-
The Khaplu valley is in Pakistan's Gilgit–Baltistan province located in the extreme north. Khaplu is the center of Ghanche district sourrounded by Ladakh's Leh district to the east (under the administration of India), Xinjiang province of China is to the northeast, the Skardu district and Kharmang is to the south, the Shigar district is to the northwest and the Indian territory of Ladakh is to the south. The mountains of Gilgit–Baltistan are located in three mountain ranges, the Karakorum, Himalayas and Hindu Kush therefore they are well renowned for their abundant biodiversity[1,2]. Mountain climate, dry climate, lowland climate and coastline climate are the four climatic zones in Pakistan. Gilgit Batistan is located in the mountain climatic region which has a cold, snowy and long winter season while a pleasant and brief summer season. Because the diversity of lichen species is massively greater in colder and moist areas, Pakistan's mountainous climate is favorable for their diversification[3]. These are complicated creatures that have a symbiotic relationship with a green alga (a photobiont) or a cyanobacterium and a fungus (a mycobiont) and they have piqued interest due to their location in the evolutionary chain of terrestrial plants[4,5]. Algae gives the fungus glucose and the mycobiont in return helps to expand the photobiont's geographical and ecological range as well as protecting and improving supply of water and nutrients. Lichens have adjusted to all types of natural environments and have worldwide localization[6]. Epiphytic lichens are being utilized as bioindicators for air pollution monitoring and detection. Lichens also have significant antimicrobial action which can be particularly useful in spoiling food and curing a range of illnesses caused by microbes[7,8].
In the past, there have been limited attempts in Pakistan to identify the variety of lichens using molecular methods such as the ITS of rDNA. The majority of Pakistan's lichen species are only superficially described based on micro and macromorphological characteristics. However, the lichen studies and biodiversity status of the Khaplu Valley have not been adequately examined thus far. DNA barcoding techniques are critical for an organism's identification and taxonomic placement. Protoparmeliopsis zareii, a new species that was not included in the checklist of Pakistan's lichen biota, has been discovered during special analyses of a recent collection of lichens and lichenicolous fungi from the Ghanche broq Khaplu Gilgit Baltistan. Recently, the first molecular data verifying the generic status of Protoparmeliopsis has been obtained[9,10]. By comparing the material to existing descriptions and DNA analysis especially the ITS and LSU regions of ribosomal DNA, the identity of the species has been further validated. The purpose of this work is to describe a new lichen genus for Pakistan, the detailed descriptions and illustrations of representative species along with a discussion are provided.
-
In order to better understand Pakistan's lichen biota, lichen specimens were collected in September 2021 from Ghanche broq, Khaplu, Gilgit Baltistan. At 4,600 m above sea level (a.s.l.), near Khaplu city, the study location is located 100 km to the east of Skardu. The most notable feature of Ghanche Broq is the breathtaking view of the four 8,000-m summits in the world as well as other well-known peaks. K2, the second-highest peak in the world, stands at 8,611 m, followed by Broad Peak (8,047 m), Gashabrum-4 (7,925 m), Gashabrum-3 (7,952 m), Hrnami Ka Peak (6,325 m), Tchogho Lingsa Peak (6,325 m), Gashabrum-2 (8,035 m), K6 (7,281 m), and K7 (6,934 m). This area encompasses 8,915 square kilometres in total[11]. Khaplu is a section of the Karakoram range and is located at 35.1611° N and 76.3319° E. In this region of the Krakoram range, which is situated between 2,600 and 5,200 m above sea level, there are dry temperate deciduous forests[12]. On a typical day in January, Khaplu has temperatures ranging from a high of −11 °C to a low of −21 °C. The studied site experiences its warmest temperatures in July, with highs of 26 °C and lows of 12 °C. In these areas, the average annual precipitation is 59.3 cm, with a humidity level of 57%.[12].
Morphological and chemical characterization
-
Under a stereomicroscope (Meiji Techno, EMZ-5TR, Japan) and a compound microscope (SWIFT M4000-D with a 9MP camera system), the specimens were analyzed macro- and micromorphologically. The thallus was cut out by hand in water for anatomical study. For each of the three samples, at least 20 measurements were taken. By performing spot tests with KOH (10%; K) and sodium hypochlorite solution (C), the secondary chemistry was examined. Utilizing Solvent System G and following industry standards, Thin Layer Chromatography was performed.
Molecular characterization and phylogenetic analysis
-
Using the 2% CTAB procedure, genomic DNA was extracted from dried materials[1]. Polymerase Chain Reaction (PCR) was used to amplify the ITS region (Internal Transcribed Spacer) and LSU (larger sub-unit) regions ribosomal DNA of the fungal equivalent of the lichen DNA using ITS1F/ITS4 and LR0R/LR5 primer pairs respectively[13]. Following the conditions: initial denaturation at 94 °C for 1 min, final denaturation at 94 °C for 1 min followed by 35 cycles of annealing at 53 °C for 1 min, initial extension at 72 °C for 1 min and a final extension at the same temperature for 7 min[14]. In 1.2% Agarose gel, the amplified DNA fragments were visible[15]. PCR products were sequenced by BGI, Hong Kong (China). Using the BioEdit sequence alignment editor v7.0.9.0, the forward and reverse sequences were put back together[16]. To align the sequences with other sequences obtained from NCBI GenBank, Clustal W was employed[17]. On the CIPRES Portal, the HYK + G + I model was selected using jModelTest[6,18]. Using RAxMLHPC2 v8.1.11 on CIPRES, a maximum likelihood analysis was implemented using rapid bootstrapping with 1000 iterations. The resulting tree was visualised with FigTree v1.4.3[3,19]. For the phylogenetic analysis of this species based on the ITS-rDNA marker, a total of 47 sequences belonging to different genera were included in the final alignment (Table 1).
Table 1. ITS sequences included in the recent phylogenetic analysis are mentioned with GenBank accession numbers and country of the taxa.
Lichen Location Accession number ITS Protoparmeliopsis achariana Austria AF070019 Protoparmeliopsis bipruinosa Austria AF159932 Protoparmeliopsis bolcana Ukraine MK672838 Protoparmeliopsis chejuensis South Korea MK672839
MK672840Protoparmeliopsis zareii Pakistan KHP05 ITS Lecanora garovaglii Austria
USA
USA
Poland
Poland
IranAF189718
KT453728
KU934537
MK084624
MK084626
MK672841Protoparmeliopsis kopachevskae Korea
South Korea
South KoreaMK672845
MK672846
MK672847Protoparmeliopsis laatokkensis China MN912366 Protoparmeliopsis muralis Austria
Romania
Germany
USA
Russia
PolandKC791770
KP059048
KT818623
KU934555
KU934560
KY379232Protoparmeliopsis nashii Austria AF159931 Protoparmeliopsis peltata USA
Iran
Kazakhstan
RussiaKT453722
KT453723
KU934746
KU934751Protoparmeliopsis pseudogyrophorica China MK672851 Protoparmeliopsis zareii Iran KP059049 Protoparmeliopsis sp. Russia KU934865 Protoparmelia badia Germany KF562191 Protoparmelia badia Austria AF070023 Lecanora sp China MW590816 Lecanora muralis Austria AF070015 Lecanora muralis Austria AF159922 Protoparmeliopsis laatokkensis China MT875031 Protoparmeliopsis laatokkensis China MN912366 Protoparmeliopsis nashii Czech Republic ON447553 Lecanora garovaglii Austria AF189718 Rhizoplaca peltata China AY530887 Rhizoplaca peltata China AY509802 Rhizoplaca melanophthalma USA JX948289 Rhizoplaca melanophthalma USA JX948294 Rhizoplaca subdiscrepans China AY509789 Rhizoplaca chrysoleuca USA HM577252 Lecanora perpruinosa Austria AF070025 Lecanora contractula Austria AF070032 Myriolecis contractula USA HQ650604 Lecanora intricata Austria AY398703 Lecanora intricata Austria AF070022 Lecanora polytropa USA HQ650643 Lecanora polytropa Korea DQ534470 Rhizocarpon geminatum China KP314320 Protoparmelia picea Germany KF562194 Protoparmelia montagnei Spain AF101277 Protoparmelia montagnei Spain AF101275 Lecanora achroa USA JN943715 Lecanora achroa USA JN943719 Lecanora tropica USA JN943718 Lecanora tropica USA JN943720 Lecanora caesiorubella USA JN943722 Lecanora caesiorubella USA JN943727 Lecanora farinacea Austria AY541261 Lecanora farinacea Austria AY541262 Lecanora flavopallida USA JN943724 Lecanora carpinea Austria AY541249 Lecanora carpinea Austria AY541248 Lecanora allophana Austria AF070031 Lecanora allophana Austria AF159939 Xanthoria alfredii Sweden EU681345 Xanthoria ulophyllodes Sweden EU681341 Xanthomendoza fallax Sweden EU681346 Oxneria fallax USA MZ922253 -
Protoparmeliopsis zareii S.Y. Kondr.S. J. (2012) Fig. 1
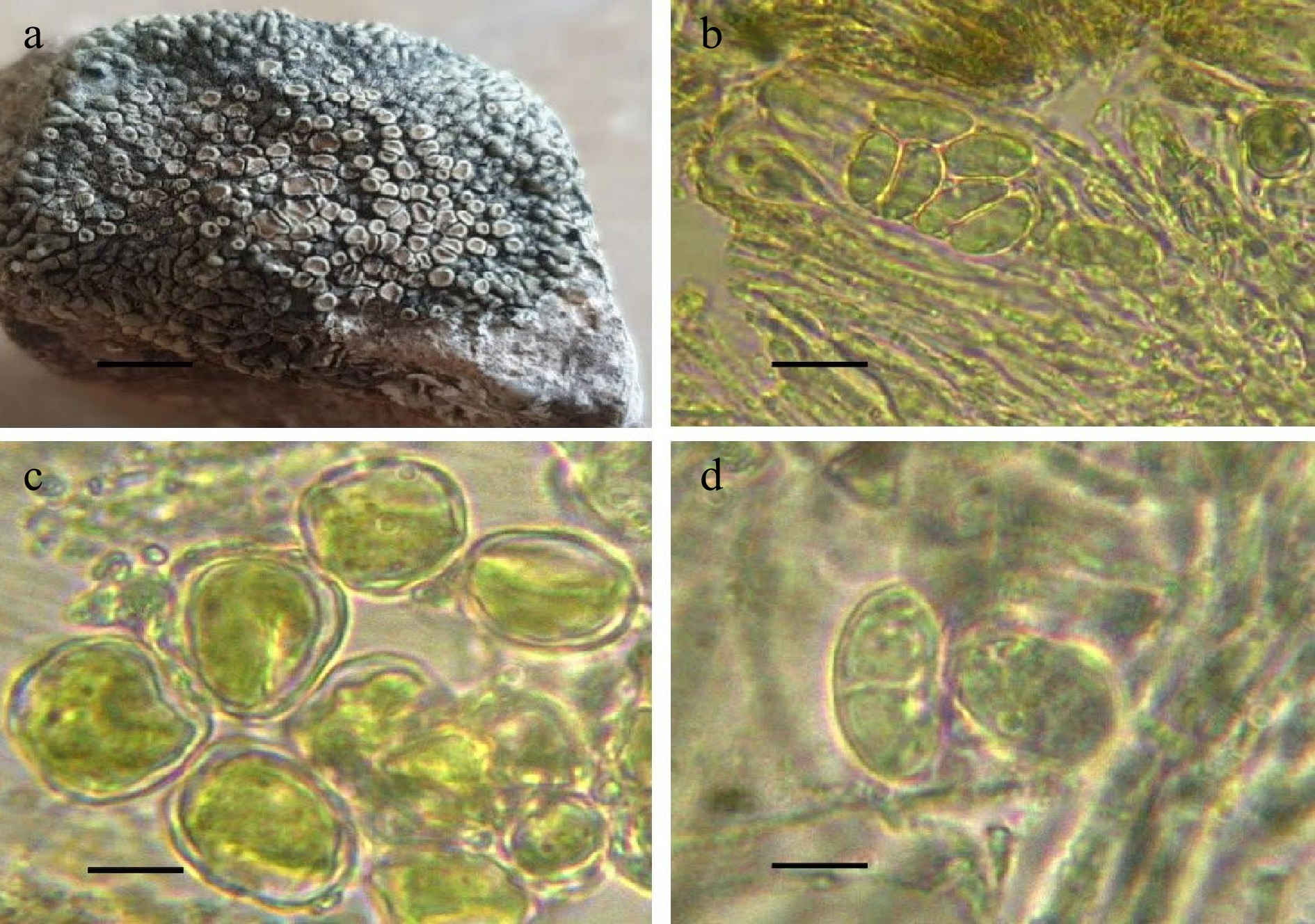
Figure 1.
Protoparmeliopsis zareii. (a) Thallus. (b) Ascus with ascospores. (c) Algal cells. (d) Ascospores. Scale bars: (a) = 1 cm, (b) = 10 μm, (c) = 100 μm, (d) = 5 μm.
Thallus Placodioid, 1.5–3.5 cm or wider, 0.5–2 mm or thicker in the center, forming compact rosettes, but frequently either confluent or dispersed and irregular, tightly to loosely linked; areolate to squamulose; central areoles (0.5–)0.7–1.5 mm wide/across; upper surface greenish-grayish often with whitish edges of thalline lobes and areoles; whitish reticulum on the upper surface of the central areoles; numerous apothecia immersed into thalline areoles; dull brown to greenish brown apothecium discs; prosoplectecnchymatous true exciple. The outermost edges, especially the extreme lobe ends, frequently have darker blue-green to black areoles. The lobes (peripheral) are approximately 4–5 mm long, 1–2 mm wide, and somewhat enlarged to 2.0–2.5 mm wide at the tips. The thallus is 1 mm thick in section; the cortical layer is 50 μm thick and uniformly split from the algal zone; the algal zone is 50 μm thick (possibly 200 μm thick) and the algal cluster is occasionally vertically extended to 200–250 μm thick.
Apothecia 0.5–1.5 mm diam., typically strongly elevated with relatively narrowing stipa to 1 mm long, ranging from buried into areoles to moderately verrucose before rising slightly but without distinguishable stipa (as in P. muralis); the disc is dull brown or greenish brown, and the thalline margin is greenish-grayish, rounded by whitish edges that are the same as the edges of the thalline lobes and areoles (frequently, young apothecia appear to be identical to thalline areoles); thalline exciple with cortical layer to 30–40 μm thick; true exciple relatively thin to 50(–100) μm broad in the lateral portion and quite thick (75–)100–200 μm thick in basal portion, thalline edge to 0.3–0.35 mm wide. lax; algal zone below true exciple in vertically elongated clusters to 50–200(–250) μm thick; epihymenium dull greyish or dull brownish with pigment granules, in K becoming hyaline; Hymenium 70–80 μm high; subhymenium 20–40 μm thick with multiple oil cells to 4.8–7.2 μm in diameter; asci 8–spored, 48–60 × 22–24 μm; ascospores hyaline, generally ovoid to broadly ellipsoid, slightly smaller (7.2–)10.8–13.2 × 7.2–9.6 μm in K while larger with huge oil droplet, (8.4)10.8–13.2(–14.4) × 8.4–9.6(–10.8) μm in water.
Spot tests: Thallus usually K-, C-, KC-, P-; cortex usually KC+ yellow; medulla usually KC- .
Material examined Pakistan: Gilgit-Baltistan: Ghanche, Khaplu, 35.1611° N, 76.3319° E; 3,900 m a.s.l., dry temperate area, cold desert, on rock; coll.: A. Bano (KHP-05), 20 September 2021.
Ecology: On silicate rocks beside Bromus tectorum L., Valeriana cymbicarpa C.A. Mey., and Malva sylvestris L. var. silvestris.
Distribution: Known so far from Ghanche Broq Khaplu, Gilgit Baltistan, Pakistan.
Phylogenetic analyses
-
The final dataset contained 605 locations. Rhizocarpon geminatum (L.) DC. was used to root the tree (Fig. 2). Protoparmelia complex, Lecanora complex, Lecanora complex, Rhizoplaca complex, Protoparmeliopsis complex, and Lecanora complex are the six clades that are represented in the tree. The specimen from Pakistan, designated Protoparmeliopsis zareii below, is grouped with the species from Iran (KP059049), both of which belong to clade V.
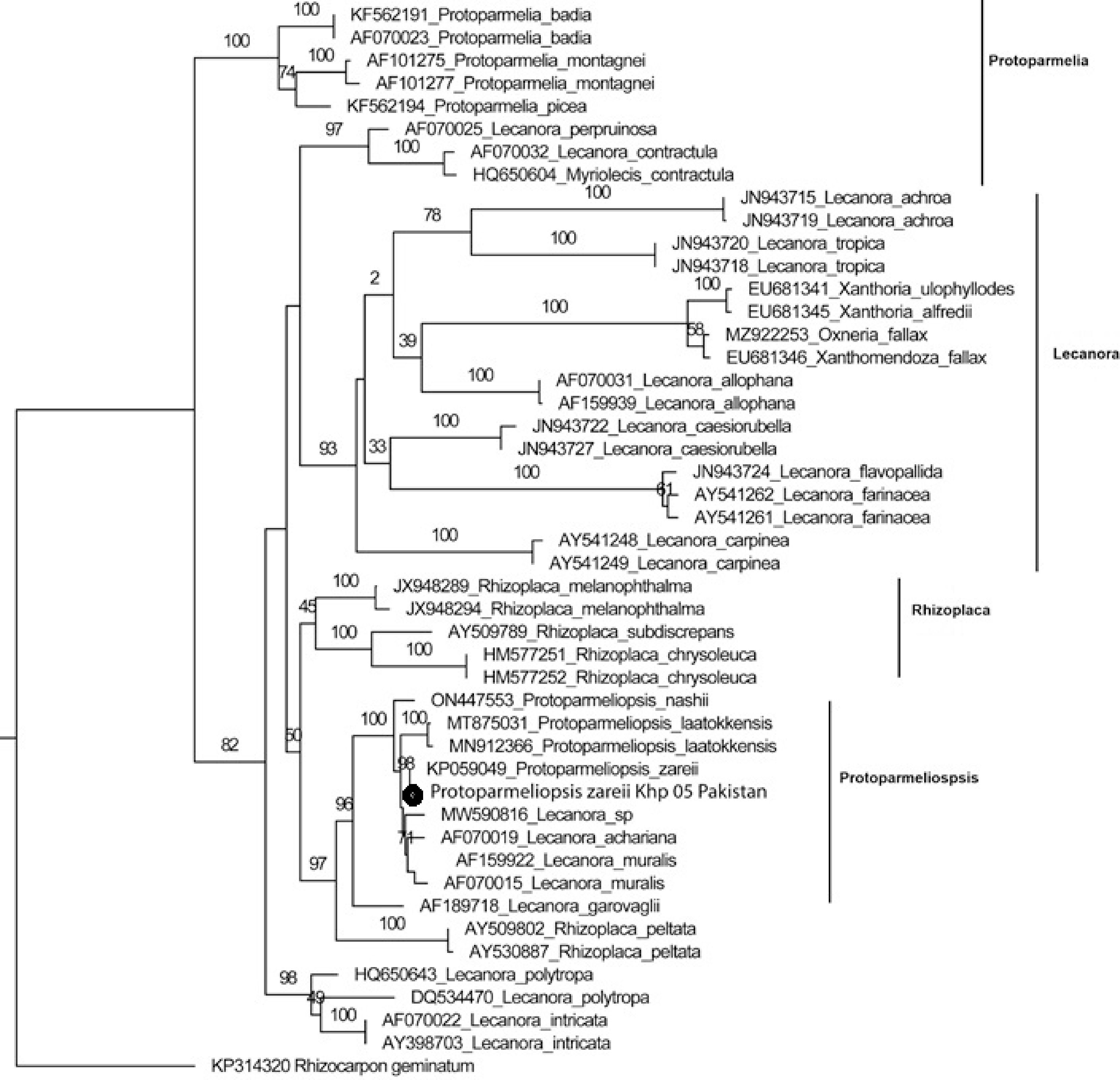
Figure 2.
The phylogenetic tree of the representatives of the family Lecanoraceae including genera Protoparmeliopsis, Lecanora, Rhizoplaca and Protoparmelia based ITS-rDNA marker sequences. All the genera are separated in the form of distinct clades in the tree. The maximum likelihood method is used in this phylogenetic analysis showing Protoparmeliopsis zareii is the same as the Iranian collection. Pakistani collection is labeled with a black circle in the figure.
For LSU marker analysis, 26 sequences have been extracted from the GenBank database and included in the final alignment (Table 2). The final dataset contained 2,317 locations and these sequences were recovered in major clades in the phylogenetic analysis (Fig. 3).
Table 2. LSU sequences used in the phylogentic analysis mentioned with acession numbers and type locality.
Lichen Location Accession number LSU Lecanora achroa Thailand
AustraliaJN939502
JN939527
JN939504Lecanora tropica Thailand
Fiji
KenyaJN939518
JN939533
JN939537Lecanora caesiorubella USA
Australia
AustraliaJN939509
JN939506
JN939508Lecanora farinacea Australia
AustraliaJN939511
JN939513Lecanora flavopallida Australia JN939514
JN939516Lecanora intricata Sweden DQ787345 Lecanora polytropa USA DQ986792 Lecanora sulphurea Sweden DQ787355 Lecanora carpinea Sweden DQ787363 Lecanora aff. Sweden AY853376 Lecanora campestris Sweden DQ787361 Lecnora contractula USA DQ986746 Lecanora perpruinosa Sweden DQ787343 Protoparmeliopsis achariana Sweden
USADQ787341
DQ973027Lecanora muralis Germany HQ660533 Lecanora muralis Sweden DQ787339 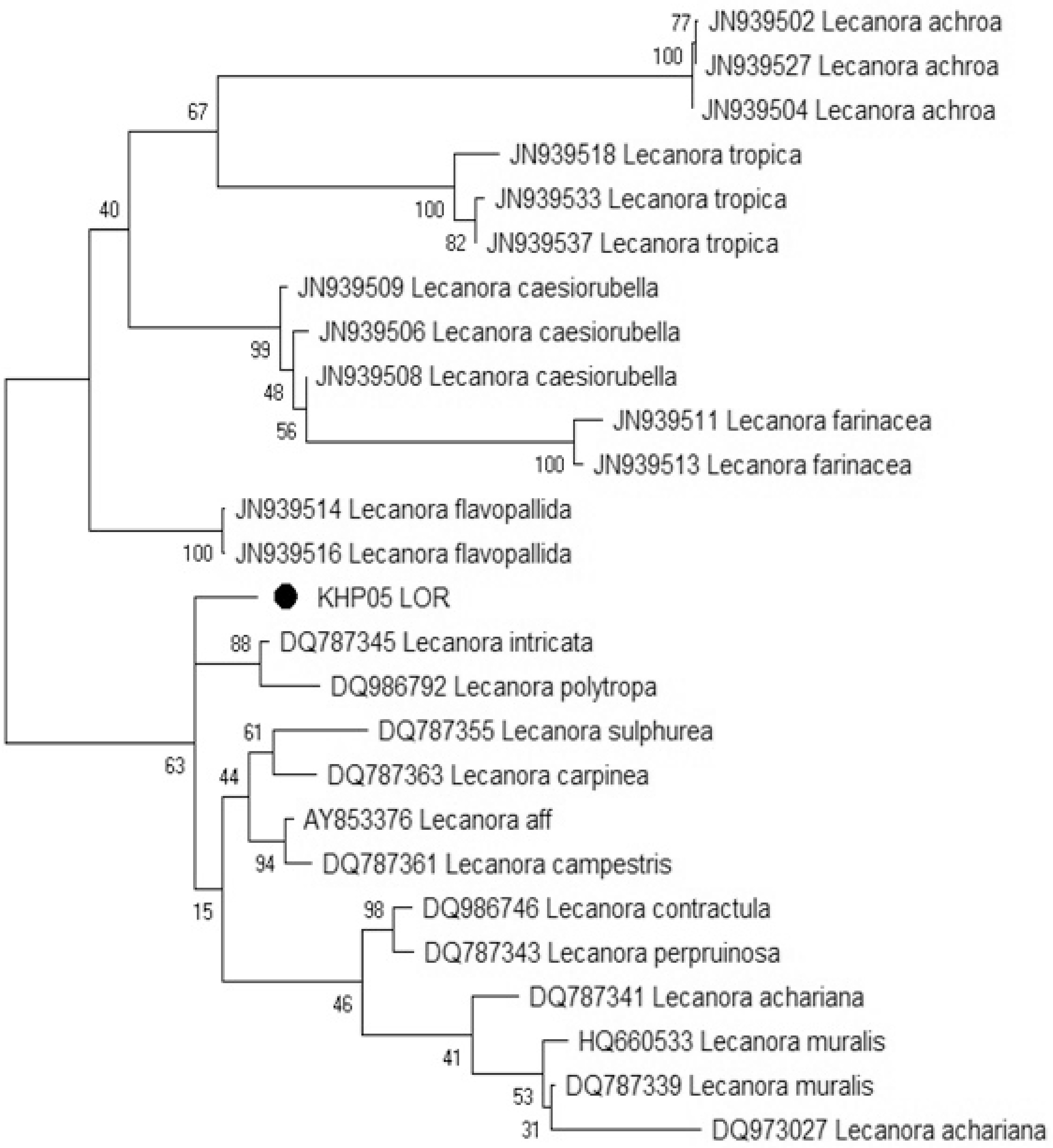
Figure 3.
Phylogenetic analysis of Protoparmeliopsis zareii (KHP05 LOR) based on LSU sequences. The evolutionary history was inferred by using the Maximum Likelihood method and Jukes-Cantor model . The tree with the highest log likelihood (–7,214.55) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 26 nucleotide sequences. Codon positions included were 1st + 2nd + 3rd + Noncoding. There were a total of 2317 positions in the final dataset. Evolutionary analyses were conducted in MEGAX.
-
Morphologically, Protoparmeliopsis zareii seems similar to P. muralis morphologically in the field, but P. zareii differs in having whitish reticulum (or distinctly fissured) on the upper surface of the central areoles as well as in having numerous immersed into thalline areoles apothecia, often aggregated in gall-lake formations, dull brown to greenish brown apothecium discs, prosoplectecnchymatous true exciple, and much wider ascospores (11–13(–14.5) × 8.5–9.5–11.0) μm vs. 7.5–15 × 5–7 μm). P. zareii differs from P. esfahanensis, with which it coexists, in that it has a larger and lighter greyish green to slightly yellowish grey-green thallus, lighter pale brown apothecium discs, a thicker thalline margin, smaller ascospores, whitish areole edges, and whitish apothecium margins. P. zareii differs from the other Asian taxa of this genus, P. Chlorophthalma, P. bogdoensis, P. kukunorensis, P. usbekicum and P. pruinosum, in lack of white pruine as well as in having a negative reaction of the thallus with K, C, KC, and P[8−10]. In possessing an areolated verruculose surface in the center of the thallus, P. zareii is somewhat similar to P. verruculiferum, occurring on soil in Central Asia (Uzbekistan). However, it differs from P. verruculiferum in that the center of the thallus has a more delicate whitish reticulum on the surface, whereas P. verruculiferum thallus is areolated and verrucose all over. P. zareii differs from brownish or yellow-brownish P. baicaliensis in that its thallus is greenish or greenish-yellow and that it reacts negatively with P. zareii differs from other migrant species of the genus, such as P. baranowii, P. sphaeroideum, and others, growing on soil in Central Asia, in that its thallus is closely attached to the substrate, it doesn't react favorably to K, C, or P, and it has an epilithic habit. P. zareii differs from another Asian endemic species, P. hieroglyphicum in that it has bigger peripheral lobes and smaller central areoles and does not have the hieroglyph-like brownish formations of conidiomata that characterise P. hieroglyphicum. Protoparmeliopsis zareii differs from several vagrant or attached species of the genus, including P. kotovii, P. verruculiferum, and P. garovaglii by not reacting favourably to K, C, or P as well as by having an epilithic lifestyle. P. zareii differs from P. riparium another Eurasian taxon that frequently grows in intermittently wet rock outcrops, in that it typically grows in dry, well-exposed locations and has a greenish or greenish-yellow thallus rather than reddish-brown or with whitish pruine.
Molecularly, the local collection has been identified as P. zareii as the ITS sequence of Pakistani sequence is exactly similar to the sequence of P. zareii reported from Iran (KP059049) in the BLAST analysis indicating they both are the same species while in the phylogenetic analysis (Fig. 2) the Pakistani sequence is clustering with the same sequence of P. zareii (KP059049) on the same branch showing similar identification of both sequences[9,20,21].
It is also noted that in phylogenetic analysis based on LSU (Fig. 3), the Pakistani sequence clustered with Lecanora muralis instead of P. zareii as the LSU sequences of P. zareii are not available in the DNA database which is why it is grouped with Lecanora muralis in the LSU based phylogenetic analysis. Further, this species has been identified as P. zareii as its con-specific nature confirmed by molecular and morpho-anatomical data.
Authors are very thankful to Higher Education Commission (HEC), Pakistan for providing funds for conduction of this study under NRPU Grant No. 7064. We also appreciate Prof. Dr. Abdul Nasir Khalid, Director, Institute of Botany, University of the Punjab, Lahore, Pakistan for providing working space in Fungal Biology and Systematics Laboratory for some of the experimental parts of this study.
-
All the authors contributed equally in this study: conception and design: Razaq A, Liaqat H; data collection: Ishaq A; analysis and interpretation of results: Bano A; draft manuscript preparation: Ilyas S; drawing and scale bar preparation: Bano A. All authors reviewed the results and approved the final version of the manuscript.
-
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Bano A, Razaq A, Ishaq A, Ilyas S, Liaqat H. 2024. Molecular identification and phylogenetic analysis of a lichen, Protoparmeliopsis (lecanoraceae, ascomycota), a new lichen genus for Khaplu valley, Gilgit-Baltistan, Pakistan. Studies in Fungi 9: e005 doi: 10.48130/sif-0024-0002
Molecular identification and phylogenetic analysis of a lichen, Protoparmeliopsis (lecanoraceae, ascomycota), a new lichen genus for Khaplu valley, Gilgit-Baltistan, Pakistan
- Received: 19 May 2023
- Revised: 11 January 2024
- Accepted: 02 February 2024
- Published online: 08 May 2024
Abstract: Protoparmeliopsis is reported for the first time from Pakistan and is represented here by Protoparmeliopsis zareii as a species record of this genus. This species has been identified using macro- and micromorphological descriptions, spot tests, ITS (Internal Transcribed Spacers), and LSU (Larger Sub Unit) of rDNA molecular markers analyses. Molecularly, in the phylogenetic analysis based on the ITS sequence of Pakistani collection of Protoparmeliopsis zareii (KHP 05) clustered with DNA sequences of Protoparmeliopsis zareii reported from Iran (KP059049) while in LSU analysis, it assembled with Lecanora muralis as the LSU sequences of P. zareii are not available in the DNA database. Morphological data was also found and the studied specimen is found to be conspecific with Protoparmeliopsis zareii.
-
Key words:
- Fungal taxonomy /
- Lichenology /
- Mycology /
- ITS /
- rDNA












