-
For many years, farmers have been urged to cultivate genetically pure seeds with a high germination rate. Releasing high-quality seeds is the first step toward sustainable food production and stable economic profits. Therefore, seed testing and certification processes have been arranged to facilitate this process[1]. In the seed certification process, four classes of seeds, including (i) nucleus, (ii) breeder, (iii) foundation, and (iv) certified seeds, should be evaluated based on some features that will be explained as follows[2]. Four features are investigated through physical purity, seed health, moisture content, and genetic purity in the four seed classes to achieve high-quality seeds (Fig. 1). Physical purity determines equality in size and shape, free of stones, weed seeds, and other crop varieties ranging from 96% to 98%. Seed health is a critical attribute that presents germination capacity and vigor with no contamination of microbes and insect damage. Besides, it would be necessary to keep the seeds' proper moisture content because it plays a crucial role in germination capability and viability. The optimum moisture level is 9%−13% which protects seeds from diseases and insects spreading[2]. Genetic purity is an important characteristic that implies preserved innate traits from parents to progenies. During the seed certification process, offspring should look like their mother plant[3]. Distinctness, uniformity, and stability (DUS) tests along with molecular markers analysis are performed to confirm that a new variety is distinct from other varieties, its features are uniform and the variety is stable with relevant phenotypic traits from one generation to the next[4]. The genetic purity of the certified seeds should be 99%, while it can be decreased by several factors comprising a mechanical mixture of different seeds, mutation, genetic drift, etc. Thus, off-type and impure seeds should be removed from pure seeds[5]. Most of the seed certification evaluation steps have produced big datasets that are difficult to be analyzed by the conventional statistical methods. For instance, image analysis is considered as a significant field of study employed in image identification, classification, and anomaly recognition[6]. Image analysis is used for the classification of different seed varieties in several crop species, including wheat[7], barley[8], rice[9], and corn[10]. The most prevalent sensing systems are multi-spectral and hyperspectral imaging, synthetic aperture radar (SAR), thermal and near-infrared (NIR) cameras, and optical and X-ray imaging[11]. Since, the mentioned multi-spectral and hyperspectral systems are costly, other cost-effective methods based on computer vision system (color, size, shape, and morphological characteristics) were employed in several crops[12−16].
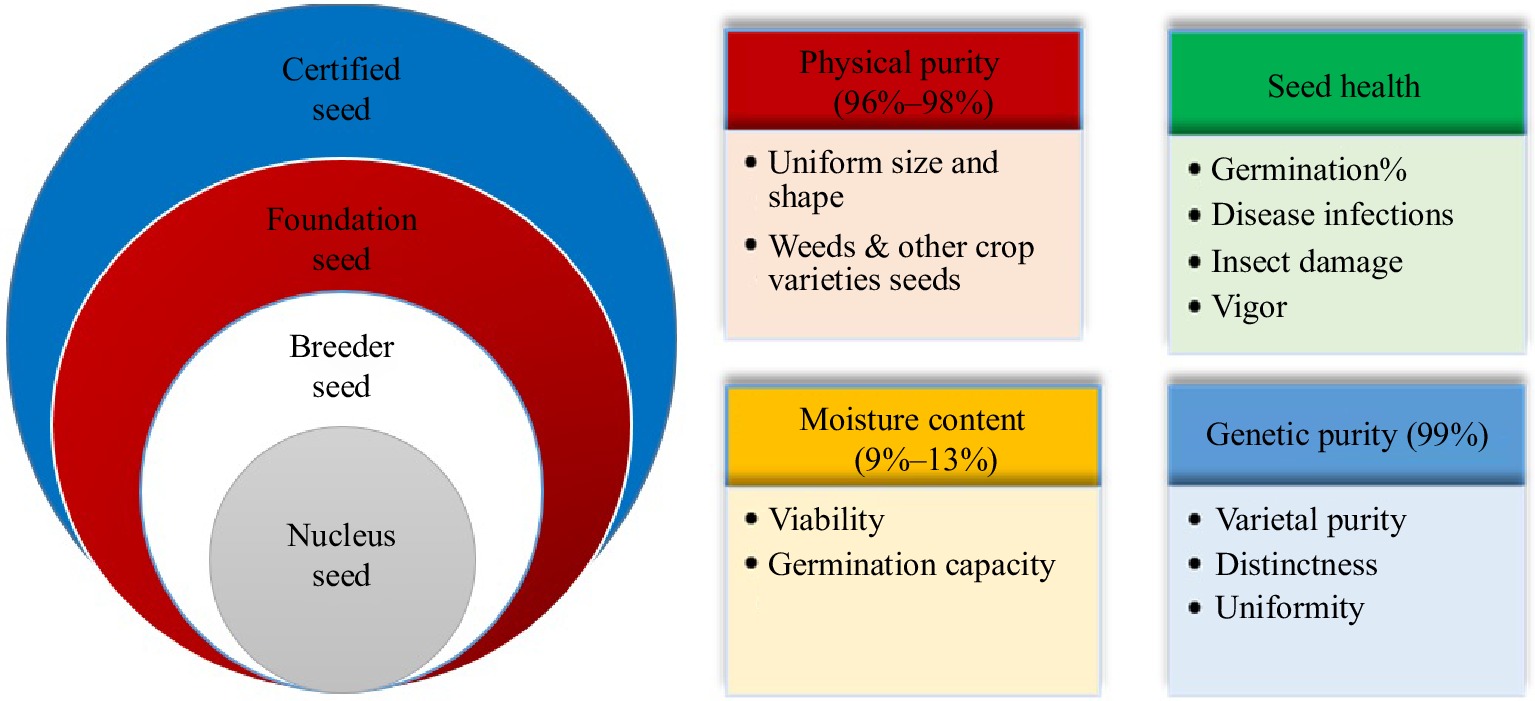
Figure 1.
The characteristics that are evaluated in different seed classes during seed certification process.
In the certification process, seed classification can be carried out based on visual characteristics with the involvement of experts. This conventional system is time-consuming, costly, and relies on experts' information. In contrast, identification of seed properties and varieties has been performed through image processing that is fast, accurate, and reliable[16]. This classification technique generates valuable information on seed quality, recognition of impurities, and outliers[17]. Images provided by sensing systems generate an outstanding amount of data[18], requiring advanced computational and mathematical analyses skills.
Artificial intelligence (AI) has a broad range of applications in agriculture, from robotics, soil, and crop monitoring to predictive analytics that provide farmers with beneficial data to achieve more from the land[19]. Machine learning (ML), as a branch of AI, is known as a reliable and effective computational approach to develop model-based methods that can increase the efficiency of breeding procedures[20]. ML algorithms such as K-means, support vector machines (SVM), artificial neural networks (ANNs), linear polarization, and wavelet-based filtering[6] can be employed in different fields of study such as image analysis.
-
In this review, the aim was to (1) briefly describe the seed processing steps, (2) review the past and current efforts to efficiently analyze big datasets derived during seed processing through ML algorithms, and (3) provide a future direction in this area to facilitate seed processing steps using novel mathematical methods. Different types of ML algorithms were exploited in different steps of seed certification. Each tested algorithm has its advantages and disadvantages. In this section, some important algorithms in the seed certification process are explained in detail.
Artificial neural networks (ANNs)
-
ANNs algorithms are one of the most broadly used algorithms in seed recognition research[21]. ANN-based modeling structures are stimulated by the human brain's neurological processing ability. It could be considered as an encouraging approach for managing the nonlinearities and complexities of complex processes like seed recognition that is replete with incalculable, noisy, fractional, and missing data. ANN-based models could make models of compound dynamic structures without an understanding of the comprehensive basic physical mechanisms in multivariate traits. There are various ANN models, though the basic principle is similar[22].
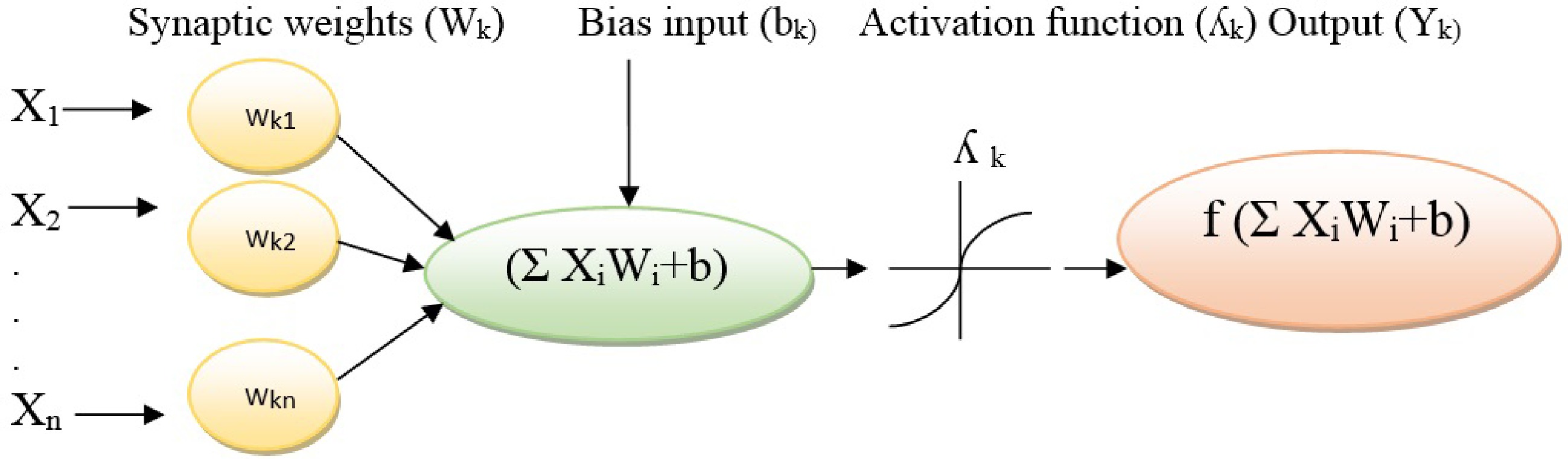
An ANNs model contains several neurons as signal-processing components that are connected by synapses as unilateral communication channels. An ANN receives input signals, processes the received signals, and finally produces an output signal. Every ANNs is linked to at least one other neuron based on its significant degree of a particular connection to the network that is called the weight coefficient[23]. The generated signals from other neurons are considered as input data of [X1, X2, ... , Xm]. The input data are multiplied by their associated synaptic weights (Wkj) and then moved to the artificial neuron. Furthermore, a bias input (bk) is considered an additional input signal to the artificial neuron. The Strength of the incoming signals is calculated by aggregating weighted input data and the bias input (Σ(weight*input ) + bias)[24]. An activation function (λk) is imposed to model that decreases the domain of incoming signals into a limited value, and finally, the non-linear output (Yk = f Σ (weight * input) + bias ) is generated. Activation functions lead to the no-linear transformation of input that makes the ANN capable of learning and performing complex tasks (Fig. 2). The most common activation functions in ANN systems are binary step, identity, softmax, sigmoid/logistic, tangent, hyperbolic tangent, and Gaussian[25].
ANN models are classified on the basis of their learning mode into supervised/unsupervised or on their structures into feed-forward or feedback recall methods[26]. Supervised ANNs uses a set of data patterns in the learning procedure that has separate input and output. However, unsupervised ANN algorithms employ a set of data patterns in the training process with only input values. Feedforward ANN models exploit unilateral information processing approaches that transmit data only from inputs to outputs. In contrast, feedback ANN models employ the bilateral stream that results in achieving insights from the prior layers letting feedback to the next layers in any neuron[26].
Synapses transfer numerical weights to diminish the error between real and simulated data in several training algorithms. However, optimizing ANN models has some complications including data compilation, data processing, topology selection, training and testing the selected model, and simulation and validation of the established ANN models.
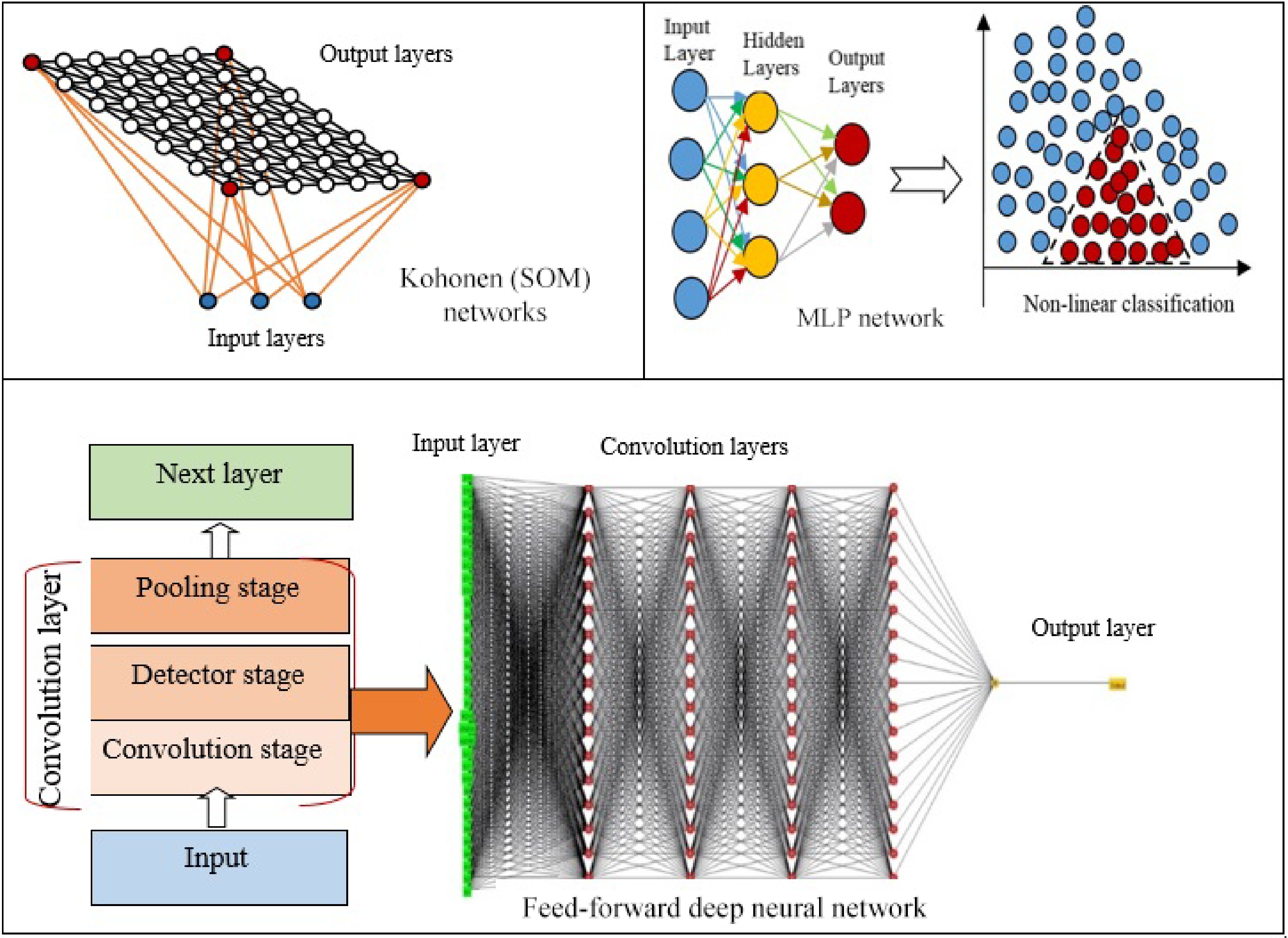
There are several types of ANNs that every have been developed into particular problems and applications. Some networks are appropriate for solving conceptual complications, whereas others are proper for data modeling and function estimation. The most popular ANNs in seed classification and recognition are Kohonen networks, multilayer perceptron (MLP), networks, deep neural networks (DNNs), and convolutional neural networks (CNNs) (Fig. 3)[22].
Kohonen network
-
Kohonen network algorithm, known as self-organizing maps (SOM), is a type of unsupervised learning algorithm, consisting of two-layer networks where the input and output layers are completely connected where similar patterns are promoted in the vicinity to one another that results in a 2D map of the output neurons (Fig. 3)[27]. Neurons that are closest to input win, and only weights of the winning neurons and their neighbors are updated. Kohonen networks map high-dimensional data into a smaller space that leads to data compression. Kohonen networks are employed for clustering (data grouping), pattern recognition, image segmentation, fuzzy partitioning, and classification[28]. Chickpea seed varieties were identified through unsupervised ANNs, a self-organizing map (SOM) that showed a better performance with 79% accuracy compared to supervised ANN with 73% accuracy. SOM can learn new things and changes with variable conditions and inputs. However, unsupervised learning algorithms do not result in expected outputs[29]. Besides, classification and identification of plants were made in leaf blade samples by the ANNs based on backpropagation algorithm (BP), KNN algorithm, Kohonen network based on SOM algorithm, and support vector machine. The training and identification time of the Kohonen network is moderately short, but the error rate of the Kohonen network is also very high due to the Kohonen network being applied without supervision (Table 1). Comparisons between four algorithms indicated that it could be effective for clustering, but it is not proper as a classifier and cannot deliver sufficient information to separate from other items[30].
Table 1. Comparisons among ML algorithms.
ML algorithms SOM MLP DA SVMs RF NB CNNs Accuracy + +++ ++ ++++ +++++ +++ ++++++ Flexibility + ++ + +++ ++++ +++ ++++++ Advantages Fast Non-linear classification Quick, inexpensive Analyzing complex networks, diminishing the generalization error, using large number of hidden units Tolerant of
highly correlated predictorsSimplicity,
fast training, decreases the number of parametersAnalysis of massive
amounts of
unsupervised data, better classification
and predictionDisadvantages Unsupervised Time-consuming, few hidden neurons Unsupervised Algorithmic complexity, development of ideal classifiers for multi-class problems and unbalanced data sets Evaluation
of pairwise interactions is difficult, future predictions require the original dataOversensitive
to redundant
or irrelevant attributes, classification
biasIt needs further developments
for big data
analysisMultilayer perceptron (MLP)
-
Multilayer perceptron (MLP) is a class of feed-forward artificial neural networks developed by[31]. MLP is a non-linear computational process that is highly efficient for the classification and regression of complex features. Furthermore, MLP was created to address non-linear classification problems that further layers of neurons employed between the input layer and the output neuron, and these neurons are called hidden layers (Fig. 3). Thus, these hidden layers process the information achieved from the input layers and process them to the output layer that develop perceptrons to resolve non-linear classification problems[32]. MLP is frequently employed for modeling and forecasting complex attributes, such as yield[33], classification seed varieties[7], weed discrimination[34], unknown seed identification[35]. This algorithm discovers the relationship between the input and output variables through some interconnected processing neurons that identify a solution for a particular problem[36]. However, MLP is time-consuming method that may result in inaccurate modeling. Besides, MLP regularly employs few hidden neurons that makes it inappropriate for modeling and predicting than other algorithms with more hidden neurons[33] (Table 1).
Discriminant analysis (DA)
-
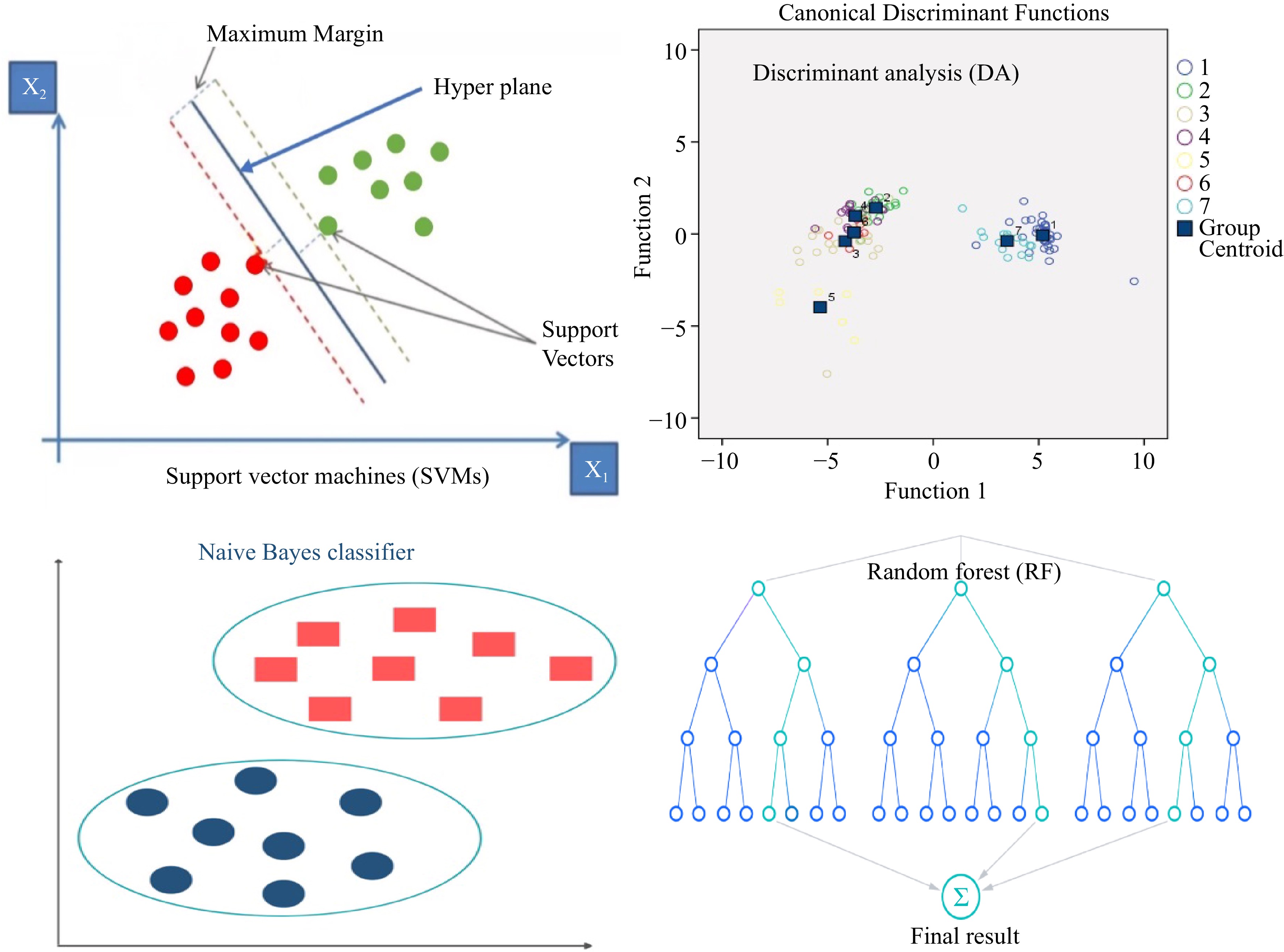
Discriminant analysis (DA) is a flexible classifier that was exploited to classify observations into two or more groups or classes, and it examines methods and degrees of the contribution of variables to group partitioning (Figs 4 & 5). It was employed for image processing by several scholars in textural analysis in wheat (Fig. 5)[37] and color analysis in castor[38]. In wheat, a stepwise discrimination system was exploited for selection and ranking of the most important textural features by LDA (linear discriminate analysis) classifier with 98.15% accuracy in top selected traits in nine cultivars[37]. Furthermore, color analysis in castor through partial least squares-discriminant analysis (PLS-DA) and LDA demonstrated that the PLS-DA model with 98.8% accuracy was more efficient than LDA. It showed that this method was straightforward, quick, beneficial, and inexpensive (Table 1)[38].
Chen et al. analyzed 28 color features of five corn varieties by step-wise discriminant with 90% accuracy[39]. In addition, the color and shape attributes of Italian landraces of the bean were investigated using LDA that showed 82.4%−100% accuracy[40]. However, another research used the shape plus texture features to recognize two weed species, rumex, and wild oat, in Lucerne and Vetch. They employed ANNs and stepwise discriminant analysis, while the ANNs presented better precision (92%−99 %) than the DA[41].
Support vector machines (SVMs)
-
Support vector machines (SVMs) introduced by Vapnik in 2000 can be considered one of the most prevailing and simple machine learning algorithms[42]. SVMs can be categorized based on the output variable to the Support Vector Classification (SVC) that classifies data, and the Support Vector Regression (SVR), which determines regression[6]. The data was separated into training and validation sets. The majority of the dataset was assigned to the training set, whereas the rest was partitioned into the validation set based on different approaches such as cross-validation (Fig. 4)[43].
SVMs are usually applicable to a two-class problem that creates a boundary between two groups in linear, non-linear, and trade-off penalty parameters to handle complexity (Fig. 4)[44]. In non-linear relationships, SVM can discover patterns and performances. SVMs have several benefits over MLP depending on the dataset, which can analyze complex networks and employ numerous learning problem formulations to solve a quadratic optimization problem[45]. Besides, SVMs showed a greater advantage over ANNs due to diminishing the generalization error by exploiting the structural risk minimization principle rather than the practical one used in ANNs[21]. Even though there are several benefits of the SVM, there are some noticeable flaws including, algorithmic complexity leading to longer training time of the classifier in large data sets, development of ideal classifiers for multi-class problems, and unbalanced data sets (Table 1)[46]. SVM was employed in several studies for discrimination and classification (Table 2). For soybean seed discrimination, several morphological and color attributes from several seed classes were analyzed by SVMs. Results showed that color traits had better discrimination ability than morphological traits with 77% and 59% accuracy, respectively[47]. When the purity of waxy corn seeds was identified by image analysis of morphological and texture features through SVM that data showed 98.2% accuracy[48]. Furthermore, an SVMs classifier was employed to detect seed defects in a large volume of corn seeds using color and texture features analysis. The results showed that the best accuracy (81.8%) was obtained through the combination of both color and texture analysis than color and texture individually[49]. The same method was exploited to identify corn varieties with the best accuracy (94.4%)[50].
Table 2. Examples of applied machine learning models in modern seed recognition and classification studies.
Plant species Type of machine learning Classifier Accuracy Features Purpose Ref. Corn Digital image MLP 98.83% Texture - spectrum hybrid Seed varietal purity [10] Wheat Digital image ANNs 85.72% Morphology Seed varietal purity [7] Wheat Digital image ICA-ANN hybrid 96.25% Color, morphology,
and textureSeed varietal purity [72] Wheat Digital bulk image LDA 98.15% Texture Seed varietal purity [37] Wheat Digital bulk image ANNs 97.62% Texture Seed varietal purity [73] Forage grass (Urochloabrizantha) FT-NIR spectroscopy & X-ray imaging RF 85% Spectrum-composition hybrid Seed germination & vigor [74] Corn FT-NIR spectroscopy PLS-DA 100% Chemical composition Seed germination & vigor [75] Pepper FT-NIR & Raman spectroscopy PLS-DA 99% Chemical composition Seed germination & vigor [76] 57 weed species Digital image NB & ANNs 99.5% Color, morphology,
and textureWeed identification [59] Wheat Video processing ANN - PSO hybrid 97.77% Shape, texture & color Physical purity & weed identification [77] Rice Digital image MLP 99.46% Morphology, texture & color Seed varieties classification [9] Rice Digital image ANNs − Morphology Seed grading [78] Rice Digital image DFA 96% Morphology Physical purity [79] Soybean Aerial imagery CNNs 65% Object detection Weed identification [80] Soybean Digital image CNNs 97% Color, texture and shape Seed deficiency [66] Soybean Digital image CNNs 86.2% Color, texture and shape Seed counting [70] Corn Digital image MLP 94.5% Color Physical purity [81] Soybean Flatbed scanner SVMs & RF 78% Color Seed grading [47] Bean Digital image RF 95.5% Color, texture and shape Seed varieties classification [16] Corn Hyperspectral image SVMs 98.2% Spectrum-texture – morphology hybrid Seed varieties classification [48] Corn Digital image SVMs 95.6% Color and texture Seed varieties classification [49] Corn Digital image GA–SVM hybrid 94.4% Color, texture and shape Seed varieties identification [50] Corn Digital image CNNs 95% Color, texture and shape Haploid and dioploid discrimination [82] Corn Digital image CNNs 95% Color, texture and shape Seed varieties identification [83] Barley Digital image DA & K-NN 99% Color, morphology & texture Seed varieties classification [8] Barley Digital image CNNs 93% Color, morphology & texture Seed varieties classification [84] MLP: Multilayer Perceptron, ICA: Imperialist Competitive Algorithm, ANNs: Artificial Neural Networks, LDA: linear discriminate analysis, FT-NIR: Fourier transform near-infrared, PLS-DA: partial least squares discriminant analysis, NB: naïve Bayes, PSO: partial swarm optimization, DFA: stepwise discriminant function analysis, CNNs: Convolutional neural networks, SVM: support vector machine, GA: genetic algorithm, DA: discriminant analysis, K-NN: K-nearest neighbors, SVMs: support vector machines. Random forest (RF)
-
Random forest (RF) was developed by Breiman[51] that is known as an effective method for seed classification[16], object recognition[52], plant phenomics, and genomics[53]. The RF is based on multiple decision trees, which are built at the same time (Fig. 4). They are constructed by bootstrapping data samples to learn, similar to bagging (bootstrap aggregation).
During training for each tree, the bootstrapped data samples are used as observations, and another randomly chosen beech of bootstrapped data samples is used as an out-of-bag observation. This is done repeatedly until every sample has been left out of one bag. Out-of-bags are used as an input for a newly built random forest with the same number of trees[54]. The RF algorithm can be extended to multiclass, sequential regression, and binary classification problems. The RF algorithm has been extended to the tree pruning problem, called 'non-Breiman random forests'. In this regard, it is possible to generate resampled trees and use them as a candidate for each node in the constructed decision tree. The selection of the best tree is made based of mean-square error[16]. Despite these advantages, RF generates a large number of candidate predictors that makes the evaluation of pairwise interactions difficult. Also, future predictions require the original data due to no possibility of replicating predictions without an actual forest[55].
RF along with SVMs were exploited to discriminate soybean varieties based on color and morphological features. Data showed RF classifiers discriminated color features better than morphological features with a better accuracy 78% better than SVMs (77%)[47]. However, three algorithms including, RF, SVMs, and K-Nearest Neighbors (KNN) were employed to classify dry beans through a computer vision system for seed certification. The results showed the better accuracy of the KNNs classifier (95%), while the accuracy of RF and SVMs was 93.1% and 93.5% respectively. The RF model accuracy exceeded 95.5% when using principal component transform was compared with the original variables[16]. Another research classified rice through image features of rice seed including, color, shape and texture. RF classifiers was employed along with SVMs, while RF classifiers through simple features showed the greatest classification with accuracy of 90.54% than SVMs[56].
Naive Bayes (NB)
-
Naive Bayes (NB) classifiers are simple probabilistic classifiers based on the Bayes hypothesis that implies the independence of pair traits[57]. In comparison with other classifiers like Neural Networks and Support Vector Machines, NB calls for quite little data for training, thus it does not include several parameters. It trains data fast and simply implemented. However, NB is oversensitive to excessive or unrelated attributes. When some attributes are extremely correlated, they take great weight in the final decision, resulting in a decrease in accuracy of prediction of correlated features and classification bias[58]. It was employed in several studies to identify and classify different seeds and plants (Table 2). NB is usually employed to recognize weed seeds based on morphological, color, and textural characteristics from images[59]. This algorithm decreases the number of parameters to nearly ideal sets in every feature[60]. The NB classifier outperformed ANN algorithms in weed seeds identification[34]. Moreover, it was exploited in the seeds classification of Kama, Rosa, and Canadian wheat varieties according to their morphological features as the second-highest accuracy classifier (94.3%), though ANN algorithms showed the highest performance (95.2%) than NB[61].
-
Deep ANNs are commonly mentioned as deep learning (DL) or DNNs[62]. They are a fairly novel part of ML with multiple processing layers to learn complex data representation through multiple levels of concepts that are known as representation learning. It transforms the representation from starting level with the raw input into a representation at a higher, moderately abstract level through g simple and non-linear modules Thus, compound functions can be learned by creating sufficient transformations. The leading benefit of DL in some cases is to extract features through the model itself. DL models have developed intensely in several sectors and industries, including agriculture. DNNs are basically ANNs with multiple hidden layers among the input and output layers and can also be supervised, partially supervised, and unsupervised[63].
Convolutional Neural Networks (CNNs)
-
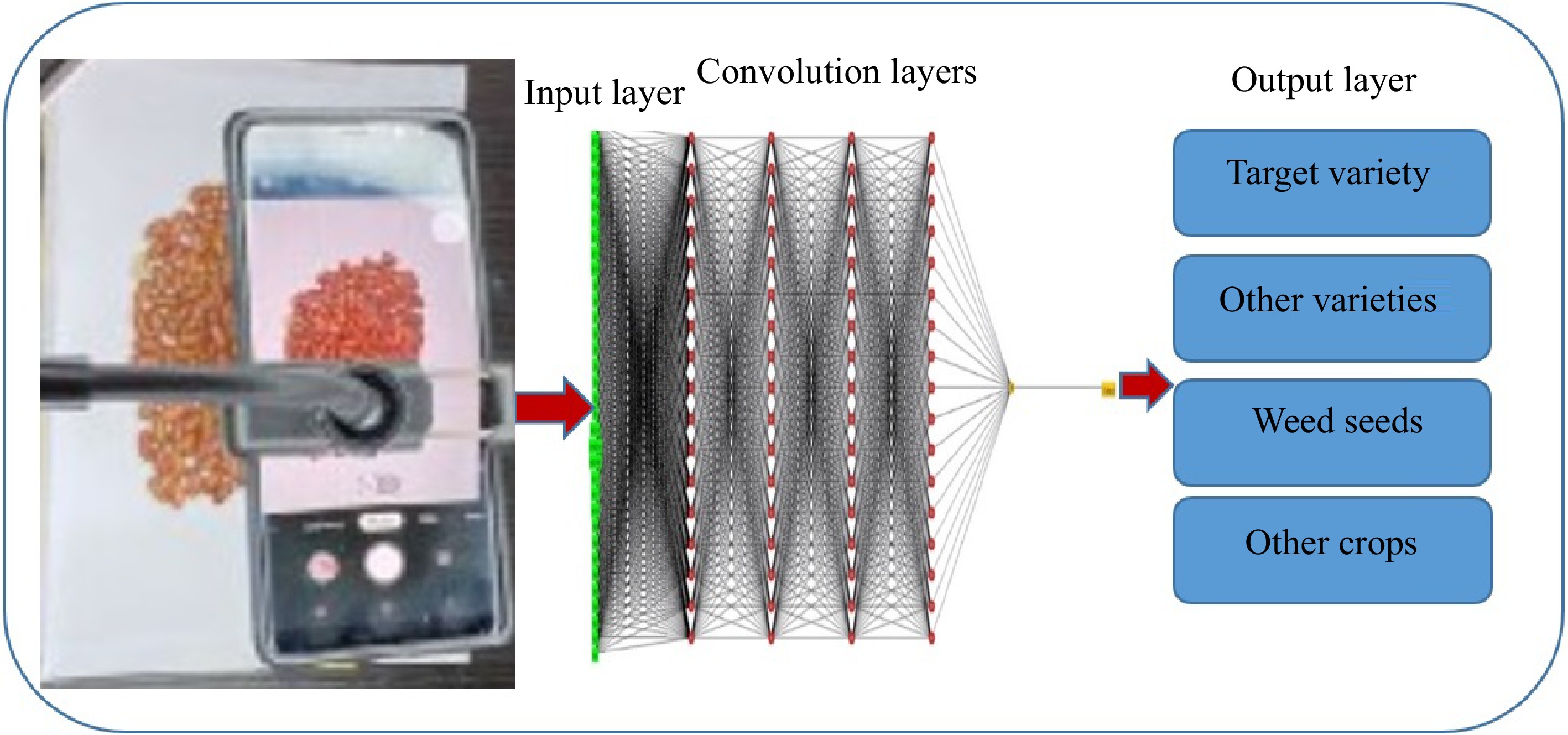
CNN is a popular DL model, which extracts feature maps via executing convolutions in the image domain. For instance, a color image makes up three 2D arrays of pixel intensities in the three color channels that data formed of multiple arrays are processed through the CNN. There are four main concepts behind CNN, including local connections, shared weights, pooling, and the application of several layers[15]. A CNN network (Fig. 6) is made up of a series of stages that every convolution layer has three stages including, convolution, detector, and pooling stages. First, in a convolutional stage, units are arranged in feature maps. Therefore, a filter bank, a set of weights, connects every unit of local patches in the feature maps of the previous layer to produce a set of linear activations. All units in a feature map are assigned the same filter bank. Next, the outcome of the aggregated local weights has proceeded over a non-linearity function that is called rectified linear unit (ReLU). This stage is sometimes called the detector stage. Finally, the pooling function offers the output of the network at a certain location with a summary statistic of the adjacent outputs. Thus, functions of the pooling stage adjust the output of the layer better[64] (Fig. 3). CNN algorithms displayed successful application in agriculture, particularly for the identification and classification of unclear feature data, like the delicate features of small seeds[65]. It was reported that a lightweight CNN could identify the defects on the full surface of seeds in seed sorting equipment. Three lightweight models (SqueezeNet, ShuffleNet, and MobileNetV2) were employed for training and deployment. Comparison between the classification results of the lightweight networks and classic networks showed that the classic network is two times greater in training time, three times longer in prediction time, and 41 times longer in training model size. Though the average accuracy of the classic networks is only 3% higher than that of the lightweight networks, the overall performance of lightweight networks is significantly better than that of classic networks. The decrease in average accuracy is due to the lower accuracy of SqueezeNet (0.89), while MobileNetV2 and ShuffleNet showed accuracies of 0.97 and 0.96, similar to the accuracy of the classic networks (0.97)[66]. Another study[67] employed SeedNet, a new kind of CNN and precise both in accuracy and training time for classification and identification of several seed images belonging to distinct families. Moreover, ten state-of-the-art CNNs were compared with SeedNet and classical machine learning approaches. As a result, SeedNet showed strong performance in both datasets and lower training time compared to the other networks. The CNNs presented robust performances compared to the traditional methods in accuracy and time training[67]. Taheri in 2021 exploited VGG16, a CNN framework model for automatic recognition of chickpea varieties through seed images taken by a digital camera and a mobile phone camera in the visible spectrum (400–700 nm) (Fig. 6). VGG-16 is an architecture of the VGGNet that contains over 15 million parameters of convolutional layers. It can extract the image features containing shape, color, and texture.VGG16 design was improved by a global average pooling layer, dense layers, batch normalization, and dropout layers. The average accuracy of the modified CNN model was over 94%. They proposed that this method can be applied in the seed industry and mobile applications as a fast and strong automated seed identification procedure[14]. Further, VGG-16 architecture was employed for the automated identification of grapevine cultivars by leaf Imaging[68]. Similarly, the VGG16 model was employed to identify and classify 14 common distinct seeds. In VGG16 architecture, the last layer was substituted with five performed layers: the average pooling layer, the flattening layer, the dense layer, the dropout layer, and the softmax layer. As a result, training and testing accuracy reached over 99%[69]. Moreover, CNN has been exploited for computing the number of seeds in soybean pods. The architecture of CNN comprises 12 convolutional layers besides an output softmax layer where every convolutional layer is characterized by its number and size filter and the stride. CNN and SVM methods were employed in the project, while results showed CNN higher accuracy (86.2%) in seed-per-pod estimation than SVM (50.4%)[70]. Another study proposed a mobile application that was developed to detect and classify seed images through serval CNN models with high accuracy. Four different CNN models, including Inceptionv3, Xception, ResNet50, and Inception ResNetV2 models were applied for the detection and classification of 15 types of seed images with 99% accuracy[71].
-
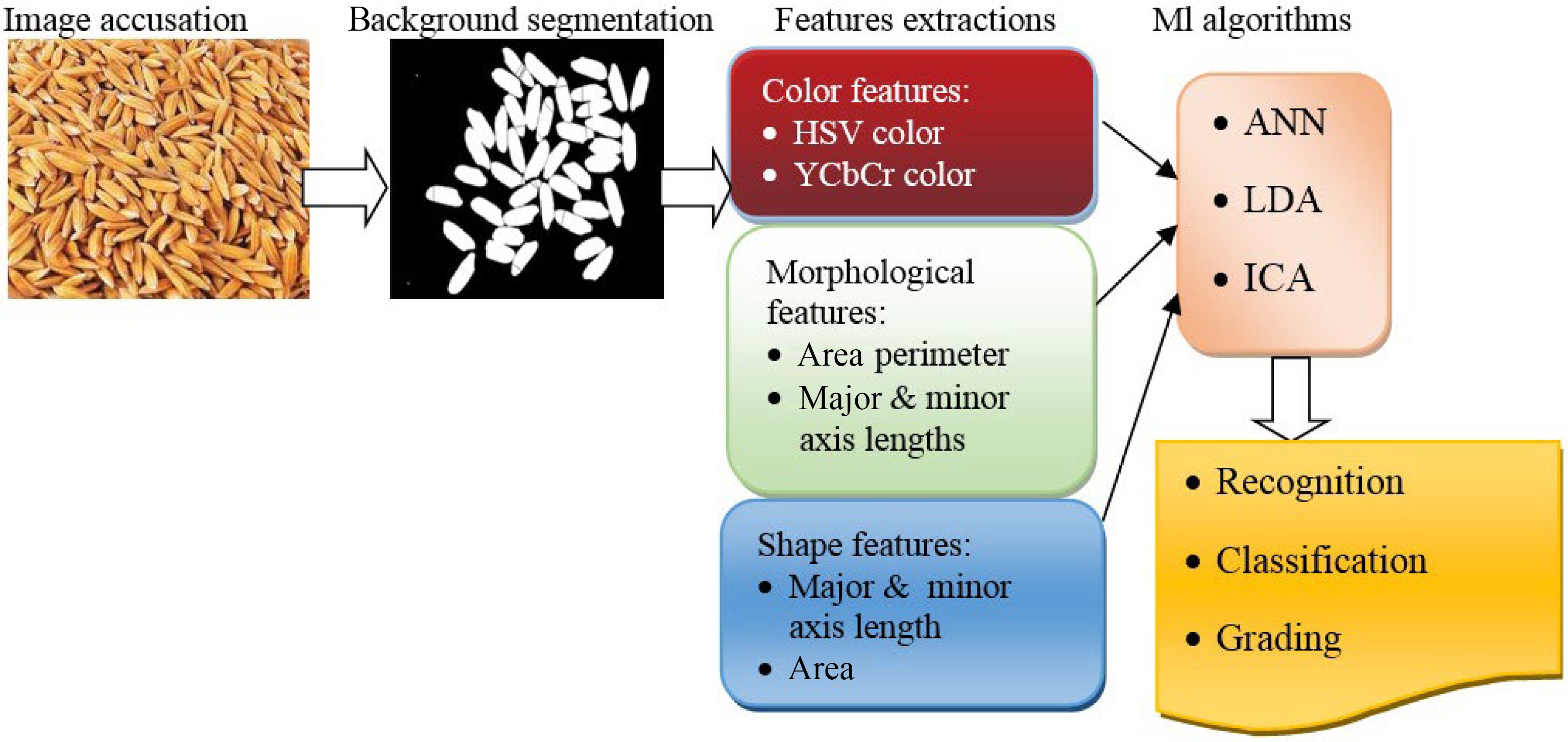
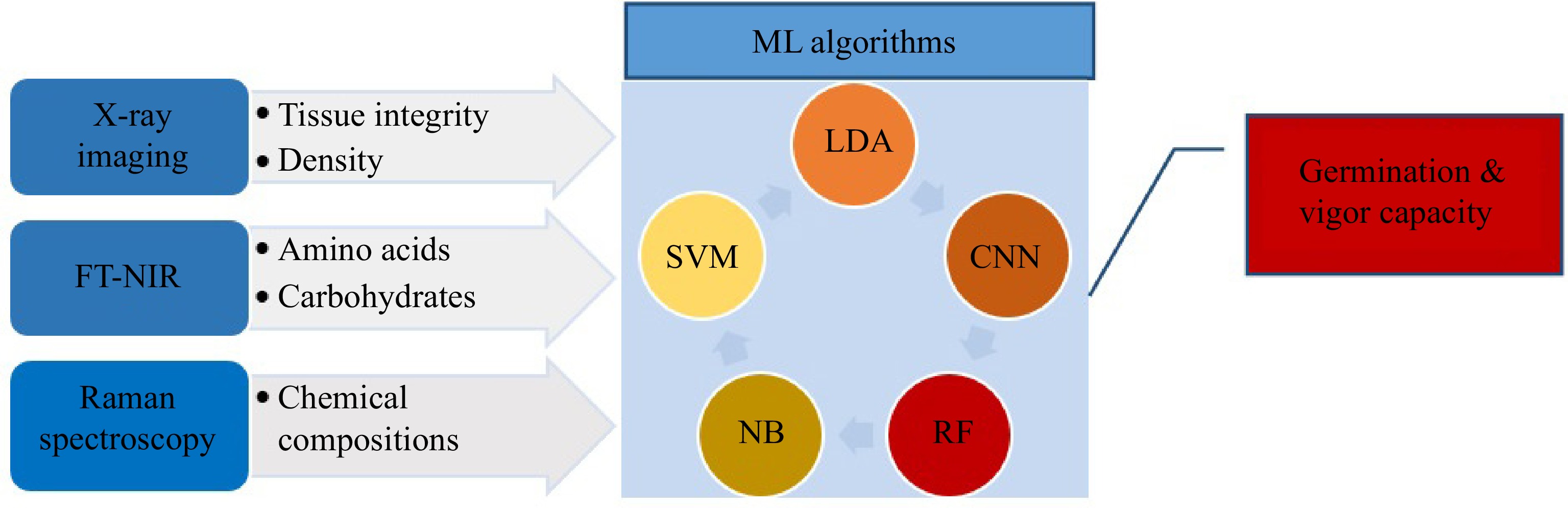
The quality of seeds are usually determined by germination and vigor tests[2]. This process is time-consuming and based on experts' experiments and knowledge[1]. Some changes in the chemical compound and internal features that lead to deficiency in germination and vigor cannot be detected visually[85]. Therefore, effective techniques like FT-NIR and Raman spectroscopy and X-ray imaging are vital for extracting information of compound features relevant to seed quality[85]. Fourier transform near-infrared (FT-NIR) spectroscopy is efficient for the identification of seed compounds through spectral measurements ranging from 780 to 2,500 nm[86]. Besides, Raman spectroscopy (RS) is an analytical spectroscopy technique that employs laser sources on seed samples to capture and analyze scattering signals by a detector[87]. Each chemical molecule has particular vibrational frequencies that lead to identifying chemical compositions and viability[88]. RS is more accurate than conventional methods for discovering seeds' viability. RS was used in the corn[75] seeds' viability test. However, X-ray imaging can detect seeds' internal attributes by a reduction in several tissues[89]. ML algorithms can classify seeds quality traits (Fig. 7). Some efficient algorithms have been exploited to predict seed germination and vigor, including linear discriminant analysis (LDA), partial least squares discriminant analysis (PLS-DA), RF, NB, and SVMs[90].
In another study for determining seed germination and vigor capabilities of a forage grass (Urochloabrizantha) by FT-NIR and X-ray techniques, the RF model was employed for analyzing the compound data of both techniques that reached accuracy at 85%. In contrast, the accuracy of the individual method was lower than combined data. However, LDA and PLS-DA algorithms showed the highest accuracy (90% for germination and 68% for vigor estimate) in the X-ray method, whereas the accuracy of 82% for seed germination was achieved by FT-NIR data with the PLS-DA algorithm[74]. The physiological quality of the soybean seed lot was evaluated successfully through RF with a higher accuracy than other classifiers: MLP and J48[91]. A study was conducted using NIR to determine seed viability in Japanese mustard spinach. PCA and SVMs were employed in the first step. Then, CNN was applied to realize the differences between viable and non-inviable seeds and categorize them automatically. Classification accuracy was 90% showing the CNN was an efficient method to classify viable seeds[92].
-
Most prediction models produced by machine learning do not display proper methodological quality, leading to a high risk of bias. There are several reasons for a risk of bias: small study size, inadequate classification and handling of missing data, and improper control on overfitting. Improvement of the design, conduct, reporting, and validation of such studies will enhance the confidence, speed and precision application of prediction models in several field of studies[93]. For example, high lot quality caused unbalanced data in soybean seed analyses. Unbalanced learning results in surpassing one class on another class is a classification process. To overcome this problem, a resample filter was applied to prevent bias of the algorithm. The accurate selection of resources reduces the dimensionality of the data and benefit to faster function of the classifier, resulting in higher accuracy. When classes are classified incorrectly by classifiers, the classes with more data had higher accuracy and precision values than the other classes in unbalanced data. A smaller sample number of classes resulted in lower values of the performance compared with larger data classes. Employing classification via regression (CVR) showed a lower amount of false positives in some classes, led to better accuracy in the rejected and intermediate classes of soybean seed lots[91].
-
The main principle to estimate accuracy is that the evaluation samples should not be the same as the training samples. Training samples are excluded from the evaluation samples of certain parameter values that decreases the probability of overtraining and led to an increase of the generalization of the classifier[94]. Several partitions are created for each sample to be applied several times for several aims. The percentage split divides data into training and testing by percentage value (70%, 30%−90%, 10% for training and testing data). It is essential to choose the best percentage due to raising uncertainties[95]. To increase the reliability of the machine learning results, cross-validation is developed to train and asses accuracy of samples several times. The k-fold cross-validation is a popular validation method that randomly divided the sample set into a series of identical sized folds. The dataset is divided into several partitions, or folds that is showed by k. For example, if a k-value of ten is applied, the dataset is divided into ten partitions. Thus, nine of the partitions are allocated for training data, while the remaining one partition is allocated for test data. The training is iterated ten times where nine partitions are specified for training data and a different partition is applied for the test set each time[94]. A study evaluated several machine learning models to assess soybean seed lot quality. Soybean lots evaluated through two methods of test sets including cross-validation (with 8, 10, and 12 folds), and percentage split (with 66% and 70%). Results revealed that the 10-fold cross-validation achieved better figures than 8 and 12 that was 90.22% classification accuracy. However, the method applying 66% of data for training reached 93.55% accuracy. Therefore, precise classification through appropriate algorithm training lead to reliable information for seed quality testing[96]. However, in other research, k = 10 cross-validation was applied to training and testing data of maize seed lots led to the high accuracy and precision of the classification for corn seed lots[10,97].
-
Rapid and robust seed variety and purity recognition are the main challenges in the seed registration and certification process. Thus, developing high-throughput analyzing procedures is required to accelerate the seed variety discrimination accurately and affordably. ML algorithms were exploited in several studies to identify seed varieties, recognize weeds and evaluate seed viability through image analysis. In terms of discussing studies in the literature and providing insights into particular subjects in seed certification procedures, this review was dedicated to two important parts, including: 1) conventional machine learning methods and algorithms and their application in seed recognition, vigor, and purity; 2) the deep learning algorithms and architectures for seed identification and classification.
Several conventional machine learning and feature engineering algorithms comprising ANN, DA, RF, SVM, PLS-DA, and NB were employed to classify and detect seeds. These algorithms showed their great efficiency in the seed recognition and certification process (Fig. 5 & Table 2). However, it still needs human involvement for feature extraction. Therefore, deep learning was introduced to automatically extract complex data features from huge volumes of unsupervised and un-categorized data. Deep learning simplifies big data analytics procedures by categorizing learning and extracting distinct levels of multiplex data, particularly for discriminative purposes such as classification and prediction. Deep learning created great opportunities to develop in situ seed identification and sorting systems. For instance, imaging data can be easily accessed through mobile phones, tablets, or action cameras (GoPro) for in situ information processing. Thus, it can open a new window for utilizing a smartphone as a fast and robust substitute for the online seed variety discrimination stage by developing a CNN model-based mobile app[14]. Some scholars presented that the CNN algorithm is trustworthy and highly effective for variety recognition[98] in farms. Developing mobile apps would be a great approach for seed producers, processors, and distributors. Besides, a real-time seed recognition system is a fast and automatic recognition system through deep learning to detect seeds deficiencies in the selection process. The mechanism evaluates the whole surface features of seeds precisely via deep learning[66]. Precise seed sorting methods will lead to increasing yield in the breeding industry. These developments can be extended to invent more mobile and online apps to detect and discriminate several features of seeds with minimum human involvement, such as seeds protein content, hardness, moisture content, and so on. These developments have evolved in the agriculture sector to accelerate food production for the projected global population in the near future. In summary, deep learning is a promising means for smarter, sustainable agriculture and secure food production.
However, the relatively low maturity of the deep-learning models demands further study. Specifically, further developments are crucial to adjust deep learning procedures for big data problems, containing high dimensionality, streaming data analysis, scalability of deep learning models upgraded formulation of data abstractions, distributed computing, semantic indexing, data tagging, information retrieval, criteria for extracting good data representations, and domain adaptation. Upcoming research should address one or more of these difficulties[99].
-
The author confirms sole responsibility for the following: study conception and design, data collection, analysis and interpretation of results, and manuscript preparation.
-
All the data presented in this study are available from the corresponding author.
-
The author declares that there is no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Ghaffari A. 2024. Precision seed certification through machine learning. Technology in Agronomy 4: e019 doi: 10.48130/tia-0024-0013
Precision seed certification through machine learning
- Received: 02 December 2023
- Revised: 05 May 2024
- Accepted: 07 May 2024
- Published online: 23 July 2024
Abstract: Original and pure seeds are the most important factors for sustainable agricultural production, development, and food security. Conventionally, seed protection and certification programs are carried out on several classes from breeders to certify seed based on a physical, biochemical, and genetic evaluation to approve seed as a cultivar. In seed industries, quality assurance programs depend on different methods for certifying seed quality characteristics such as seed viability and varietal purity. Those methods are mostly conducted in a less cost-effective and timely manner. Combining machine learning (ML) algorithms and optical sensors can provide reliable, accurate, non-destructive, and quick pipelines for seed quality assessments. ML employs various classifiers to authenticate and recognize varieties through K-means, Support Vector Machines (SVM), Discriminant Analysis (DA), Naive Bayes (NB), Random Forest (RF) and Artificial Neural Networks (ANNs). In recent years, progress in ANN algorithms as deep learning simplifies big data analytics procedures by categorizing learning and extracting distinct levels of multiplex data. Deep learning opened a new door for developing a smartphone as a fast and robust substitute for the online seed variety discrimination stage through developing a Convolutional neural networks (CNNs) model-based mobile app. This review presents machine learning and seed quality assessment areas to recognize and classify seeds through long-standing and novel ML algorithms.