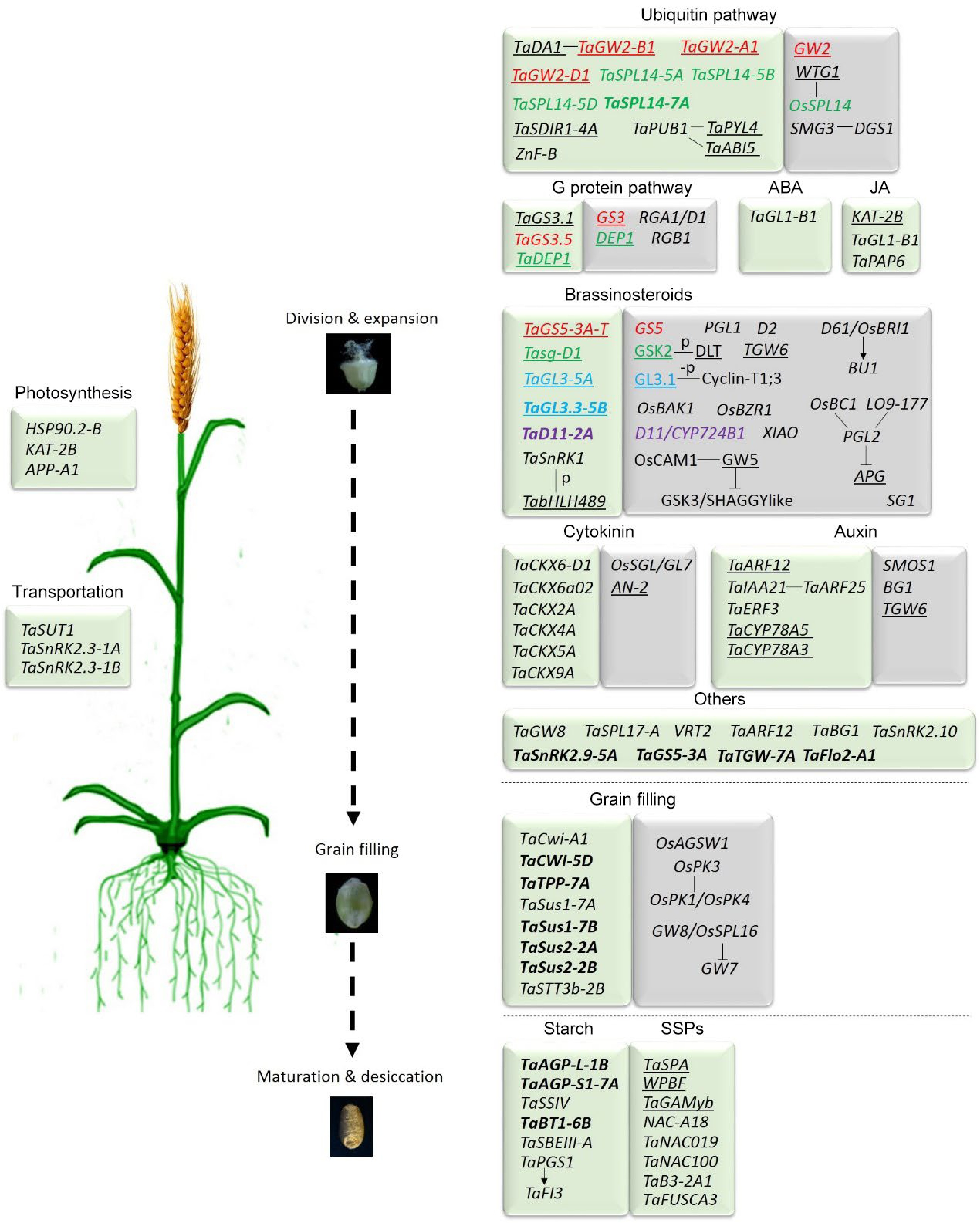
Figure 1.
Genes and genetic pathways regulating grain size in bread wheat and rice. The components without underlines are positive regulators of grain size, and those with underlines are negative regulators. The short connecting lines represent the proteins that physically interact. The bond genes were selected during wheat breeding. The genes with the same colors are homologous gene between wheat and rice. References for the individual genes are listed in Supplemental Tables S1 & S2 for wheat and rice genes, respectively.
-
During photosynthesis, solar energy is utilized and drives the accumulation of plant biomass and sink storage, such as in crop grains[27]. Genes related to chloroplast and chlorophyll synthesis play important roles in photosynthetic efficiency and capacity and influence wheat grain size. Recently, three genes related to chlorophyll content and the photosynthetic rate have been identified as grain size regulators[20, 21, 28]. Map-based cloning identified keto-acyl thiolase 2B (KAT-2B) as the causal gene for a large-grain mutant. Analysis of KAT-2B overexpressing lines revealed that KAT-2B can positively regulate grain size, weight, and yield. In addition, the leaf area and chlorophyll content increase in KAT-2B-overexpressing lines, indicating that the photosynthetic capacity is enhanced[21]. HSP90.2-B is the causal gene for the CO2 ASSIMILATION RATE AND KERNEL-ENHANCED1 (CAKE1) gene cloned from durum wheat[20]. It encodes a cytosolic molecular chaperone folding nascent preprotein that is crucial for the localization of nuclear-encoded photosynthesis units in chloroplasts. Mutation of HSP90.2-B led to a decreased photosynthetic rate, decreased grain size, and an 80% decrease in grain yield[20]. Similarly, the CAKE2 gene has been mapped to ASPARTIC PROTEASE1 (APP-A1), and a premature stop mutation increases the photosynthetic rate, grain size, and grain weight. The APP-A1 protein can degrade PsbO which is an important member of photosystem II[28]. By producing carbohydrates, photosynthesis is the first step in the source-flow-sink. Identification of genes that enhance photosynthetic capacity and efficiency is a constructive and effective way to increase sink storage at the source.
-
The long-distance transport of photosynthetic products is an essential bridge connecting the source and the sink and is key to the growth and development of sink tissues, such as grains and fruits. Sucrose is one of the main photosynthetic products and is remobilized by sucrose transporters (SUTs) in the vasculature leading to the grain. Among the 14 wheat sucrose transporter genes TaSUTs, TaSUT1 homeologous genes on chromosomes 4A, 4B, and 4D are expressed predominantly in the stem, leaf sheath, rachis, lemma, and developing grain, and their expression levels are significantly correlated with grain size and weight[29]. The peduncle is the key connection between the spike and the stem and plays important roles in the transportation of water-soluble carbohydrates[30]. Association analysis revealed that TaSnRK2.3-1A and TaSnRK2.3-1B are significantly associated with the length of the peduncle and penultimate node, as well as with grain size and weight. Additionally, one haplotype of TaSnRK2.3-1B was shown to be associated with increased grain weight and decreased plant height, whereas another was associated with increased grain weight, increased stem water-soluble carbohydrate contents, and decreased plant height; thus these two haplotypes were considered elite[31].
-
Genes involved in grain development may directly determine grain size. Cloning and functional analysis of such genes have been a key research direction for grain size[24, 26]. Since the large polyploid wheat genome substantially hinders the use of forward genetic methods and molecular functional identification, many grain size regulatory genes in wheat were studied by reverse genetic analysis of genes homologous to rice grain size regulators or by association analysis, which revealed homologs of rice grain size regulators. Here, we organized the genes related to grain size in wheat according to known regulatory pathways.
Ubiquitin-proteasome pathway
-
Ubiquitin‒proteasome degradation is extensively utilized by plants to regulate development. Several grain-size regulatory genes were found to encode ubiquitin‒proteasome degradation pathway proteins, including E2, E3 and their regulators. GRAIN WIDTH2 (GW2) encodes a RING-type E3 ubiquitin ligase in rice[32]. Downregulation of three homeologous TaGW2 genes by RNA interference (RNAi) leads to significant increases in grain width and grain weight[33]. TaGW2-B1 has stronger effects on grain size regulation than TaGW2-D1 does, and TaGW2-B1 and TaGW2-D1 double gene knockdown plants have significantly larger grain sizes than single gene knockdown plants[34]. TaDA1 negatively regulates grain size in wheat. Downregulation of TaDA1 increases grain size and grain weight. In addition, TaDA1-A physically interacts with TaGW2-B1 and subsequently affects grain size in an additive way[35].
Salt and drought-induced RING finger1 (SDIR1), is a RING-type E3 ubiquitin ligase that has been identified as a key player in the response to both salinity and drought stresses[36]. The grain size of the TaSDIR1-4A-silenced lines increased, and one haplotype showed a significant association with increased grain size and greater grain weight[37]. Another E3 ligase TaPUB1 can interact with TaPYL4 and TaABI5, both of which are components of the ABA signaling pathway, and subsequently induces the degradation of TaPYL4 and TaABI5. TaPUB1 overexpressing plants exhibit larger grains, whereas the corresponding RNAi lines exhibit smaller grains[38]. Similarly, ZnF-B, which is also a RING-type E3 ligase, positively regulates grain size[39].
Rice OsOTUB1 is a deubiquitinating enzyme, whose mutant exhibits decreased grain width and thickness[40]. Rice OsSPL14 is degraded by OsOTUB1, which controls grain development[41]. In wheat, upon cleavage by miR156, the triple mutant of TaSPL14-5A, -5B and -5D exhibited reduced grain size and weight[42]. For another TaSPL14 member TaSPL14-7A, the preferred haplotypes are correlated with increased grain size and weight, which underwent positive selection during wheat breeding worldwide[43].
G protein signaling
-
The G protein signaling pathway plays important roles in various developmental processes in plants and animals. Generally, the G protein complex has three subunits, Gα, Gβ and Gγ. In rice, both the Gα-encoding gene RGA1 and the Gβ-encoding gene RGB1 positively regulate grain size[44−46]. Rice GRAIN SIZE3 (GS3), which encodes a noncanonical Gγ subunit, was identified as the causal gene that determines grain length[47, 48]. Alternative splicing of the heterotrimeric G-protein encoding gene TaGS3 controls wheat grain size. Different alternative splicing isoforms of TaGS3 have different functions related to grain size control[49]. The splicing isoforms TaGS3.2–3.4 have no effect on grain size, whereas the splicing isoform TaGS3.1 overexpression line has deduced grain weight and grain length. In contrast, upregulation of TaGS3.5 can significantly increase grain weight and grain length[49].
Rice DENSE AND ERECT PANICLE1 (DEP1) shares homology with rice GS3, and a gain-of-function mutant of DEP1 with a truncated ORS domain shows decreased grain size, grain weight, and dense and erect panicles and increased grain number per panicle and grain yield per plant[50]. The TaDEP1 locus was also identified within a key QTL correlated with grain thickness by GWAS. It has been proposed that the function of TaDEP1 is conserved, and rice DEP1 is a regulator of grain weight and size[51].
Phytohormones
-
The phytohormone brassinosteroids (BRs) regulate multiple plant growth and development processes[23, 52]. The BR signaling component rice OsGSK2, which encodes an ortholog of Arabidopsis BIN2, negatively regulates grain size[53], and phosphorylates DWARF AND LOW-TILLERING (DLT, also known as OsGRAS32, DWARF62, and GRAIN SIZE6)[54]. Wheat Tasg-D1 is an ortholog of OsGSK2 and plays a conserved role in negatively regulating grain size and weight[55].
The causal gene of a rice grain length QTL, qGL3/GL3.1, is OsPPKL1, which encodes a Ser/Thr phosphatase with a Kelch-like repeat domain[56−58]. OsPPKL1 dephosphorylates cyclin-T1;3 and subsequently regulates cell division in spikelets. A reduction in OsPPKL1 activity results in increased grain length and grain yield without affecting grain quality. In fact, the qgl3 allele has been utilized in breeding[56]. The wheat homologous gene TaGL3-5A harbors a SNP in the 11th exon that leads to an amino acid change, and the resulting TaGL3-5A-G allele is correlated with increased grain size and grain weight[59]. OsPPKL2 and OsPPKL3 are two OsPPKL1 homologs in the rice genome[56]. OsPPKL1 and OsPPKL3 decrease grain length, whereas OsPPKL2 increases grain length. TaGL3.3-5B is the homolog of OsPPKL3, with one allele associated with large grain size and under selection in breeding[60]. Taken together, the results of these studies in rice have shown that BR signaling plays important roles in regulating grain development, not only in grain size and grain filling but also grain number per panicle. Wheat homologous genes are frequently found in seed trait QTLs, suggesting possible conservative roles in seed development. Wheat TaD11-2A positively regulates the endogenous BR content, and TaD11-2A mutants exhibit dwarfism and small grains. In addition, wheat TaD11-2A is significantly associated with plant height, grain size, and grain yield per plant, with an elite haplotype that has been positively selected during wheat breeding[61].
In addition to BR, auxin also plays important roles in determinating grain size. Rice THOUSAND-GRAIN WEIGHT 6 (TGW6) encodes indole-3-acetic acid (IAA)-glucose hydrolase, which catalyzes the transformation of IAA-glucose back to IAA and regulates early endosperm development. TGW6 negatively regulates grain size and weight without affecting grain quality[62]. Wheat TaIAA21 negatively regulates grain length, width, and weight. TaIAA21 can physically interact with TaARF25 and subsequently positively regulates grain size and weight in tetraploid wheat. TaARF25 further positively regulates the expression of TaERF3, whose mutants exhibit reduced grain size and weight. Therefore, TaIAA21 is a negative regulator of grain size and weight via interaction with the TaARF25–TaERF3 module in wheat[63].
Overexpression of TaCYP78A5 specifically in the maternal integument significantly enhanced grain size, grain weight, and grain yield per plant in field trials. TaCYP78A5 is involved in the auxin synthesis pathway and increases auxin accumulation in the ovary. TaCYP78A5 expression prolongs the proliferation of maternal epidermal cells and increases the number of seed coat cells through auxin mediated flowering delay, thereby promoting grain enlargement[64]. Similarly, upregulation of TaCYP78A3 expression can increase grain size by regulating the number of cells in the seed coat[65].
Rice OsCKX2 controls grain number[66], and several wheat homologs of OsCKX2 are associated with grain size traits. An 18-bp deletion in the 2nd intron of TaCKX6-D1[67] and a novel allele of TaCKX6a02[68] are associated with grain size and weight. In addition, TaCKX2A_2, TaCKX4A_2, TaCKX5A_3, and TaCKX9A_2 are also significantly associated with increased grain weight in the Chinese wheat micro-core collection (MCC) and a landrace wheat population[69].
PYLs, which are abscisic acid (ABA) receptors, respond to plant drought stress. Overexpression of TaPYL1-1B improved drought tolerance and increased grain size and weight. The elite allele TaPYL1-1B confers not only drought tolerance but also increases grain size and yield[70].
TaGL1-B1 has been identified as a regulator of grain length by GWAS. A mutant of TaGL1-B1 exhibits dramatically shorter grain lengths, and TaGL1-B1-overexpressing lines exhibit increased grain lengths. Moreover, TaGL1-B1 physically interacts with TaPAP6 which positively regulates grain length. The content of jasmonic acid (JA) is significantly increased in both the TaGL1-B1- and TaPAP6-overexpressing lines, suggesting that the JA pathway may influence grain length[71].
Other regulators of grain size
-
Among the genes affecting grain size, some are not associated with the abovementioned pathway. Wheat TaGW8 is an ortholog of rice GW8/OsSPL16 that increases grain size and grain yield by promoting cell division and grain filling[72]. The TaGW8-B1a haplotype is associated with greater grain size and greater grain weight, which is in contrast with the TaGW8-B1b haplotype, which lacks a 276-bp indel in the first intron[73]. Using GWAS, wheat TaSPL17 was identified as a positive regulator of grain size and grain number by regulating spikelet and floret meristem development, which in turn leads to increased grain yield per plant[74]. An elite TaSPL17 haplotype results in wider and longer grains and greater grain weight, accompanied by more spikelets per spike[74].
Polish wheat (Triticum turgidum ssp. polonicum, previously known as Triticum polonicum) is a subspecies of tetraploid wheat with long glumes, lemmas and grains. Map-based cloning identified a MADS-box TF encoding gene VEGETATIVE TO REPRODUCTIVE TRANSITION2 (VRT2) as the causal gene underlying the long-glume P1 locus[75, 76]. Ectopic expression of VRT-A2 in hexaploid bread wheat causes longer glumes and grains, leading to larger grain sizes, confirming that the VRT-A2 expression level affects glume and seed length[75].
Two members of the wheat TaSnRK2 family are reportedly involved in the regulation of grain size[77, 78]. Two favored TaSnRK2.9-5A haplotypes are associated with higher grain weight and have been positively selected in wheat breeding in China[77]. The TaSnRK2.10 haplotype is significantly associated with larger grain size[78].
Using GWAS, wheat TaARF12 was identified as a plant height and spike length regulator[51]. Knockout of TaARF12 can significantly increase grain number per spike and result in higher grain weight[79].
Wheat TaGS5-3A has two alleles that are discriminated by a G/T SNP in the coding region, and the TaGS5-3A-T allele was associated with larger grain size and higher grain weight in a population analysis[80, 81]. Schizosaccharomyces pombe expressing TaGS5-3A-T was shown to exhibit higher total serine carboxypeptidase activity than TaGS5-3A-G[80]. In addition, transgenic experiments in rice showed that TaGS5-3A-T overexpression lines are superior to TaGS5-3A-G overexpression lines in grain width, length and weight[80]. Furthermore, population analysis indicated that TaGS5-3A-T is a favored haplotype under positive selection[80].
TaTGW-7A transcription levels were found to be positively associated with grain size and grain weight using the Chinese mini-core collection with 501 varieties. The elite allele TaTGW-7Aa exhibits higher expression levels and has been under strong and positive selection during wheat breeding[82].
TaFlo2-A1, an ortholog of rice FLOURY ENDOSPERM2, is associated with grain size and weight in bread wheat[83]. Population genetics analysis was used to identify the TaFlo2-A1b allele as a positively selected haplotype[83].
Ectopic expression of TaBG1, the homologous gene of rice BIG GRAIN1, leads to a larger grain size, but transgenic lines exhibit a trade-off in grain number per plant, resulting in no significant overall increase in gain yield[84].
-
The grain filling stage is a key step that determines grain weight and grain size. During this stage, starch, protein, and other organic matter produced through photosynthesis are assimilated in wheat grains. The duration and velocity of grain filling are two factors that affect grain size and weight. Cell wall invertases catalyze the irreversible hydrolysis of sucrose to glucose and fructose and play a key role in sink strength. Based on the nucleotide polymorphisms at the TaCwi-A1 locus, one haplotype was found to be associated with lower grain weight, whereas another was found to be associated with higher grain weight[85]. Similarly, a TaCWI-5D haplotype was shown to be distinctly associated with greater grain weight and is strongly selected during wheat polyploidization, domestication, and breeding[86]. TaSus converts sucrose to fructose and UDP-glucose during seed development. Both TaSus1 and TaSus2 are associated with grain size regulation[87, 88]. The trehalose-6-phosphate phosphatase-encoding gene, TaTPP-7A, was identified as a putative grain size regulator that is expressed specifically in developing grains and significantly influences grain filling. Identification of TaTPP-7A-overexpressing wheat plants confirmed that TaTPP-7A is a key regulator of source-flow-sink interactions and sucrose allocation. Population genetic analysis revealed that TaTPP-7A is a domestication- and breeding-selected target gene[89].
As an important posttranslational protein modification, asparagine N-glycosylation, which is catalyzed by the oligo saccharyl transferase (OST) complex, plays essential roles in plant development[90]. STAUROSPORINE AND TEMPERATURE SENSITIVE3 (STT3) is one of the subunits in the OST complex that catalyzes the subunit encoding oligosaccharyl transferase[91]. Overexpression of TaSTT3b-2B distinctly increases grain size and weight by increasing the expression of genes encoding starch synthase, and sucrose synthase[92].
-
Starch and seed storage proteins (SSPs) are the main components of wheat grains; they account for approximately 70% and 13%, respectively, of the weight of ripen grains and play essential roles in determining grain weight, size and quality[93]. Several key genes involved in starch synthesis, including ADP-glucose pyrophosphorylase (AGPase), starch synthase (SS), starch-branching enzyme (SBE), and debranching enzyme (DBE), have been studied in wheat. AGPase catalyzes the conversion of glucose-1-Pi to ADP-glucose, which is a rate-limiting enzyme in the starch synthesis pathway[94]. The AGPase tetrameric complex comprises two large subunits (LSUs) and two small subunits (SSUs)[94]. A haplotype of TaAGP-L-1B and a haplotype of TaAGP-S1-7A are associated with larger grain size and greater weight, respectively, in modern wheat cultivars, and their additive effects on the determination of grain size have been detected in wheat populations[95]. Starch synthesis IV (SSIV) is an important starch synthesis isoform, and its mutation reduces the number of starch granules per chloroplast in wheat. An elite SSIV allele is significantly associated with grain size and weight[96]. BRITTLE1 (BT1) is responsible for the transport of ADP-glucose which is essential for starch synthesis in Arabidopsis[97]. Knocking down the expression of TaBT1-6B in wheat decreases grain size and starch content[98]. TaBT1-6B-Hap1 and TaBT1-6B-Hap2 are two useful alleles under selection during modern wheat breeding[98]. In addition, a polymorphism in a starch-debranching enzyme-encoding gene, TaSBEIII-A, is associated with grain size and grain weight[99]. T. aestivum Positive Regulator of Grain Size 1 (TaPGS1) is a bHLH transcription factor (TF) encoding gene that is strongly expressed in the endosperm at 10-20 d postanthesis in wheat. Ectopic expression of TaPGS1 increases grain weight (up to 13.81% in wheat and 18.55% in rice) and grain size with decreasing starch granule size (smaller and tighter) in the proteinaceous matrix[100]. Furthermore, TaPGS1 can regulate the expression of TaFI3 by binding to the E-box motif in its promoter region.
SSPs are also essential determinants not only of wheat grain weight but also of the end-use quality of wheat flour[101, 102]. SSPs include glutenin and gliadins. Based on their mobilities in sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‒PAGE), glutenin can be further divided into high-molecular-weight glutenin subunits (HMW-GSs) and low-molecular-weight glutenin subunits (LMW-GSs)[102]. Gliadins consist of α/β-, γ-, ω- and δ-gliadin based on their different primary structures[103]. The expression and abundance of SSPs are regulated by the complicated network of TFs[104]. Wheat storage protein activator (TaSPA), a bZIP TF, is expressed specifically in grains and binds to the GCN4-like binding motif in the promoter of LMWG-1D1[105]. Wheat prolamin-box binding factor (WPBF) is a DNA-binding TF with one finger (Dof) that can bind to the promoter regions of gliadin-encoding genes, and LMW-GS and HMW-GS-encoding TaGlu-1By8 and -1Dx2 loci[106, 107]. TaGAMyb, which is a companion of the GCN5-like histone acetyltransferase, regulates the expression of the HMW-GS 1Dy encoding gene by binding to its promoter directly[108].
The starch content in grains directly determines grain size and weight. However, the increase in SSPs usually occurs at the cost of grain weight. Therefore, the coordination of the starch content and SSPs content is crucial for balancing of grain size, weight and quality. Three NAC (NAM-ATAF-CUC) TFs, NAC-A18, TaNAC019 and TaNAC100, are coordinators of starch and SSP accumulation[109−111]. Ectopic expression of NAC-A18 in rice dramatically decreased the starch content and increased the SSP content, grain size and weight. Moreover, NAC-A18 can upregulate the expression of TaLMW-D6 and TaLMW-D1 by binding directly to their promoters and suppressing the expression of TaGBSSI-A1 and TaGBSSI-A2. TaNAC019 binds to specific motifs in the promoters of TaGlu-1Bx, TaGlu-1By, TaGlu-1Dx, TaSuSy1, and TaSSIIa and positively regulates their expression. Compared to those of the wild type, the triple mutant of three homeologous TaNAC019 exhibits lower gluten contents and smaller grain sizes with shorter grain widths and lengths[109]. In contrast, TaNAC100 negatively regulates the expression of GLU-1 through binding to its promoter. Overexpression of TaNAC100 suppresses Bx14, By15 and Dx2 expression and further decreases their protein levels[110]. Similarly, the TF TaB3-2A1 binds to the cis-element CCRM1-1 in the promoter of GLU-1. TaB3-2A1 overexpression significantly reduces the contents of HMW-GSs and other seed storage proteins but increases the starch content, leading to increased grain size, including grain length and width[112]. Another B3-superfamily TF, TaFUSCA3, interacts with TaSPA and subsequently activates Bx7 by binding to its promoter[113].
Although the abovementioned genes independently and coordinately regulate the contents of starch and SSPs, the intricate mechanisms underlying their regulatory pathway still need to be determined to accumulate additional genetic resources for genome-wide target editing.
-
Grain size is a key agronomic trait in crop breeding. In recent decades, there has been exciting progress in research on grain size development in rice. Studies in wheat suggest that some grain size regulators have conserved functions in rice and other plants[32, 34, 35, 114−116]. However, many of the observed associations in wheat remain unconfirmed by experiments. Often, a homolog of the rice grain size regulatory gene is found in a grain trait QTL by GWAS in wheat. It is often difficult to experimentally validate whether a favorable haplotype indeed promotes a grain trait in wheat, as often only a single SNP exists between two haplotypes, and we still cannot edit the genome precisely to reconstruct the haplotype in the desired genetic background. On the other hand, overexpression of an allele could lead to misleading results. Because of these difficulties, we sometimes know much about a wheat homolog of a rice grain regulator, including its interacting proteins and binding promoters, in the case of TFs; however, no solid data confirming its role in regulating a seed trait, such as for its rice homolog, have yet been obtained. Therefore, our understanding of wheat grain traits heavily relies on knowledge from rice, and the proposed roles of genes often need validation. For grain trait regulators that are not conserved in rice or have not yet been found in rice, we know little about these regulators in wheat. In addition, as an allopolyploid crop, bread wheat exhibits complex genetic regulation. For example, the effect of TaGW2-B1 on grain size is significantly greater than that of TaGW2-D1, and the grain sizes of double mutants are substantially greater than those of single mutants[34]. The interactions among subgenomes need to be elucidated, especially by utilizing improved genetic transformation and editing technology.
Like for other crops, maintaining a balance in yield components are essential for improving wheat yield. Many seed trait QTLs or genes exhibit intensive genetic trade-offs between grain size/weight and seed number, which substantially impedes the utilization of these genes[117]. Only a few genes, such as TaGSNE in wheat and OsOTUB1, OsSGL, and OsAGSW1 in rice, have been found to simultaneously increase both grain size and number and hold promise to improve wheat yield[40, 118, 119]. Recently, overexpression of an α-expansin gene specifically in early developing grain led to a significant increase in grain size without a negative effect on grain number, resulting in an increase in yield under field conditions[120]. Similarly, targeted expression of TaCYP78A5 in integument also enhances grain weight without negative effects on grain number per spike. Therefore, modifying the expression of genes in specific organs, developmental periods, or biogenesis steps may be a feasible way to overcome the trade-off between the grain size/weight and grain number.
Additionally, grain size and weight usually have a negative correlation with grain quality, as indicated by the protein content and end-use quality[4]. Therefore, synergetic improvement of grain yield and end-use quality is also a goal for wheat breeding. In rice, both OsPPKL1 (qGL3/GL3.1) and TGW6 influence grain size but have no significant influence on grain quality[56, 62], which are potentially useful for improving grain weight and quality. Additionally, some TFs can regulate the contents of starch and SSPs by binding directly to the promoters of related genes, such as TaNAC019 andNAC-A18[109, 111]. Therefore, identifying additional genes that regulate grain size development and SSPs is still an effective way to coordinately improve grain size, weight, and quality.
-
The authors confirm contribution to the paper as follows: draft manuscript preparation: Wei B, Jiao Y. Both authors approved the final version of the manuscript.
-
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
This work was supported by the Key R&D Program of Shandong Province, China (ZR202211070163); the Shandong Provincial Natural Science Foundation (ZR2021ZD30 and ZR2022ZD22), the National Natural Science Foundation of China (32072061, 31921005 and 32230010), and National Key R&D Program of China (2019YFA0903900 and 2023YFE0101100).
-
The authors declare that they have no conflict of interest. Yuling Jiao is the Editorial Board member of Seed Biology who was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of this Editorial Board member and the research groups.
- Supplemental Tables S1 List of genes regulated grain size in wheat.
- Supplemental Tables S2 List of genes regulated grain size in rice.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press on behalf of Hainan Yazhou Bay Seed Laboratory. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Wei B, Jiao Y. 2024. Grain size control in wheat: toward a molecular understanding. Seed Biology 3: e007 doi: 10.48130/seedbio-0024-0007
Grain size control in wheat: toward a molecular understanding
- Received: 06 October 2023
- Revised: 20 March 2024
- Accepted: 19 April 2024
- Published online: 13 May 2024
Abstract: Grain size is a major determinant of bread wheat (Triticum aestivum) yield, which has a broad impact on worldwide food security. Not surprisingly, grain size underwent extensive artificial selection during wheat domestication and breeding. Recent advances in wheat molecular genetics and genomics have facilitated the elucidation of the molecular basis underlying grain size. Grain size determination is the cumulative result of source strength, photoassimilate remobilization, and sink strength. Here, we systematically review the recent progress in the cloning and molecular mechanisms of genes that regulate grain size in wheat following the source-to-sink flow. In addition, we discuss possible strategies for overcoming the trade-off between grain size and grain number, as well as synergetic improvement of grain yield and grain quality.
-
Key words:
- Grain /
- Grain size /
- Wheat /
- Yield














