-
Food production and security has been one of the most important issues worldwide, especially as the world's population has rapidly increased[1] and the global climate has changed[2] in the past decades. Global climate changes, including, but not limited to, elevated temperature, increased erratic rainfall, declined water table, and raised drought incidence, are threatening the world's food production and security. Therefore, it has been a major research subject in agriculture on how to feed the world. It has been the consensus that development and application of genetically improved crop varieties and livestock strains is the most efficient, economical, and environmentally friendly approach to continuously increase or sustain the world's food production and security. To improve the ability, efficiency, and productivity and/or accelerate the process of developing improved crop varieties and livestock strains, several molecular methods have been developed[3], including marker-assisted selection[4], genetic engineering[5], RNA interference or RNAi[6], genomic selection[7,8], gene or genome editing[9], and gene-based breeding[10−16]. As most agronomic traits important to food production, such as yield, quality, and biotic and abiotic resistances or tolerances, are complex traits each controlled by numerous genes while marker-assisted selection, genetic engineering, RNAi, and gene editing can often manipulate at a time only a single or few genes controlling an agronomical trait, they are more suitable to improve elite varieties with only a single or few undesirable traits influenced by the unfavorable alleles of one to a few genes[3]. Therefore, only genomic selection and gene-based breeding have been the method of choice that can assist at development of or develop brand new varieties with complete intellectual properties[3]. Given that genomic selection has been demonstrated to be efficient only for genome-wide assisted progeny selection and it is impossible for genomic selection to help develop a superior new variety from a progeny pool without superior individuals, gene-based breeding has become the method of choice to develop brand new varieties with complete intellectual properties through the entire breeding process. In this review article, gene-based breeding, therefore, is introduced, including what is gene-based breeding; concept, research, development, and potential; advanced research and development; and perspectives. Research, development, and application of gene-based breeding for improved crop varieties and livestock strains is promising to improve current best varieties substantially and continuously, thus helping feed the world.
-
GBB can be a revolutionary technology for plant and animal breeding that allows breeders to develop superior pure-line and hybrid varieties by design according to breeding objectives based on the genes controlling breeding objective trait(s) and facilitated by artificial intelligence (AI) technology through the entire breeding process, including parent selection, crossing design, and progeny selection[3] (Figs 1 & 2). Studies have demonstrated that the single nucleotide polymorphism (SNP)/insertion or deletion (InDel) markers that have significant biological impacts on objective trait(s) (Fig. 3a, c), the number of favorable alleles (NFAs) including heterosis genotypes (Fig. 3b, d), and the transcript expressions of the genes controlling objective traits (Fig. 3e, f) were well suited and could be used for GBB either separately or jointly. Because the genes controlling breeding objective traits are used for all steps of parent selection, crossing design, and progeny selection (Figs 1 & 2), the results of GBB can be highly predictable, the practice of GBB can be proceeded in a manufactory manner in greenhouse or phytotron, and the progeny selection of GBB can be highly reliable and the process of GBB for development of the selected progeny into new varieties can be dramatically accelerated, given that the genotypes of the genes controlling objective and other agronomic traits in progeny are known and high-throughput (e.g., 1,000,000 plants) genotyping of the genes is affordable and feasible. Therefore, GBB can be multiple-fold more productive, more predictable, more accelerated, and more cost-efficient for development of superior pure-line or hybrid varieties than current breeding methods (Figs 1 & 2).
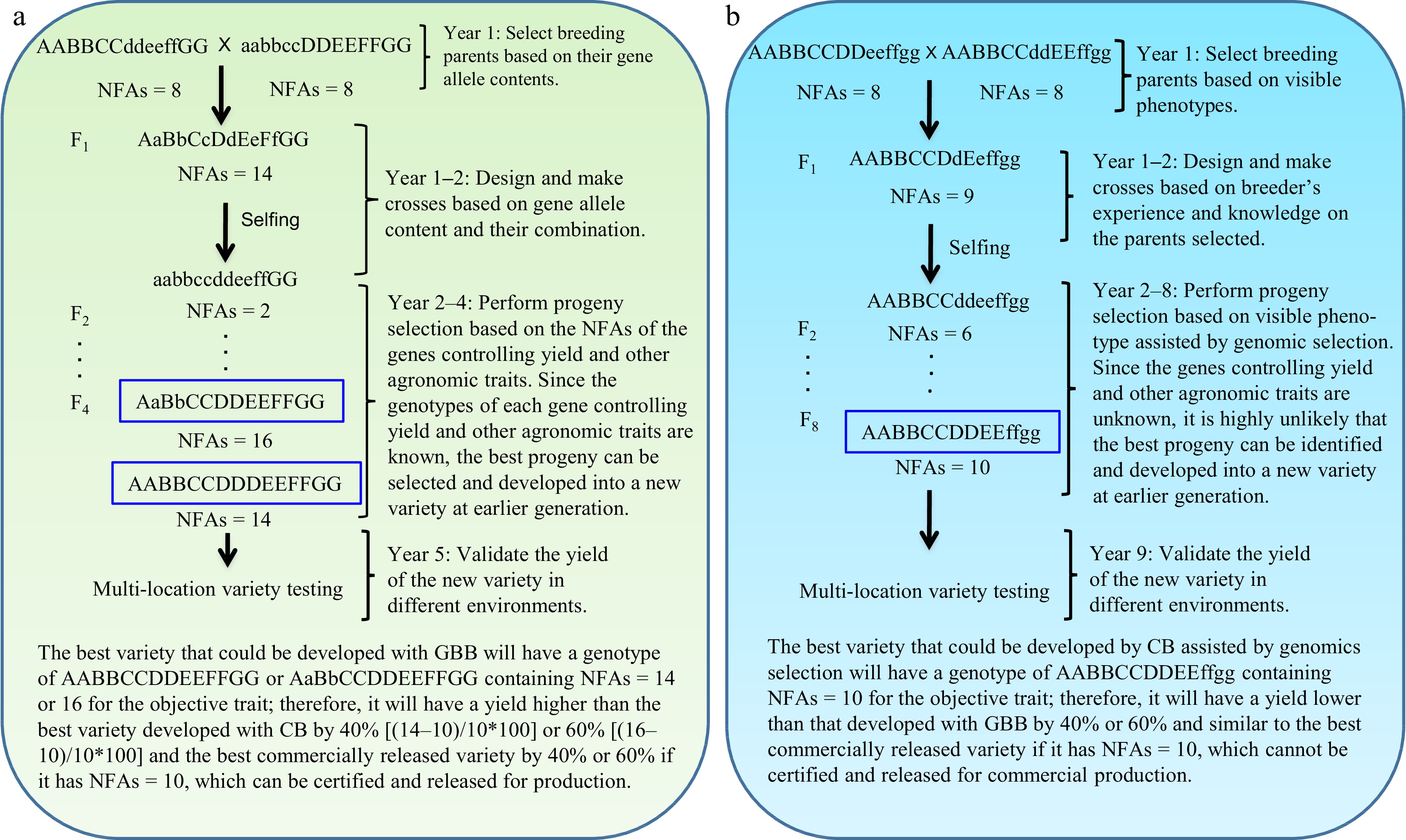
Figure 1.
Gene-based breeding (GBB) for pure-line varieties using (a) the total NFAs of the genes controlling breeding objective trait (yield) vs (b) the current breeding (CB) assisted by genomic selection. NFAs of the genes controlling yield are used for parent selection, crossing design, and progeny selection. Where a GBB variety is released for commercial production will be determined by multi-location variety testing. Assuming that yield is controlled by seven genes, with AA and BB having over-dominant effects (NFAs = 3 for Aa or Bb); CC, EE, and GG having additive effects (NFAs = 1 for Cc, Ee, or Gg); and DD and FF having complete dominant effects (NFAs = 2 for Dd or Ff) on yield. The total NFAs of the genes in an individual plant is the sum of the NFAs of individual genes for the plant. Therefore, the parents selected, with each having NFAs = 8, for GBB and CB, have similar phenotypes for the objective trait, but they have different contents of the genes controlling the objective trait. As yield is controlled by more than one thousand genes[13], it is impossible for breeders to identify the most desirable breeding lines as breeding parents and optimally combine most, if not all, of them into a new variety with CB. GBB can do so because it allows identification of the breeding parents best meeting the breeding objectives and allows optimal crossing design that can maximally combine the favorable alleles and heterosis genotypes into a new variety. The current best commercial variety is assumed to have NFAs = 10.
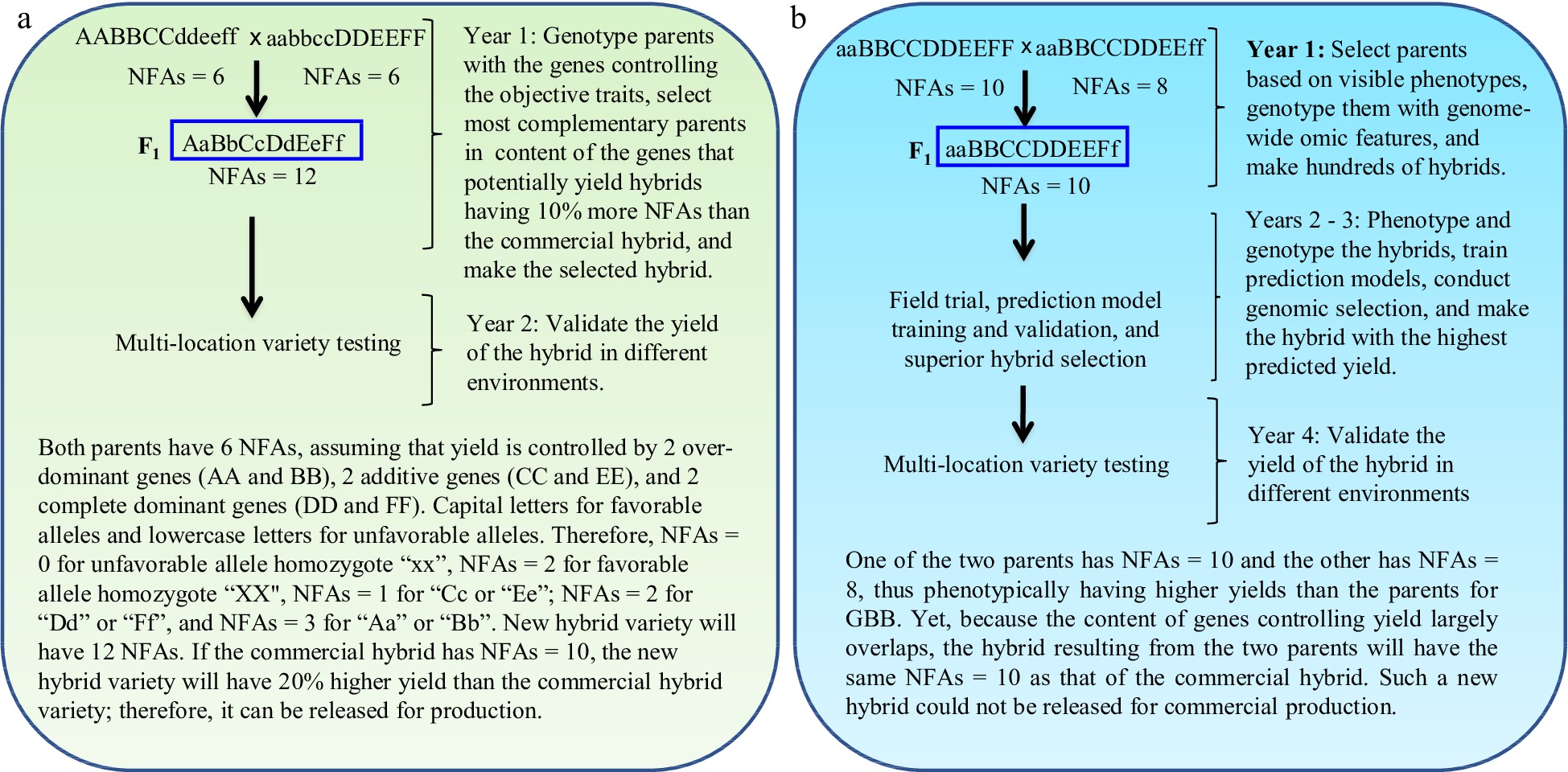
Figure 2.
Gene-based breeding (GBB) for hybrid varieties using (a) the total NFAs of the genes controlling breeding objective trait (yield) vs (b) the current breeding (CB) assisted by genomic selection. The NFAs of the genes controlling the objective trait are used for parent selection, crossing design, and progeny selection. Simplifying for example that yield is controlled by six genes, with capital letters for favorable alleles and lowercase letters for unfavorable alleles. The total NFAs of the genes in a hybrid is the sum of the NFAs of individual genes for the hybrid. The parents selected for GBB, with each having NFAs = 6 for the objective trait, have lower grain yields than those selected for CB, with each having NFAs = 8 or 10. As yield is controlled by more than one thousand genes[13,14], it is impossible for breeders to identify the most desirable inbred lines as breeding parents and optimally combine most, if not all, of them into a new hybrid variety with CB. GBB can do so because it allows identification of the breeding inbred line parents best meeting the breeding objectives and allows optimal crossing design that can maximally combine the favorable alleles and heterosis genotypes into a new hybrid variety. The current best commercial hybrid variety is assumed to have NFAs = 10.

Figure 3.
Prediction accuracies of agronomic traits using genic datasets of the genes controlling breeding objective traits for GBB. (a) Prediction accuracies of cotton fiber length using the SNPs/InDels contained in 226 GFL (Gossypium fiber length) genes[12]. The 226 SNPs/InDels were selected from the 740 SNPs/InDels, with one SNP per GFL gene. (b) Prediction accuracies of cotton fiber length using the total NFAs of the 226 GFL genes versus those predicted with 3,000 genome-wide random SNPs[15]. (c) Correlation of predicted cotton fiber lengths between the NFAs and the SNPs of the 226 GFL genes[15]. (d) Correlation of predicted cotton fiber lengths between the NFAs and the transcript expressions of the 226 GFL genes[15]. (e) Prediction accuracies of maize inbred line grain yields using the expressions of the transcripts of the ZmINGY (Zea mays inbred grain yield) genes responsible for grain yield[13,27]. (f) Prediction accuracies of maize hybrid grain yields from their parents using the expressions of the transcripts of the ZmF1GY (Zea mays F1 hybrid grain yield) genes responsible for hybrid grain yield vs those with the expressions of 122,029 genome-wide random transcripts[14]. For the abbreviations of prediction models, see Liu et al.[12,15] & Zhang et al.[13,14].
-
GBB was first initialized and reported in 2014 by Liu et al.[10] & Zhang et al.[11] based on the following hypotheses and facts:
(1) Genes are the primary determinants of genetic performances of all traits, thus being at the central position of plant and animal breeding, despite modulation of the performance of traits by epigenetic factors, such as small RNAs and methylation, and variable environments (G × E interaction). Therefore, a variety having a desirable gene content, especially their favorable alleles, heterosis genotypes, and desirable networks, will perform best when grown in an environment to which it is best adapted. In fact, when the expressions of the gene transcripts controlling breeding objective traits are used for GBB, epigenetic factors and G × E interaction are included because gene transcript expressions are a consequence of not only gene activities, but also gene × gene interaction, gene × non-gene element interaction, G × E interaction, and epigenetic factors such as small RNAs and methylation.
(2) The molecular basis of breeding is to continuously incorporate the favorable alleles and heterotic genotypes of the genes controlling objective traits into new varieties.
(3) Most agronomic traits, such as yield, quality, yield and quality component traits, and biotic/abiotic stress tolerances, are controlled by numerous genes, probably over 1,000 genes[10−14] (e.g., GenBank acc. No.: MW082098–MW082571), which makes it impossible or difficult for breeders to simultaneously manipulate such large numbers of genes for efficient breeding and to incorporate most, if not all, of their favorable alleles and heterosis genotypes from parents into new varieties.
(4) Both pure-line varieties, e.g., bread wheat and soybean, and hybrid varieties, e.g., maize, rice, most vegetable crops, and most fruit trees, are used in food production. The current breeding procedures, whether for pure-line or hybrid varieties, all include parent selection, crossing design, and progeny selection, followed by multi-location variety testing; therefore, GBB could be performed in a manufactory manner in greenhouse or phytotron. Where a GBB variety is grown can be determined by multi-location variety testing, thus dramatically accelerating the breeding process, increasing breeding efficiency, and reducing breeding cost.
GBB has been first tested in cotton for pure-line variety breeding using fiber length, a typical quantitative agronomical trait with a high heritability of 0.83−0.90, as a breeding objective trait[10,12,15,16] and in maize for inbred line and hybrid variety breeding using grain yield, an extremely complex agronomical trait with a low or moderate heritability of 0.41–0.62, as a breeding objective trait[11,13,14]. In summary, the following have been learned from these studies. First, since the genes controlling breeding objective traits are used, GBB is efficient not only for progeny selection, but also for parent selection and crossing design that are critical to success of breeding. Therefore, the most desirable parents can be selected to approach the breeding objectives; an optimal crossing design can be performed to maximally incorporate the favorable alleles and heterosis genotypes of the genes controlling breeding objective traits from breeding parents into new progeny; and the best individual in the objective traits can be accurately identified from the progeny pool and rapidly developed into a new elite variety or breeding line at earlier generations. It is essential for a successful breeding program to have breeding parents that are the most complementary in content of the genes controlling breeding objective traits and that potentially yield progeny with performance better than the currently released commercial varieties and to have a cross that can maximally combine the favorable alleles and heterosis genotypes of the genes from parents into progeny containing individuals more superior than the currently released best commercial variety. It is impossible for a breeder to identify superior individuals in a progeny pool and to develop them into superior varieties if no superior individual exists in the progeny pool. For example, two higher-yielding parents can be selected for breeding in a current breeding (CB) program that may be assisted by genomic selection, without a comprehensive knowledge of the genes underlying yield. But if they have the same or similar set of alleles for the genes controlling yield, no genetic improvement may result from crossing two seemingly higher-yielding or better parents (Fig. 2). On the other hand, two seemingly phenotypically mediocre lines but possessing complementary allele combinations may be selected as the parents for GBB if detailed knowledge of their favorable alleles and heterosis genotypes is available, and when crossed, can produce superior progeny, from which superior varieties can be developed[3] (Figs 1 & 2). In comparison, genomic selection, a genomics-assisted method developed for genome-wide assisted progeny selection[7], has been extensively studied in over the past 20 years for progeny selection in breeding programs based on genome-wide random SNP markers[17−22], genome-wide gene expressions[23], or genome-wide metabolites[23,24]. Nevertheless, although it has been shown that genomic selection was efficient for progeny selection, no study has been reported, to the best of my knowledge, about its efficiency for parent selection and crossing design. Second, for progeny selection GBB predicted the cotton fiber length phenotypes of breeding progeny and maize inbred line grain yield and F1 hybrid grain yield from parents, for instance, at a prediction accuracy (r, correlation coefficient between predicted and observed phenotypes) of 0.83–0.86[12−16] (Fig. 3). These prediction accuracies of GBB were higher than those of genomic selection for the same or similar types of populations with cross-validation schemes using tens to hundreds of thousands of genome-wide random DNA markers, gene expressions, or metabolites by 116% for cotton fiber length[12], 63% for maize inbred line grain yield[13], and 27%–406% for maize F1 hybrid grain yield from parents[14]. Furthermore, the phenotypes of breeding objective traits can be predicted with genic SNP/InDel markers, NFAs, and expressions, individually or jointly, for GBB's progeny selection. When the phenotypes of the traits predicted with two or all three of the genic datasets were jointly employed for progeny selection, the top 10% plants selected by GBB were consistent up to 100% with the top 10% plants selected based on the phenotypes determined by standard replicated field trials[12,13] (Table 1). Third, development of varieties with GBB can be designed according to breeder's objectives, including selecting parents that can potentially result in progeny having more NFAs than the current best commercial variety (Fig. 1), designing crosses that can maximally combine the favorable alleles and heterosis genotypes of the genes from parents into progeny, and accurately identifying the individuals that have more NFAs than the current best commercial variety. Therefore, the results of breeding efforts can be much more predictable with GBB than with current breeding methods. Fourth, since it is based on genes controlling breeding objective traits for parent selection, crossing design, and progeny selection, GBB can be practiced in a manufactory manner in greenhouses or phytotron before multi-location variety testing, thus accelerating the breeding process, reducing breeding cost, and increasing genetic gain per unit time dramatically. Fifth, the varieties developed by GBB can be readily fine-tuned by gene or genome editing, if they have undesirable traits, because the genes controlling objective traits are known, including the SNP/InDel mutations, expression variation, and interactions or networks of the genes and their impacts on performance of objective traits. Sixth, it is often necessary for all genomic selection projects to train and validate prediction models with a portion of the targeted breeding population, known as a training population[8,18,25], which is costly and slows the breeding process. In comparison, when the NFAs of the genes are applied for GBB, a training population and a prediction model are unnecessary for progeny performance prediction and selection because the total NFAs of the genes controlling objective traits were correlated with the performance of the objective traits in both cotton and maize (r = 0.85, p < 0.0001)[13−16] and can be directly used for progeny selection. Seventh, when the total NFAs of the genes controlling breeding objective traits are used for GBB, the computation for phenotype prediction of the objective traits that is necessary by means of super-computing facility is dramatically simplified. A simple calculator is sufficient to calculate the total NFAs of the genes controlling a breeding objective trait in an individual or a hybrid, which is the sum of the NFAs of individual genes controlling the trait in the individual or hybrid (Table 2). Eighth, a simple and rapid method can be developed for high-throughput genotyping of genes by sequencing the genes only controlling the objective traits for GBB, with which > 1,000,000 progeny individuals can be genotyped with more than 2,000 genes that control 10–30 agronomic traits by an Illumina sequencer (HiSeq 4000) run at a cost of <
${\$} $ Table 1. Progeny selection of gene-based breeding (GBB) for top 10% plants with the highest grain yields predicted with ZmINGY genes vs phenotypic selection (PS) of conventional breeding for top 10% plants with the highest grain yields determined by replicated field trials for inbred line breeding in maize[13].
Consistency of GBB with PS ZmINGY genic datasets I II III I + II I + III II + III I + II + III Field trials, Halfway, Texas, 2010 40.0% 50.0% 66.7% 100.0% 66.7% 100.0% 100.0% Field Trials, College Station, Texas, 2010 41.2% 33.3% 55.6% 80.0% 100.0% 100.0% 100.0% I. Number of favorable alleles (NFAs) of 27 SNP/InDel-containing ZmINGY genes; II. SNPs/InDels of the 27 SNP/InDel-containing ZmINGY genes; III. The transcript expressions of the 150 key ZmINGY genes. Note that when the grain yields of the plants predicted with two or all three genic datasets of the ZmINGY genes were jointly used for progeny selection, the top 10% plants selected with the highest grain yields predicted with the genes were consistent up to 100% with those selected with the highest grain yields determined by replicated field trials. Halfway, Texas and College Station, Texas represent two different agricultural ecosystems and climate zones in the USA. Table 2. Statistics of NFAs of each gene and total NFAs of all genes controlling an agronomical trait[15].
Statistical analysis (ANOVA and LSD) Effect Genotype of a gene AA Aa aa AA > Aa > aa Additive 2 1 0 Aa = AA > aa complete dominant 2 2 0 Aa > AA > aa Over-dominant 2 3 0 Allele 'A' is the favorable allele over allele 'a', when 'AA' is larger than 'aa' (p ≤ 0.05). The total NFAs of all genes controlling an agronomical trait is calculated with the following formula: y = ∑xi, where y is the total NFAs of the genes controlling an agronomical trait, x is the NFAs of individual genes controlling the trait, which is 0, 1, 2, or 3, in individual 'i'. 
Figure 4.
Genetic potential of current cotton varieties or advanced breeding lines having a fiber length of 33.8 mm predicted using the total NFAs of the 226 GFL genes contained in cotton advanced breeding plants or lines[16]. (a) Training linear and non-linear models using the fiber lengths of 198 advanced breeding lines determined by multiple replicated field trials and the total NFAs of their 226 GFL genes controlling fiber length. (b) Predicting the genetic potential of the current cotton variety or advanced breeding line with the longest fiber length using the trained linear and non-linear models, respectively. The current cotton variety or advanced breeding line with the fiber lengths (33.8 mm) of the current best cotton varieties only contained 52% of the 728 total NFAs of the 226 GFL genes and that they could potentially be further improved by up to 118% for the linear model or 73% for the non-linear model, if all 728 NFAs of the 226 GFL genes are incorporated into a new variety through GBB.
-
As GBB is a recently developed novel technology for plant and animal breeding, additional research and development are needed to obtain a robust GBB system for improved pure-line or hybrid variety development in a crop or livestock species. Although the AI technology has been already incorporated into GBB, improvement of the AI is necessary to further empower GBB for variety development. Research and development of GBB in cotton and maize have shown that development of a robust GBB system for a crop or livestock species could be accomplished within 5–10 years, from beginning to a robust GBB system. For instance, it took only five years for Liu et al.[10,12] & Zhang et al.[11,13,14] to genome-wide clone and functionally characterize most, if not all of, the genes controlling fiber and oilseed yields and fiber yield and quality component traits in cotton and the genes controlling grain yield and grain yield and quality component traits in maize, respectively, using a novel technology, designated artificial intelligence (AI)-gExpress, and approximately
${\$} $ -
GBB can be a revolutionary technology for breeding in all field crops, vegetable crops, fruit trees, and livestock for either pure-line varieties (or strains) or hybrid varieties, but only a preliminary GBB system has been established to date in maize and cotton. Additional research is necessary to develop the GBB in maize and cotton into robust GBB systems that are suited for enhanced breeding across environments and across populations in different breeding programs. Importantly, research and development of GBB in crops and livestock can also promote research, development, and application of molecular precision agriculture such as gene-based agriculture for enhanced food production, and transition of human medicine from current phenotypic medicine to genotypic medicine for improved medicine, such as gene-based health, gene-based clinics, and gene-based medicine. The invention of the AI-gExpress technology for genome-wide high-throughput cloning of genes controlling biological traits or processes, genome-wide high-throughput coning of the genes controlling cotton fiber length[10,12] and the genes controlling maize grain yield[11, 13,14], and the research in utility and efficiency of the cloned genes for GBB[12−16] have, for the first time, made it feasible and possible to genome-wide high-throughput clone most, if not all, of the genes controlling biological traits or processes in a species and have, for the first time, revealed that the actual number of genes controlling a biological trait or process much larger than that expected and that identified by QTL mapping and genome-wide association studies. This finding indicated that a quantitative or complex trait is much more complicated than we expected. This knowledge is crucial to the development of new methods and/or improvement of current methods for plant and animal breeding and human medicine for which the number of genes controlling the objective trait and their interactions should be considered. The three genic datasets of the genes controlling breeding objective traits used for GBB, including their NFAs, their SNPs/InDels significantly influencing the performance of objective traits, and the expressions and networks of their transcripts responsible for the phenotype and performance of the objective traits, are essential to research, development, and application of molecular precision agriculture. For instance, agricultural production practices, such as fertilization, irrigation, disease, pest and weed control, and drought and heat/cold stress mitigation, could be carried out based on and/or by regulating the activities and/or networks of genes controlling agronomical traits important to agricultural production. For human medicine, the SNPs/InDels that significantly influence human health or lead to human genetic diseases and/or the expressions and networks of gene transcripts responsible for human diseases are useful for human health, clinics, and medicine. The methodologies developed for GBB, such as the method for high- or moderate-throughput genotyping of individuals with genes controlling biological traits or processes, and the high- or moderate-throughput, high quality, rapid, and economical method for RNA extraction, can be used for both molecular precision agriculture and human precision medicine, including health, clinics, and medicine.
-
The author confirms sole responsibility for the following: study conception and design, data collection, analysis and interpretation of results, and manuscript preparation.
-
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
This work was supported in part by the Agriculture and Food Research Initiative Competitive Grant of the USDA National Institute of Food and Agriculture (2013-67013-21109), the Texas A&M AgriLife Research Crop Improvement Program (06–124329–85360 and 06–124215–85360), and the Texas Corn Producer Board (TCPB) (408116–85360).
-
The author declares that there is no conflict of interest. Hongbin Zhang is the Editorial Board member of Tropical Plants who was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of this Editorial Board member and the research groups.
-
Received 19 December 2023; Accepted 15 January 2024; Published online 20 February 2024
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press on behalf of Hainan University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Zhang HB. 2024. Gene-based Breeding (GBB), a novel discipline of biological science and technology for plant and animal breeding. Tropical Plants 3: e005 doi: 10.48130/tp-0024-0005
Gene-based Breeding (GBB), a novel discipline of biological science and technology for plant and animal breeding
- Received: 19 December 2023
- Accepted: 15 January 2024
- Published online: 20 February 2024
Abstract: Gene-based breeding (GBB) is an innovative technology and science for plant and animal breeding. Studies have shown that GBB is extremely powerful, predictable, accelerated, and cost-efficient for both pure-line and hybrid variety breeding. Moreover, the concepts, principles, techniques, and methodologies developed and used for GBB are also applicable to molecular precision agriculture, such as gene-based agriculture, and molecular precision medicine in humans as well as in animals, such as gene-based health, gene-based clinics, and gene-based medicine. Therefore, research, development, and applications of GBB for plant and animal breeding are promising to promote substantial crop and livestock genetic improvement, enhanced agriculture production, and improvement and transition of current phenotypic medicine to genotypic medicine in humans and animals.
-
Key words:
- Gene-based Breeding /
- Molecular breeding /
- Genes controlling agronomic trait /
- Plant /
- Animal












