-
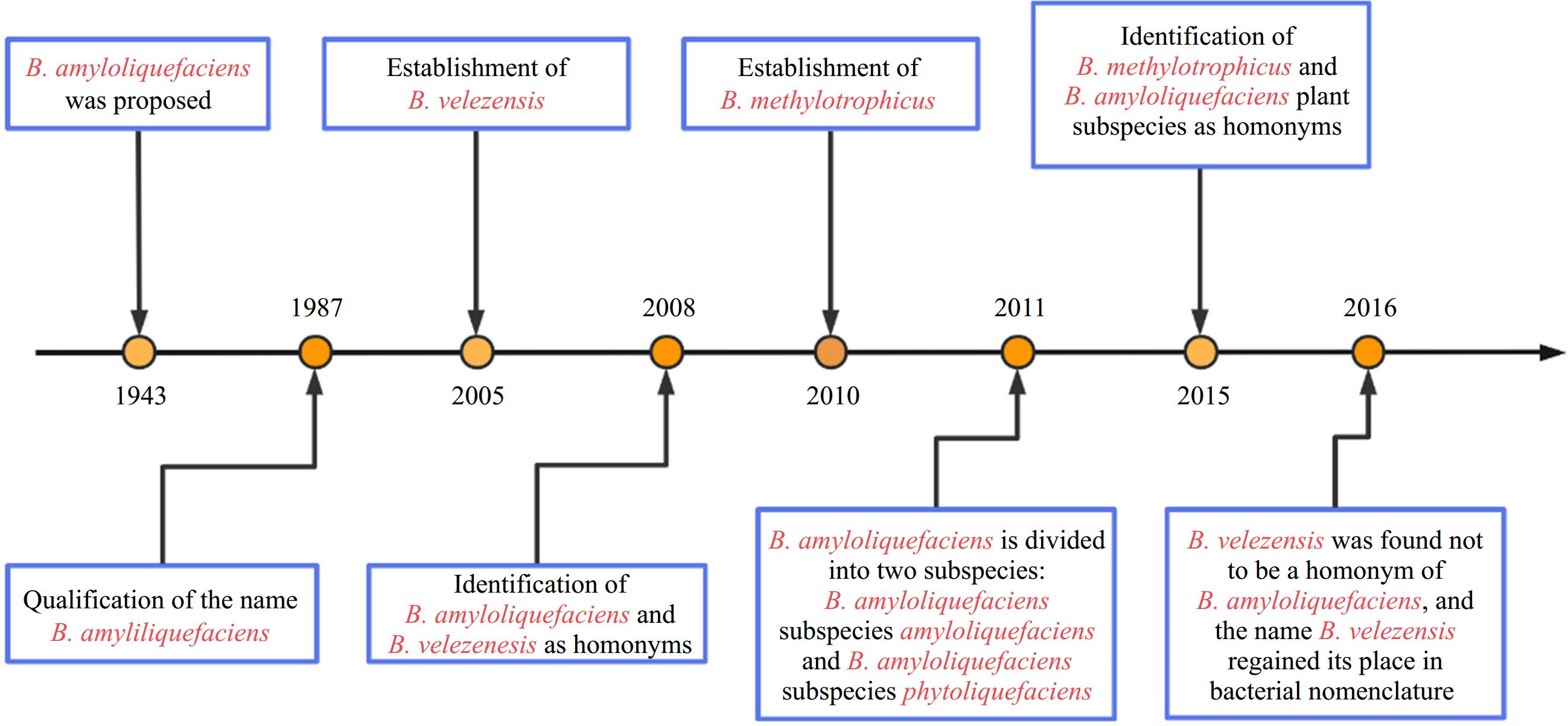
Bacillus spp. represent a diverse and extensive group of aerobic or facultative anaerobic, rod-shaped, endospore-forming, gram-positive bacteria widely distributed in the environment. These bacteria have been extensively studied as biocontrol agents because of their extreme resistance, antagonistic activity, and excellent environmental adaptability[1]. According to the latest edition of Bergey's Manual of Systematic Bacteriology, 142 species have been identified within the genus Bacillus and their number is still growing[2]. With a wide range of genomic DNA G+C contents among its species, the genus Bacillus is currently recognized as an evolutionarily and phylogenetically heterogeneous group consisting of highly diverse organisms[2]. For many years, distinguishing these species based on conventional phenotypic methods has posed significant challenges. Furthermore, phylogenetic analysis targeting the 16S rRNA gene also proved insufficient for discriminating species within the Bacillus complex because of its highly conserved nature (Fig. 1)[3]. Differentiation of Bacillus species relies primarily on the significantly low DNA relatedness values experimentally determined by DNA–DNA hybridization and distinct fatty acid profiles[4]. In addition to the 'original members', B. subtilis, B. licheniformis, and B. pumilus described by Gordon et al. in 1973[5], numerous novel species within the B. subtilis species complex have been documented in recent decades: B. amyloliquefaciens[4], B. atrophaeus[6], B. mojavensis[7], B. vallismortis[8], B. sonorensis[9], B. velezensis[10], B.axarquiensis[11], B. tequilensis[12], B. aerius, B. aerophilus, B. stratosphericus, B. altitudinis[13], B. safensis[14], B. methylotrophicus[15], B. siamensis[16], B. xiamenensis[17], B. vanillea[18], B. paralicheniformis[19], B. glycinifermentans[20], B. gobiensis[21], and B. nakamurai[22]. B.vanillea and B. methylotrophicus could not be confirmed as valid species and were later identified as heterotypic synonyms of either B. siamensis[22], or B. velezensis[23]. B. subtilis is subdivided into three subspecies: B. subtilis subsp. subtilis, B. subtilis subsp. spizizenii[24], and B. subtilis subsp. inaquosorum
(Fig. 2)[3].
Figure 1.
Phylogeny of selected strains of species from B. amyloliquefaciens, B. atrophaeus, B. mojavensis, B. vallismortis, B.axarquiensis, B. tequilensis, B. aerius, B. aerophilus, B. stratosphericus, B. altitudinis, B. safensis, B. methylotrophicus, B. siamensis, B. xiamenensis, B. vanillea, B. paralicheniformis, B. glycinifermentans, B. gobiensis, and B. nakamura clades reconstructed from a neighbor-joining analysis of core-genome sequence data.
-
B. velezensis is an aerobic, gram-positive, endospore-forming bacterial species commonly found in soil[25]. In addition, B. velezensis is abundant in various plant tissues, such as roots[26], leaves[27], stems[28], and even bulbs[29]. This species promotes plant growth and demonstrates potential as a potent biocontrol agent against plant pathogenic fungi that can serve as an alternative to chemical fungicides. Furthermore, B. velezensis produces various secondary metabolites with broad-spectrum antimicrobial activities, which promote both plant and animal growth[30].
B. velezensis was initially isolated in 1999 but officially reported and named in 2005. B. velezensis strains CR-502T and CR-14b were first isolated from environmental samples collected in the mouth of the Vélez River at Torredelmar, in the province of Málaga, Spain[10]. Phylogenetic analysis based on RNA polymerase beta-subunit gene sequences and the core genome revealed that B. velezensis belongs to a conspecific group consisting of B. velezensis, B. methylotrophicus, and B. amyloliquefaciens subsp. Moreover, in 2016, several other Bacillus species previously classified as B. amyloliquefaciens subsp. plantarum, B. methylotrophicus, and B. oryzicola were re-classified as strains of B. velezensis[19,23,31].
-
In recent years, there has been an increasing number of reports on B. velezensis due to its significant application in agriculture and biotechnology. It is widely employed as a biocontrol agent for promoting plant growth through phytohormone production and suppressing competitive plant pathogenic microbes with an extensive range of secondary metabolites[32]. The corresponding genomes are presented in Table 1.
Table 1. B. velezensis genomic data.
Strain Genome size C+G content Ref. FZB42 3,918,589 46.4 [33] S3-1 3,929,772 46.5 [34] M75 4,007,450 46.6 [35] GH1-13 4,071,980 46.2 [36] LS69 3,917,761 46.4 [37] 9912D 4,241,576 45.99 [38] 157 4,013,317 46.41 [32] SGAir0473 4,184,178 45.96 [39] 5RB 3,910,395 46.5 [40] HAB-2 3,894,648 46.6 [41] GUMT319 3,940,023 46.6 [42] KMU01 3,932,437 46.5 [43] K26 4,047,350 46.15 [44] SC60 3,962,671 46.46 [45] AF_3B 3,949,216 46.3 [46] DMB06 4,157,945 46.2 [47] US1 4,132,553 46 [48] Q12 4,182,261 46.1 [48] HMB26553 4,204,437 46.4 [49] The genome of the plant-associated strain B. velezensis FZB42 first sequenced in 2007 revealed nine large gene clusters. Five of these clusters encode biosynthetic enzymes belonging to the extensive enzyme complexes of non-ribosomal peptide synthetases (NRPSs) and are involved in the synthesis of non-ribosomal lipopeptides (LPs)[33]. Based on the whole-genome analysis, B. velezensis C4341 was predicted to encode 13 putative gene clusters responsible for the biosynthesis of antimicrobial metabolites. Among these clusters, eight encode NRPSs that synthesize LPs (one bacillibactin, one iturin, five surfactin, and one fengycin), and five that synthesize ribosomal peptides (RPs) (one microcin and four lantipeptides) that play crucial roles in the biocontrol process[27]. Chen et al.[32] reported the complete genome sequence of B. velezensis 157, which consists of a circular 4,013,317 bp chromosome and one circular 8,439 bp plasmid with 3,789 protein-coding sequences located on the chromosome, as well as 27 rRNA genes, 86 tRNA genes, and 11 sRNA genes. They also identified a unique protein with an acyltransferase domain essential for synthesizing polyketide synthase (PKS), which is potentially associated with secondary metabolite biosynthesis, transport, and catabolism of secondary metabolites. In addition, eight gene clusters encoding various NRPSs and antimicrobial PKS were found[32]. Whole-genome sequencing of B. velezensis also revealed a multitude of biosynthetic gene clusters encoding genes responsible for the synthesis of antifungal compounds[44]. B. velezensis B-4 contains 12 gene clusters related to the synthesis of antimicrobial metabolites[50]. Xu et al.[41] reported a high-quality genome sequence of HAB-2 using third-generation sequencing technology, obtaining a genome size of approximately 3,894,648 bp and identifying 13 gene clusters involved in the synthesis of antifungal and antibacterial secondary metabolites. A comparative analysis of the FZB42 and HAB-2 genome sequences showed that the two strains had very similar genome sizes, HAB-2 had one more complete prophagosomal region [Pr 2, 46 CDS], whereas FZB42 had one incomplete gene cluster and fewer complete gene clusters than HAB-2. In conclusion, previous whole-genome sequencing studies of B. velezensis have revealed an abundance of biosynthetic gene clusters encoding enzymes catalyzing the synthesis of compounds with antifungal properties[51], such as surfactins[52], fengycins[53], and bacillomycin D[54], which could control growth of pathogenic fungi.
-
Interactions of plants with beneficial microbiota can induce systemic resistance, enhance nutrient acquisition through processes such as nitrogen fixation, phosphorus solubilization, and secretion of siderophores[55], compete for niches within the rhizosphere, promote mycorrhizal functioning and alter the microbial community structure in the rhizosphere. These interactions can ultimately improve plant fitness by promoting growth, alleviating stress, and protecting against pathogens, so they are crucial for plant health[56]. Plant growth-promoting bacteria have been demonstrated to recruit beneficial taxa and suppress soil-borne pathogens. Recruitment of beneficial species can attenuate diseases and promote the growth of plants as well as reshape the structure and functionality of the soil microbiome[57].
Cooperative-competitive trade-offs among microbial interactions are often the consequence of balancing benefits, such as the sharing of public goods and cross-feeding, and costs, including resource competition and stress tolerance[58]. Sun et al.[57] demonstrated that the application of rhizosphere probiotic B. velezensis SQR9 not only promotes plant growth independently but also induces significant enrichment of Pseudomonas spp. among soil microorganisms through its metabolites, which were attracted by root exudates and colonized the rhizosphere forming biofilms on plant roots in conjunction with B. velezensis strain SQR9. In metabolic modeling, metabolomics analysis, and functional gene knockout experiments of synthetic flora, it has been certified that a PGPR (B. velezensis SQR9) recruits indigenous beneficial bacteria (Pseudomonas spp.) and establishes cooperative interactions with them via cross-feeding. This mutualistic relationship expands the niche breadth of bacterial flora in the plant rhizosphere, driven by metabolic interdependencies between these two plant probiotics. Consequently, their long-term coexistence leads to synergistic enhancement of plant probiotic effects. Similarly, numerous researches have demonstrated that the assembly of multi-species biofilms at the root-microbe interface, consisting of Bacillus spp. and Pseudomonas spp., can effectively confer protection against pathogen infection in plants[59]. The underlying mechanisms involved in pathogen control may include quorum sensing (QS) signaling, interactions mediated by siderophores, as well as systemically induced metabolite root exudation (SIREM), among other factors (Fig. 3)[60].
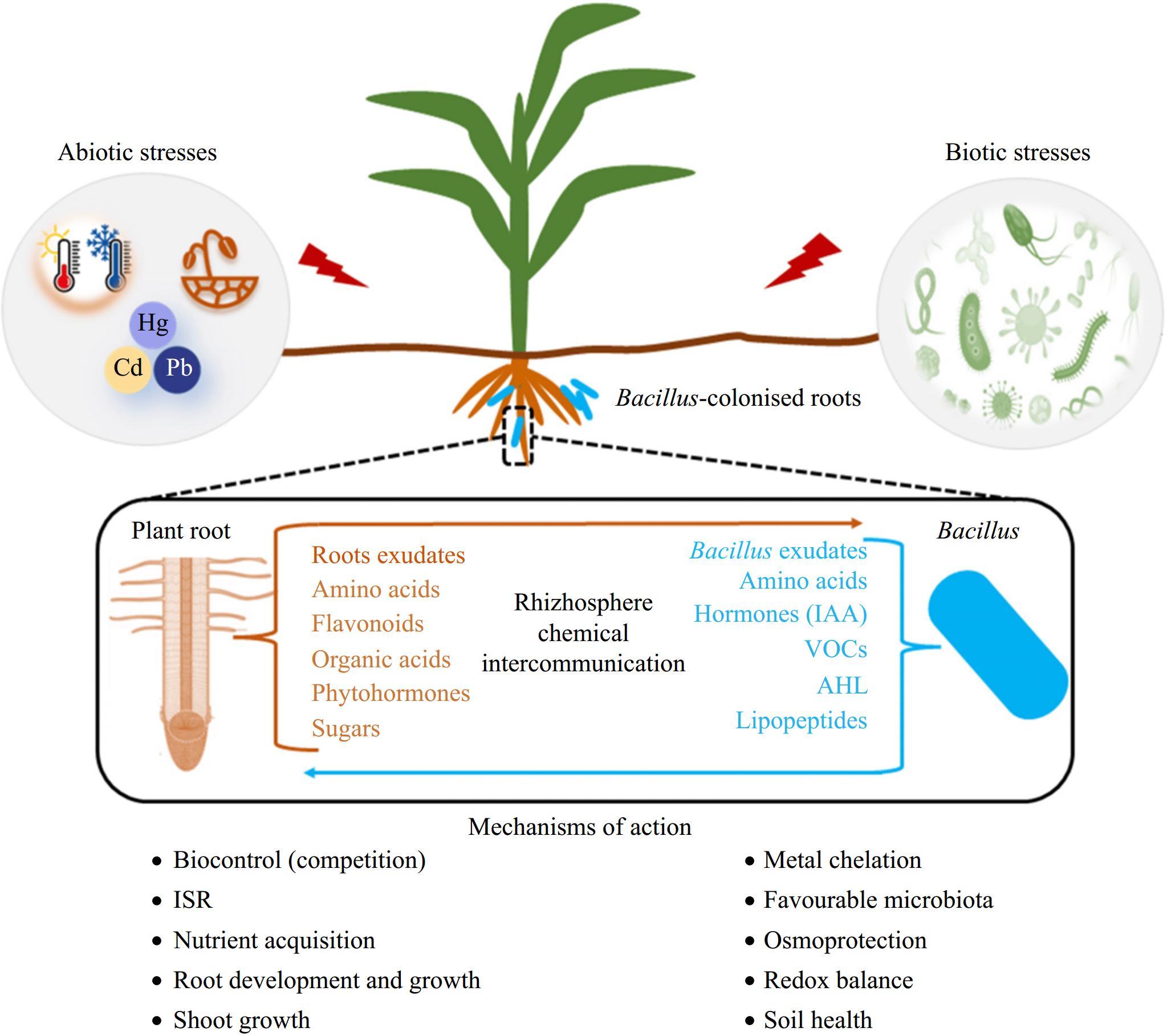
Figure 3.
An overview of mechanisms employed by Bacillus spp. in the mitigation of biotic and abiotic stresses[61].
Biosynthetic gene clusters (BGCs) are responsible for producing various secondary metabolites, which contribute to competition interference between microorganisms and generally provide resistance against self-produced antibiotics to protect host cells[62]. Consequently, closely related species can benefit more from public goods than the cost of resource competition and exhibit a relatively weak antagonistic tendency, primarily driven by nutrient competition costs[63]. By conducting comparative genomic analysis on 4000 strains within the Bacillus genus, Xia et al.[64] identified a consistent distribution pattern and phylogenetic relationship among these strains, which corresponded to the distribution of gene clusters responsible for secondary metabolite biosynthesis. Furthermore, they discovered a significant positive correlation between antagonistic assessments among representative Bacillus strains and genetic and BGC distance, particularly in strains with abundant BGCs. Finally, the mutant experiments demonstrated that the inhibition of the typical beneficial Bacillus SQR9 on different Bacillus strains, which were all derived from the unique BGC of the antagonist bacteria and the absence of the target bacteria and the common BGC of the two bacteria did not play an inhibitory role. As a consequence, metabolic similarity facilitates the negative correlation between antagonism and phylogenetic distance. The relationship between interference competition and phylogenetic distance can promote microorganisms to balance cooperation and competition economically, thereby playing a crucial role in regulating community assembly and evolution[65]. In conclusion, the experiments indicate that Bacillus spp. species collectively play a crucial role in determining root function by exerting specific influences on establishment of root microbial communities.
-
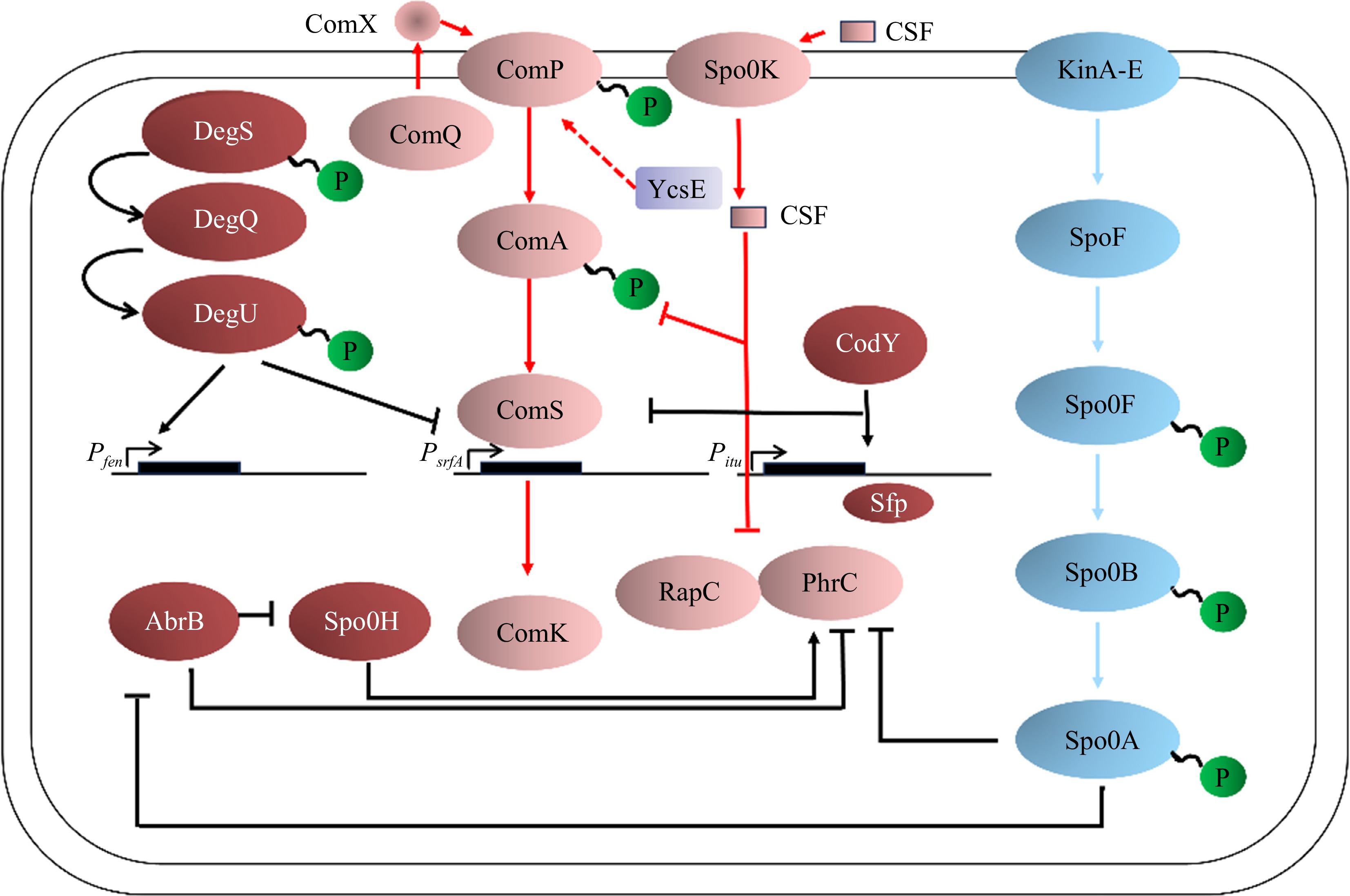
The signaling system is an important system for organisms to sense and transduce internal and external signals and to survive in changing environments[66]. The two-component signaling system (TCS) is a common signaling module in bacteria and consists of a histidine kinase (HK) and a homologous response regulator (RR)[67].
The two-component system of Bacillus is an important part of the phosphate signaling pathway that regulates several important traits. The major two-component regulatory system includes Spo0A-Spo0F, ComA-ComP, and DegU-DegS, which control spore formation, competence acquisition, and surfactin production of Bacillus[68]. The Rap-Phr system is a well-known quorum sensing system for Bacillus. Bacillus spp. form spores when exposed to harsh environmental conditions such as nutrient deprivation[69]. When the intracellular metabolite ATP is suddenly reduced, the histidine kinase KinA-E undergoes autophosphorylation and transfers a phosphate residue to Spo0F, an important intermediate response regulator[70]. Spo0F~P phosphorylates Spo0B and finally transfers the phosphate group to Spo0A[71]. High levels of Spo0A~P activate the expression of genes required for sporulation[72].
DegU and DegS form a pair of two-component regulatory systems that affect many cellular processes, including motility, quorum sensing, biofilm generation, and spore formation. DegU and DegS control many cell population differentiation processes, particularly the differentiation of extracellular protease-producing cells[73]. DegS phosphorylates DegU after sensing external signaling molecules. DegU-P induces aprE expression and secretion of extracellular proteases[74]. The induction of exogenous enzyme expression requires high levels of DegU phosphorylation. Conversely, high levels of DegU-P inhibit exercise[75]. The regulation of the DegU gene depends on the number of phosphorylated proteins in cells, and the low levels of DegU-P activate motility[76]. Finally, conditions that produce high levels of DegU-P inhibit motility and induce the production of extracellular proteases. Thus, DegU can be considered a molecular switch for cell differentiation (Fig. 4)[77].
-
In Gram-positive bacteria, oligopeptides are generally used as quorum-sensing signal molecules and autoinducing peptides (AIP), which secrete and process peptide signal molecules through the specific ATP-binding cassette (ABC)[80]. Peptide signaling molecules are recognized by homologous two-component sensor kinase proteins that interact with response regulator proteins in the cytoplasm, and their signals are amplified in a cascade to ultimately regulate important physiological activities of the bacteria[81]. In Bacillus, research on quorum sensing is relatively complete in B. subtilis, B. cereus, etc., while research on B. velezensis has not yet achieved in-depth results. The QS system of B. subtilis is mainly based on cell density and the development of extracellular signal molecules about competence and induction of sporulation. There are two QS signaling molecules related to the QS system in B. subtilis that are involved in the regulation of cell density, secretion of extracellular signaling molecules, and sporulation, named ComX pheromone and competence and sporulation factor (CSF)[82].
The ComX pheromone is generally a 10-amino acid peptide[83] formed by the 55-tryptophan residue produced by the comX gene and modified by isoprene. It is related to the ComQXPA system (consisting of four regulatory genes, comX, comQ, comP, and comA) and is transported out of the cell membrane by the ABC transporter under the action of ComQ[84]. Under the appropriate cell population density and nutritional conditions, the extracellular ComX pheromones could be continuously accumulated to a certain concentration and interact with the sensor kinase ComP in the ComP/ComA two-component regulatory system and then resulting in the phosphorylation of the ComP-ComA complex to promote the expression of the open reading code group ComS in the srfA promoter, thereby activating the transcription factor ComK and maintaining the stability of the ComK protein to induce the group of B. subtilis into the competent state, and the expression of various physiological and biochemical genes of the bacteria is initiated[85]. Details are shown in Fig. 5.
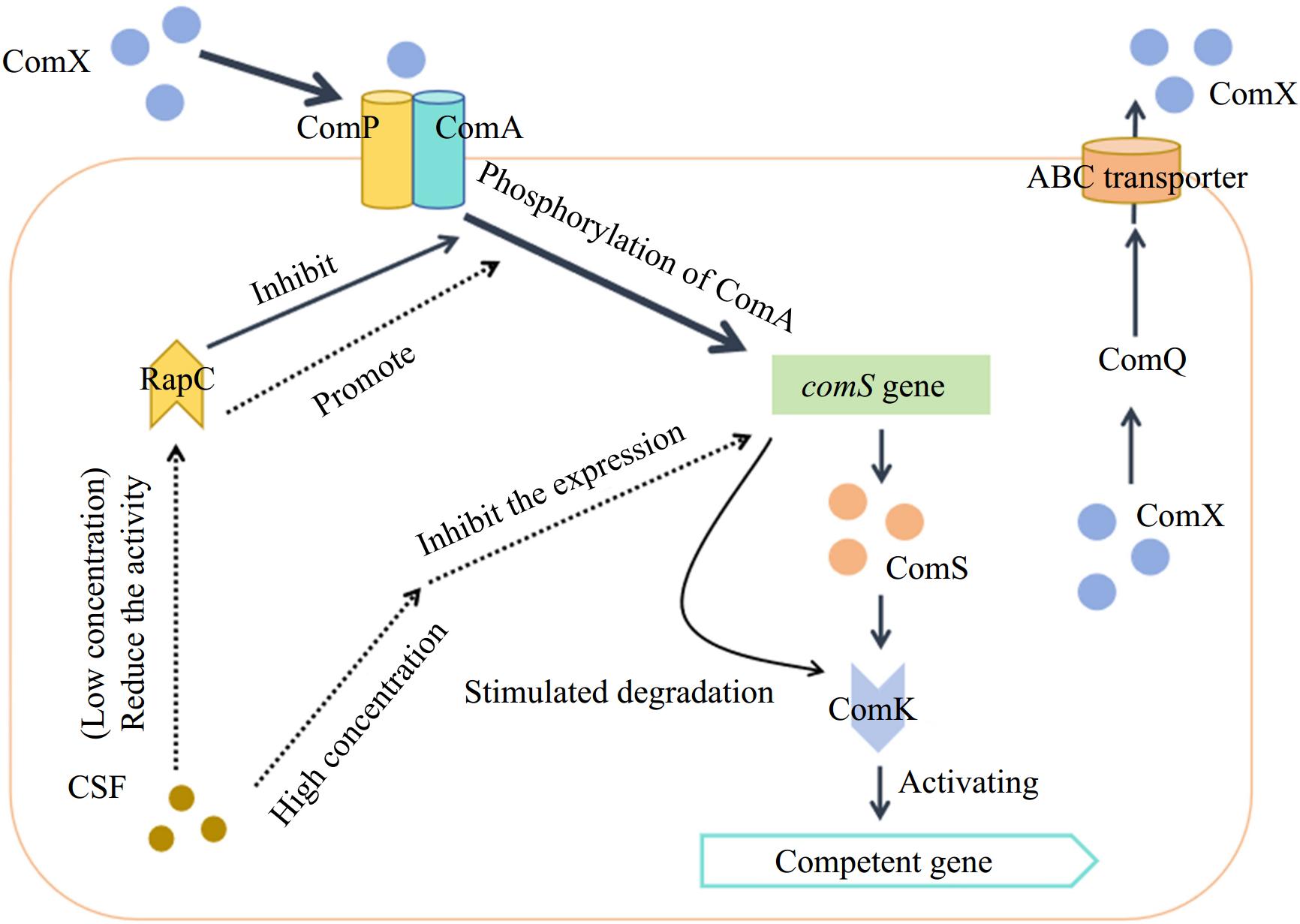
Figure 5.
QS system in B. subtilis[86].
In this process, CSF acts as a regulator of the ComQXPA system depending on its concentration. It could reduce the activity of phosphatase RapC (the phosphatase that inhibits the phosphorylation of ComA) at lower concentrations to increase the concentration of phosphorylated ComA and promote the expression of ComS[87]. Conversely, at higher concentrations, it could inhibit the expression of genes encoding ComS and promote the degradation of ComK protein, thereby promoting sporulation in the B. subtilis population[87].
As a signaling molecule of cell density, the concentration of the Bacillus pheromone ComX increases with cell growth[88]. When its concentration reaches a certain threshold, it binds to the membrane protein histidine kinase ComP and induces the autophosphorylation of ComP. Phosphorylated ComP then transfers the phosphate group to the regulatory protein ComA[89]. Phosphorylated ComA binds to the specific promoter region of the surfactin synthase gene srfA, activates the activity of RNA polymerase, and causes srfA to initiate transcription[90]. We have found that the histidine kinase ComP is also involved in the synthesis of fengycin or (and) iturin and that the phosphatase YcsE can also modulate the synthesis of lipopeptides, thereby affecting bacteriostatic activity (unpublished). The transcriptional expression of the comS and ComS genes is embedded in the srfAB gene, which determines that the Bacillus cells begin to form a competent state[91]. Therefore, the synthesis of surfactin is closely linked to the competence formation. However, the regulation of the QS system has some differences from B. subtilis in B. cereus. At present, it has been confirmed that three sets of QS systems: PlcR-PapR, LuxS, and Rap-Phr, exist in the genome of B. cereus[92], and there are relatively more studies on the PlcR-PapR system. Under environmental stimulation, the papR gene expresses and synthesizes the PapR protein, which is secreted extracellularly through the oligopeptide penetration system (Opp) and binds to the extracellular leader signal peptide to synthesize active PapR heptapeptides, which is then transported back into the bacterial cells through the Opp system. The PapR heptapeptides then interact with PlcR to form a complex, and the complex binds to the PlcR target (palindromic structure) on the DNA, thereby activating the expression of various virulence factors and PlcR regulators, which contain 48 genes[93]. PapR is also regulated by cell density. High cell density increases the intracellular concentration of PapR, which may promote its interaction with PlcR[94], but the molecular mechanism of transcriptional regulation between PapR and PlcR is still unclear.
In addition, previous reports indicate that the ComP/ComA two-component regulatory system also regulates the QS system in B. subtilis and affects the formation of its competence cells[95]. Quorum sensing is a widespread phenomenon in bacteria. It means that bacteria can adjust their physiological and biochemical properties by sensing the density of bacterial populations. It mainly depends on sensing the change in bacterial population density to regulate metabolism-related genes and eventually makes themselves able to adapt to the external environment[96]. For PGPR such as Bacillus, quorum sensing is essential for their rhizosphere colonization and beneficial effects. For example, QS is associated with biofilm formation, which influences the ability of bacteria to colonize the rhizosphere. At the same time, the synthesis of substances in bacteria such as extracellular degradation enzymes and antibiotics is also regulated by the QS system, and it also has a regulatory effect on the metabolism and transport of fatty acids[85].
Quorum sensing in B. velezensis has not been systematically examined. At least 76 genes in B. velezensis are related to the quorum sensing system, 114 genes are related to the two-component system, and 135 genes are related to ABC transporters[97]. According to Na et al.[86], B. velezensis has a DMB05 ComQXPA system similar to that in B. subtilis, and comP in this strain is truncated. The ComQXPA system regulates protease activity of B. velezensis DMB05, which may be related to its quorum-sensing mechanism. Thus, the quorum sensing regulation mechanism may also be similar to that of B. subtilis, but this aspect has not been elucidated in detail[86]. Yang et al. identified luxS, an important quorum-sensing regulator gene, in B. velezensis VJH504, which plays a key role in quorum sensing mediated by 2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (known as autoinducer-2 or AI-2) derived from the spontaneous rearrangement of the precursor 4,5-dihydroxy-2,3-pentanedione[98]. The formation of the latter compound is catalyzed by luxS, which is a S-ribosylhomocysteinase. AI-2 is a quorum-sensing signaling molecule in both gram-positive and gram-negative bacteria; it has a highly conserved structure between species[99], and may also act as an interspecies quorum sensing signal. AI-2 can enhance root colonization by promoting biofilm formation and affecting the motility of B. velezensis SQR9[100]. In particular, it may be similar to the ComX pheromone and regulate B. velezensis biofilm formation by influencing γ-polyglutamic acid synthesis as described by Xiong et al.[100].
In addition, B. velezensis could also be a quorum-quenching (QQ) bacterium that inhibits pathogens and reduces the need for antibiotics. The acyl-homoserine lactonase AiiA is one of the well-known QQ enzymes widely found in Bacillus that recognizes and degrades acyl-homoserine lactones, signal molecules produced by gram-negative bacteria. Quorum sensing inhibition is achieved by intervening in biofilm formation and AHLs/Lux-mediated pathogenic toxin release[101]. For example, Sun et al.[102] found multiple QQ enzymes in B. velezensis strain DH82 regulated the pathogenicity of Vibrio parahaemolyticus. Therefore, B. velezensis could be a novel biological control agent in the future.
-
To optimize the production of a wide variety of secondary metabolites and structurally diverse antagonistic substances, Bacillus strains are characterized by their genomic composition dedicated to synthesizing such compounds. Typically, strains exhibiting robust bacteriostatic abilities allocate a significant portion of their genomic nucleic acid sequences to the synthesis of these substances. For instance, in B. velezensis strain FZB42, genes related to antimicrobial substances constitute approximately 9% of its entire genome, highlighting its capacity in this regard. In contrast, non-plant-associated members of the B. subtilis species complex dedicate only around 5% of their genome to antimicrobial substance synthesis[33].
The active substances produced by Bacillus can be categorized based on their molecular weights and biosynthesis pathways. They include large-molecule proteins and small-molecule compounds, synthesized via ribosomal and non-ribosomal pathways, respectively. Among these active substances, lipopeptides play a crucial role and are synthesized through the non-ribosomal pathway, which also yields polysaccharides[103], and polyketides[104]. In contrast, the ribosomal synthesis pathway produces bacteriocins, enzymes, and other macromolecular proteins[105]. This diverse arsenal underscores the adaptive versatility of Bacillus in ecological niches, particularly in competitive and hostile environments.
Non-ribosomal peptide synthetases (NRPs)
-
Non-ribosomal peptide synthetases (NRPS) are versatile enzymes found in microorganisms like bacteria and fungi, essential for synthesizing secondary metabolites and oligopeptides via a specialized pathway distinct from ribosomal synthesis. These enzyme systems, comprising multifunctional protein complexes produce bioactive compounds known for their inhibitory properties[106]. A typical NRPS is modular in structure, each module containing three core domains: Adenylation (A), which activates amino acids; Thiolation (T), responsible for transporting amino acids and nascent peptides; and Condensation (C), facilitating peptide bond formation[107]. Among the bioactive products, lipopeptides (LPs) are prominent antibiotics Bacillus bacteria synthesize during growth and development. LPs are characterized by their cyclic structure formed by β-fatty acid chains that confer lipophilicity and amino acid peptides that provide hydrophilicity, crucial for their role as effective lipopeptide antibiotics[108].
Lipopeptide compounds (LPs) produced by Bacillus are categorized into three families based on their amino acid sequences: surfactins, iturins, and fengycinsc[108]. Bacillus species differ in their ability to produce LPs: some species can produce three different types of LPs, whereas others are able to produce only one type of LPs[109]. The significant heterogeneity of LPs in terms of the type, amino acid sequence, pattern of peptide cyclization, as well as the nature, length, and branching of fatty acid chains account for the fact that LPs of the same type produced by the same strain are not a single compound, but rather multiple structural analogs[110].
Among the B. velezensis, the biosynthetic gene cluster of the model strain FZB42 is the most well-studied, there are nine huge clusters of anti-biomass synthesis genes in its genome, srf, bmy, fen, dhb, bac, mln, bae, dfn, and nrs, which account for about 10% of the entire genome[111]. Five of the nine gene clusters (srfABCD, bmyCBAD, fenABCDE, dhbABCDEF, and nrsABCDEF) are responsible for encoding the corresponding NRPSs, which comprise a large complex of enzymes responsible for the synthesis of the three major lipopeptide subfamilies, surfactins, iturins, and fengycins[112].
Surfactin family
-
The surfactins are a class of biosurface-active molecules with amphiphilic structures formed by the linkage of β-hydroxy fatty acid chains (C13-C16) and hydrophilic heptapeptide rings (Fig. 6). The amino acid sequence of the peptide ring is Glu-Leu-Leu-Val-Asp-Leu-Leu[114]. B. velezenses produces small amounts of surfactin (< 10% of its biomass), which not only inhibits the growth of other bacteria but also acts as a signaling molecule in inter- or intra-species interactions[115]. The synthesis of Bacillus LPs is usually controlled by several genes. The surfactins are synthesized by complex interactions of non-ribosomal peptide synthetases encoded by the srfA manipulator[116], and the associated synthetic gene is srfABCD, which is approximately 32 kb in size. The srfA manipulator consists of four open reading frames (ORFs): srfAA, srfAB, srfAC and srfAD. Of these, srfAA, srfAB, and srfAC encode modular enzymes responsible for the integration of seven amino acids into the peptide ring[116]. On the other hand, srfAD encodes the thioesterase/acyltransferase structural domain that regulates the initiation of surfactant biosynthesis[117]. In addition, surfactins are regulated by the sfp gene, and the failure of the model strain B. subtilis 168 to produce surfactin is due to a mutation in its sfp gene[118].
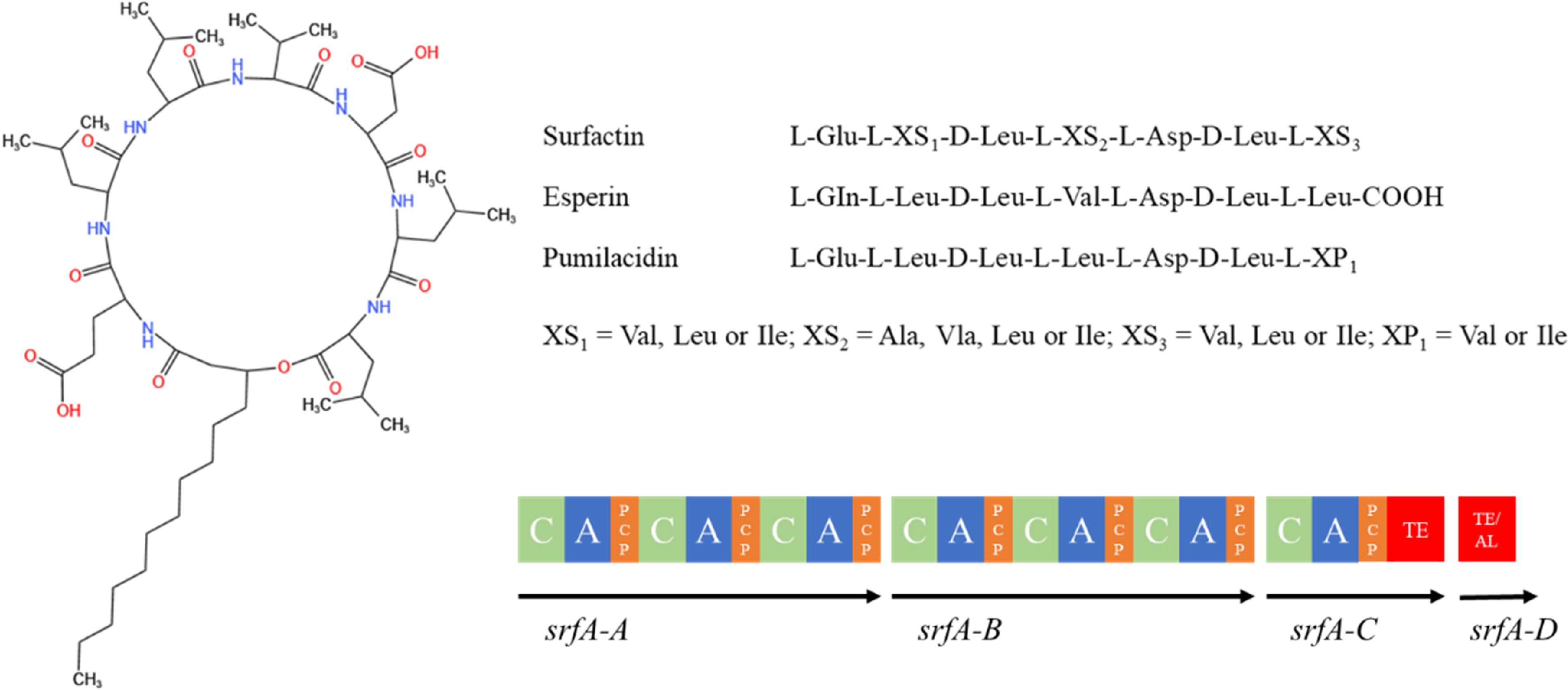
Figure 6.
Structure of surfactin family compounds synthesized by Bacillus species[113]. AL: acyl-coenzyme A ligase structural domain; ACP: peptidyl carrier protein structural domain; C: condensate structural domain; PCP: peptidyl carrier protein structural domain; A: adenylyl domain; TE: thiolipase structural domain.
The mechanism of action of lipopeptide compounds is largely dependent on their amphiphilicity and their ability to interact with the membrane structure of the target organism[119]. In contrast, compounds of the surfactin family as a class of very powerful surfactants, are highly susceptible to interactions with the lipid bilayer of the target, thereby interfering with the target's biofilm structure in a dose-dependent manner. High concentrations of surfactin lead to the formation of irreversible pores in the membrane structure, and these pores further lead to the complete disruption and solubilization of the lipid bilayer[113]. C15Surfactin A produced by B. velezensis HN-2 had antimicrobial activity against Xanthomonas oryzae (Xoo) and could effectively inhibit the infestation of rice by Xoo, and SEM and TEM observations showed that C15Surfactin A caused serious damage to the cell wall structure of Xoo cells[120].
The surfactin was also found to play a very important role in plant-beneficial bacterial interactions. The surfactin can regulate biofilm formation, motility, and colonization of B. velezensis, and through colonization, B. velezensis can feed on inter-root secretions by sensing pectin molecules and subsequently stimulate the bacterium to undergo surface-active pigment production[121]. Zhang et al.[122] found that surfactin has a more critical role in the formation of biofilm in B. velezensis compared to other Bacillus species. Moreover, since the synthesis of surfactin is affected by cell density, the swarming effect affects the production of this antimicrobial substance by limiting cell growth[123]. Compared with general chemical surfactants, surface active agent has a variety of biological activities, and can inhibit the growth of bacteria, fungi, mycoplasma[124], which mainly acts on the phospholipid bilayer of the cell membrane of the pathogenic bacteria, change the permeability and functionality of the cell membrane, to inhibit the growth of the pathogenic bacteria, and is not easy to produce drug resistance[125].
Iturin family
-
Iturin is a class of LPs that strongly inhibit the growth of fungi, and is structurally similar to Surfactin in that it consists of a peptide ring of 7 α-amino acid residues linked to β-amino acid residues of a fatty acid chain of 14-17 C atoms (Fig. 7). The amino acid sequence of the peptide ring is Asn-Tyr-Asn-Gln-Pro-Asn-Ser[126]. This family contains a variety of subtype compounds such as iturin A, C, D, E, bacillomycin D, F, L, and Lc as well as mycosubtilin, bacillopeptin, and others[127]. One of the most representative compounds, iturin A[128], is a cyclic structure consisting of a β-amino fatty acid containing 13−17 carbon atoms and seven amino acid residues, and its cyclic structure is formed by the condensation of the carboxyl group of Ser at position 7 and the amino group of the β-amino fatty acid, and by interchanging Ser at position 6 with Asn at position 7, mycosubtilin[129], which has better antimicrobial activity than iturin A, is formed. The basic structure of bacillomycin is similar to that of iturinA, which is composed of two parts, a β-amino fatty acid chain (C15-C18) and a cyclic heptapeptide, with amino acids differing at positions 1, 4, 5, 6, and 7, and is classified as bacillomycin D, F, L, and Lc[130].
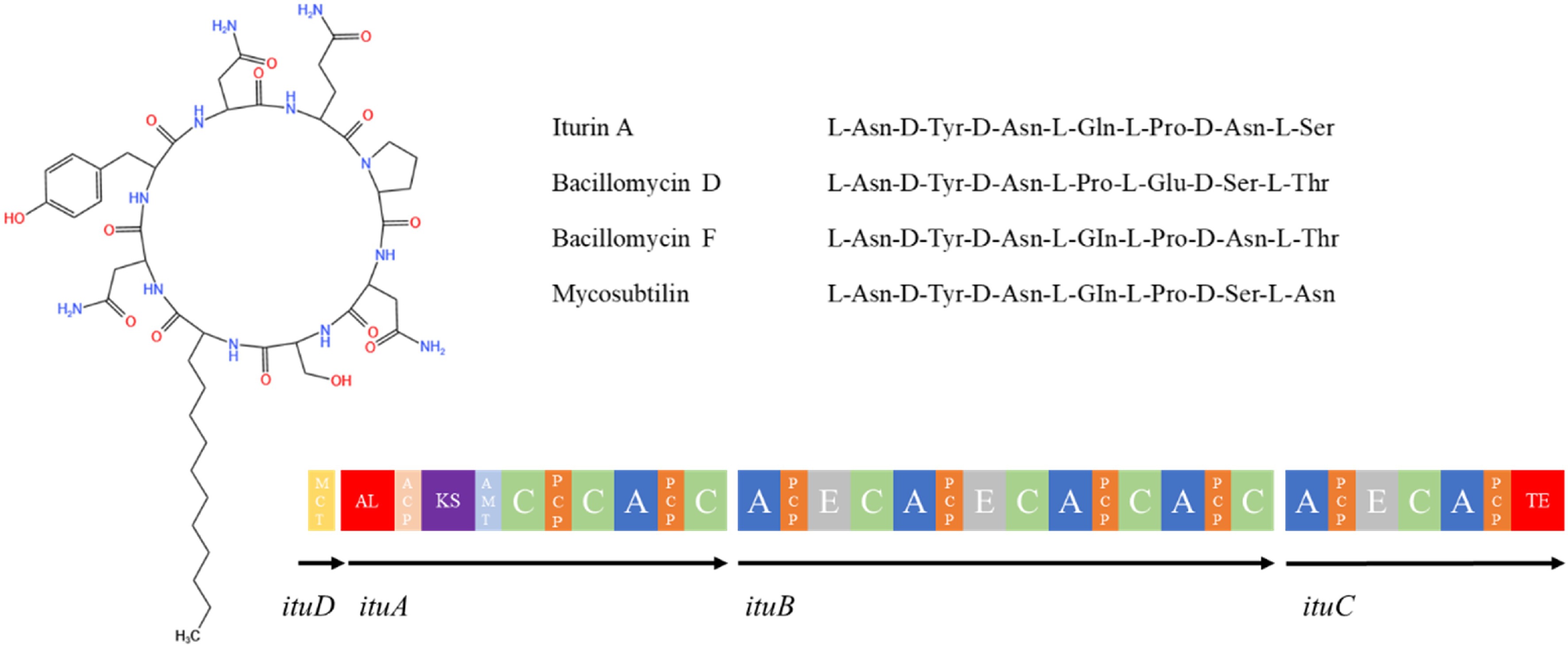
Figure 7.
Structure of iturin family compounds synthesized by Bacillus species[113]. MCT: malonyl coenzyme A transacylase structural domain; AL: acyl-coenzyme A ligase structural domain; ACP: peptidyl carrier protein structural domain; KS: β-ketoacyl synthase structural domain; C: condensate structural domain; PCP: peptidyl carrier protein structural domain; A: adenylyl domain; E: differential isomerism structural domain; TE: thiolipase structural domain.
The lipopeptide antimicrobials of the iturin family are regulated by different genes[131]. The iturin A is encoded by the itu gene cluster and is responsible for its synthesis. The itu gene cluster consists of four ORFs[132]: ituA, ituB, ituC, and ituD. The bacillomycin D is synthesized by the bam/bmy gene cluster[133], which is found in B. subtilis AU195[134] and B. amyloliquefaciens FZB42, respectively, and which encodes a multifunctional complex of enzymes including a fatty acid synthase, an aminotransferase, and a peptide synthase. In addition, DegU and a transmembrane protein, YczE, also post-transcriptionally regulate bacillomycin D production[133]. The mycosubtilin is synthesized by the myc gene cluster, and the gene regulation of this LP was first elucidated in the strain B. subtilis ATCC6633, which synthesizes mycosubtilin. The expression of the myc manipulator is ComA-dependent and is associated with group ringing induction. AbrB inhibits myc expression and knockdown of the abrB gene enhances myc expression[135].
The iturin family is of increasing interest because of its broad-spectrum antifungal activity, limited antibacterial activity, and low toxicity and hypoallergenic. The compounds of this family were found to inhibit the growth of several plant pathogenic fungi such as Colletotrichum sp.[54], Penicillium digitatum[136], Fusarium graminearum[137], etc., but the inhibition of phytopathogenic bacteria was summarized to a lesser extent, but not without significant results[138]. A new cyclic lipopeptide, bacillomycin DC, a potent fungicide with good activity against C. gloeosporioides, was isolated from B. velezensis HAB-2 by silica gel and Sephadex LH-20 column chromatography by Jin et al.[120].
The earliest research opinion is that the mechanism of bacterial inhibition of ichthyobactin-like antimicrobial peptides is based on osmotic perturbation, which forms pores on the surface of the pathogenic bacterial membrane leading to the leakage of cytoplasm causing the death of the bacterium. Aranda et al.[139] while studying the hemolytic effect of iturin A, iturin A can form ion-conducting pores on the surface of the lipid bilayer thereby increasing K permeability. The antimicrobial properties exhibited by Iturin A led to alterations in the external morphology of the yeast, possibly due to an osmotic imbalance driven by changes in cell membrane permeability[140].
Fengycin family
-
The fengycin consists of β-hydroxy fatty acids and a small peptide of 10 amino acid residues. The fatty acid chain consists of 14-18 carbon atoms and the peptide chain contains 4 D-amino acids and 6 L-amino acids (Fig. 8). The sequence of amino acid composition in the peptide chain is Glu-Orn-Tyr-Thr-Glu-Ala-Pro-Gln-Tyr-Ile[141], and the Ile carboxylate group on the 10th position in the peptide chain condenses with the hydroxyl group on the Tyr on the 3rd position to form a ring structure. There are two main types of fengycin (fengycin A and fengycin B), fengycin A when Ala is at position 6 of the peptide chain, and fengycin B when Val is at position 6 of the peptide chain[142].
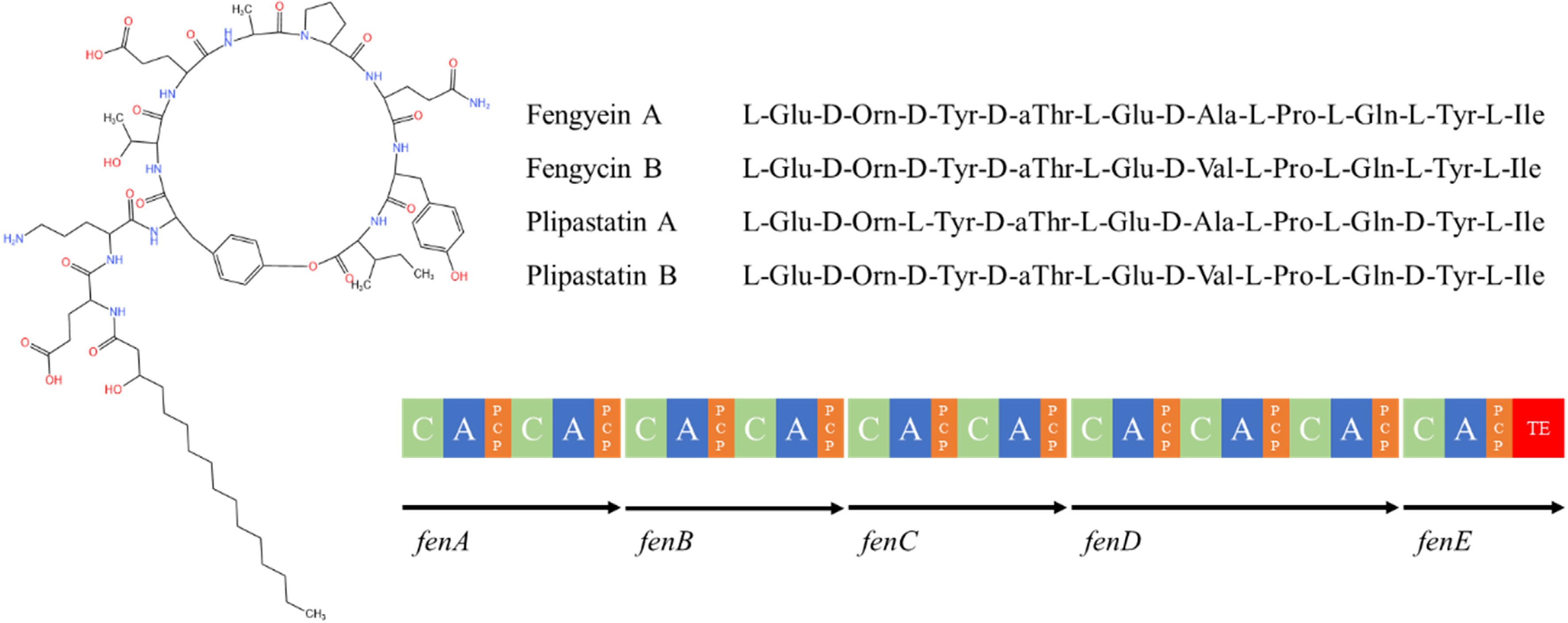
Figure 8.
Structure of fengycin family compounds synthesized by Bacillus species[113]. C: condensate structural domain; PCP: peptidyl carrier protein structural domain; A: adenylyl domain; TE: thiolipase structural domain.
The manipulator of fengycin consists of five reading frames fenA-E[143]. The fenC is the initiation module responsible for the activation and assembly of the first and second amino acids. The promoter of fengycin is located 86 bp upstream of the fenC transcriptional start site, and its UP element, a 17 bp sequence including A and T, is upstream of the promoter-35 region and the RNA polymerase binding site and is critical for promoter activity[144]. The fenB is responsible for the assembly of the last amino acid and is located at the C-terminus of the peptide synthase gene cluster; knockdown of fenB prevents the release of synthesized fengycin[145]. Endogenous factors, such as the two-component regulatory factors (ComA/ComP), Sigma A factor, and signaling proteins (DegU, DegQ), also regulate the expression of the fengycin synthase genes, and up-regulation of sigA increases fusogenic production[146].
Fengycins cause cell death of target microorganisms mainly by altering cell permeability through interaction with fungal cell membranes and can be utilized to control a variety of plant diseases[147]. Chen et al.[148] showed that crude lipopeptide of B. pelliculatus FJAT-46737 possessed a significant antagonistic activity against a variety of pathogens including P. chrysogenum, Escherichia coli, and F. spinosum and detected Fengycins as the main antifungal component of the crude lipopeptide. Fengycins were detected as the major antifungal components of the crude lipopeptide, while the rich source of organic nitrogen promoted the production of fengycin and Surfactin. Adeniji et al.[53] demonstrated that B. beleriensis NWUMFkBS10.5 was able to produce fengycin, iturin, and surfactin, which had a significant antimicrobial effect on two maize fungal pathogens, F. graminearum, and F. culmorum.
Ribosomal peptides synthetases (RPs)
-
Ribosomal peptide synthetases (RPs) are linear precursor polypeptides synthesized by the ribosome pathway through the ribosome, followed by post-translational modifications and protein hydrolysis to complete the synthesis of the substance[149]. Antagonistic substances synthesized through this process are mainly bacteriocins[150], large molecules of antimicrobial proteins[151] and Bacteriocins are small molecular weight proteins produced by bacteria such as Bacillus that are antagonistic to other microorganisms (especially pathogenic bacteria) and are resistant to temperature, pH, and proteases, and their unique target of action makes them less likely to develop resistance. Typically, when Bacillus grows to the end of the logarithm of growth, it produces several types of bacteriocins, such as wool-sulfur antibiotics such as subtilin, sublancin 168[152], and subtilisin[153], as well as small-heat-stable peptides[154], and other non-wool-sulfur antibiotics. In addition to bacteriocins, many Bacillus species secrete cell wall-degrading enzymes[155] that can inhibit plant pathogenic fungi during growth and metabolism, including chitinase[156] and glucanase[157]. The cell walls of pathogenic fungi often contain chitin and β-1,3 glucan, and the chitinase secreted by Bacillus is not only a protein related to the plant disease process but also can play an inhibitory role by destroying the cell walls of pathogenic fungi. Rajaofera et al.[158] identified 19 different volatile organic compounds (VOCs) including alkanes, alkenes, alcohols, and organic acids from B. atrophaeus HAB-5. Among these identified compounds, chloroacetic acid, and tetradecyl esters followed by octadecyl and hexadecanoic acid methyl esters showed antifungal activity against C. gloeosporioides.
Polyketide synthase (PKSs)
-
In B. velezensis strain FZB42, three gene clusters encode PKSs: mlnABCDEFGHI, which regulates macrolactin synthesis; baeBCDE, acpK, baeGHIJLMNRS, which directs bacillaene synthesis; and dfnAYXBCDEFGHIJKLM, which directs difficidin synthesis[159]. Bacillaene is synthesized by the synergistic action of PKSs and NRPSs, which are responsible for binding of malonyl derivatives and amino acids, respectively, and both act as parts of a multi-enzyme complex to synthesize various secondary metabolites with therapeutic potential through different building blocks[160]. In B. velezensis, three LPs and three PKS-type polyketides are biosynthesized via the 4′-phosphopantethelyltransferase (sfp) pathway, whereas bacilysin production is regulated by the bac gene cluster[137]. Jin et al.[161] identified an sfp homolog, lpaH2, encoding a phosphopantethenyltransferase in B. velezensis HAB-2, and when this gene was disrupted, the bacterium could not produce LPs
-
Bacillus, as a PGPR, can colonize plant roots and can promote plant growth through direct or indirect mechanisms of action, not only through the production of bacteriostatic substances for disease control, but also through the induction of systemic resistance in the plant, so that the plant can be recognized at the early stage of invasion of certain pathogens, and thus rapidly make the plant body's 'immune response'. Plant resistance is induced in two ways: systemic acquired resistance (SAR) induced by pathogenic bacteria and induced systemic resistance (ISR) mediated by non-pathogenic microorganisms (rhizobacteria)[162].
Systemic acquired resistance (SAR)
-
The SAR pathway is caused by pathogens that cause plants to acquire systemic resistance. This system protects the plant from pathogens, where the immune system generates systemic signals when the plant is invaded by pathogenic microorganisms, which are then transmitted to the entire plant, thus triggering the system's immune resistance and preventing the pathogen from furthering its own. The immune system is then transmitted to the entire plant, thereby triggering the system's immune resistance and preventing further attack by the pathogen. SAR is mainly based on the salicylic acid (SA) pathway[163]. The significant accumulation of pathogenesis-related proteins (PRP/PR) is a hallmark of the occurrence of system-acquired resistance (SAR). SAR is a defense mechanism induced by pathogenic microorganisms during the co-evolution of plants and pathogens[164]. SA, a phenolic compound, is a phytohormone in the SAR pathway. In the SAR pathway, SA regulates the level of systemic resistance, and studies have shown that SA is not a signaling molecule, but is indispensable for the distal transport of signals[165].
Induced systemic resistance (ISR)
-
The induced systemic resistance (ISR) pathway is responsible for plant self-resistance induced by beneficial flora, usually plant growth-promoting root bacteria. The ISR pathway is similar to the systemic acquired resistance (SAR) pathway in that it protects the uninfected parts of the plant by enabling systemic resistance to infection. The existence of a salicylic acid (SA)-independent ISR pathway, which does not involve the accumulation of pathogenesis-related proteins or SA but rather relies on the pathway regulated by jasmonic acid (JA) and ethylene (ET), was investigated in Arabidopsis thaliana[166]. The combination of ISR and SAR increases plant resistance to pathogens through both pathways and provides better protection against infection than activation of either ISR or SAR alone[167]. Beneficial microorganisms, including Bacillus, generally induce the production of ISR, triggering a series of defense responses in plants that increase the plant resistance level and thus help resist pathogen infection to some extent. LPs produced by B. amyloliquefaciens S499 were found to activate the ISR of tomato (Solanum lycopersicum) and increase the expression of the defense-related lipoxygenase genes LOXD (lipoxygenase D) and LOXF (lipoxygenase F)[34]. B. cereus EPL1.1.3 and Serratia nematodiphila TLE1.1 induced tomato plants to generate ISR through the production of JA, thereby facilitating resistance to infection with Ralstonia syigisub sp.[168]. B. subtilis DZSY21 can inhibit the maize leaf spot pathogen (Bipolaris maydis) by activating the SA and JA pathway-dependent ISR signaling pathway to induce systemic resistance[169]. When cotton plants were treated with B. subtilis IAGS-174 and B. megaterium ZMR-4, the activities of defense-related enzymes such as peroxidase and polyphenol oxidase increased, inducing ISR[170]. After pretreatment with B. subtilis HN-Q-8, potato plants became more resistant to early blight owing to the upregulation of the activity of defense enzymes and stimulation of the JA/ethylene pathway[171]. Cao et al.[172] found that treatment with B. subtilis SQR9 enhances maize resistance to Gibberella stem rot (GSR) by activating the maize ISR.
Reactive oxygen species (ROS), defense-related enzymes and genes
-
ROS, which mainly includes hydrogen peroxide (H2O2), superoxide anions (O2-), hydroxyl radicals (OH·), singlet oxygen (1O2), and lipid peroxides, are highly oxidizing substances in plants. When plants are subjected to biological stress, ROS plays important roles in signal recognition and transduction[173]. They play a dual role as second messengers in biological stress responses; however, above a certain level, they can damage plants. Excessive ROS production disrupts the structure and function of plant cells, decreasing photosynthetic capacity. Using DAB staining experiments, Guo found that B. subtilis Ba168 increased ROS levels in the model plant Nicotiana benthamiana[174]. Further, glycoside hydrolase 43 protein H1AD43 produced by B. licheniformis BL06 also induced ROS production in N. benthamiana[175].
There are many defense enzymes in plants that are involved in many physiological and biochemical reactions. Following infection by pathogens, these defense enzymes rapidly undergo a series of changes and play an important role in the induction of plant resistance. These plant resistance-related defense enzymes mainly include catalase (CAT), polyphenol oxidase (PPO), peroxidase (POD), superoxide dismutase (SOD), chitinase, and β-1,3-glucanase, etc. Phenylalanine ammonia-lyase (PAL) activity is associated with antibacterial activity and the synthesis of phytoprotectants and is a rate-limiting enzyme for phenylpropanoid metabolism. SOD, as an antioxidant enzyme, enhances plant disease resistance mainly by converting excess O2- in the plant to H2O2, thereby reducing oxidative damage caused by reactive oxygen species. The activity of the POD enzyme reflects the metabolic process in the plant body. Unlike SOD, its way of enhancing plant resistance is by accelerating the corkification of plant tissues while promoting the synthesis of lignin and phytoprotectants in the plant body, thereby enhancing plant resistance[176]; CAT is mainly found in peroxisomes, which can scavenge free radicals such as H2O2 and reduce excess reactive oxygen species in plants through various antioxidant enzymes[177]. Yu et al.[178] extracted total organic acids from the culture medium of B. cereus AR156, in which oxalic acid (OA) can induce programmed cell death. Pretreatment with low concentrations of OA can increase the accumulation of ROS-scavenging enzymes in tomato leaves and promote the expression of defense-related genes in the JA pathway. Inoculation of B. tequilensis PKDN31 and B. licheniformis PKDL10 strains can induce systemic resistance of tomato to F. oxysporum by increasing the accumulation of defense enzymes such as POD, and PAL[179].
Blueberry fruits treated with B. subtilis KLBC BS6 showed significantly increased activity of disease-resistance enzymes, such as PAL and POD, which effectively reduced the incidence of infections caused by Botrytis cinerea[180]. Wang et al.[181] showed that treatment with B. amyloliquefaciens HG01 of loquat fruit affected by postharvest anthracnose significantly increased the relative expression levels of disease-related genes as well as those of two defense enzymes, which indirectly induced fruit resistance. Peanuts treated with B. subtilis CB13 showed a significant increase in SOD activity after 12 h, initiating a faster defense response against peanut root rot[182]. After treatment of Korla fragrant pear fruits with B. subtilis Y2, antioxidant enzyme activity levels increased, which to some extent improved ROS scavenging[183]. The synthesis of defense enzymes, including CAT, POD, and SOD, significantly increased in lupin seedlings treated with B. subtilis CtpxS2-1. CtpxS2-1 increases the expression of SAR-related defense genes through the secretion of LPs[184]. B. subtilis F21 was found to enhance basal immunity of watermelons against wilt disease by increasing the expression of plant defense-related genes and activity of defense enzymes (such as CAT, POD, and SOD)[185].
The expression of defense genes can be one of the markers used to detect plant resistance. Non-expressed gene 1 (NPR1) is a key regulatory gene that plays an important role in inducing plant resistance. NPR1 is located in the intermediate regulatory position, downstream of SA accumulation, and upstream of PR gene expression[186]. Compared with plants infected with potato virus Y alone, potato plants treated with B. subtilis (soil application) before virus inoculation showed significantly increased relative expression of the PR-1 gene, which can induce systemic resistance and alleviate the damage caused by that virus[187]. In plants inoculated with B. subtilis K47, B. cereus K46, and Bacillus M9, an increase in the transcript levels of the defense-related genes CcNPR1, CcPR10, and CcCOI1 can be observed[188]. Microbe-associated molecular pattern (MAMP) gene products, including flagellin (Flg) and elongation factor thermos (EF-Tu) unstable from B. velezensis VB7, upregulate defense-related genes are involved in signal transduction, oxidation, and peptide production, and trigger the immune response of tomato (Shivam) plants to groundnut bud necrosis virus (GBNV)[189]. After treatment with B. velezensis SDTB022, the activity of defense enzymes such as POD and PAL were significantly increased in tobacco. The expression of the tobacco defense genes PR1a, PR3, and PR5 was up-regulated and the SA-dependent signaling pathway induced SAR throughout the entire plant[190].
In our preliminary experiments, it was found that B. velezensis HN-2 significantly increased the activity of the antioxidant enzymes CAT, POD, and SOD, upregulated the expression of defense-related genes PR1 and NPR1, and upregulated the expression of JA pathway genes in tobacco plants, making them more resistant to inoculation with pepper vein mottle virus, indicating that ISR was activated after pretreatment with B. velezensis HN-2 (unpublished). Following inoculation of tobacco plants with tobacco mosaic virus, it was found that B. velezensis HN-2 increased ROS generation in plants. The expression levels of the antioxidant enzymes CAT, POD, and SOD as well as those of the defense genes NPR1, PR1, PR3, and PAL were significantly upregulated, which enhanced plant resistance and delayed viral infection to some extent (Fig. 9).
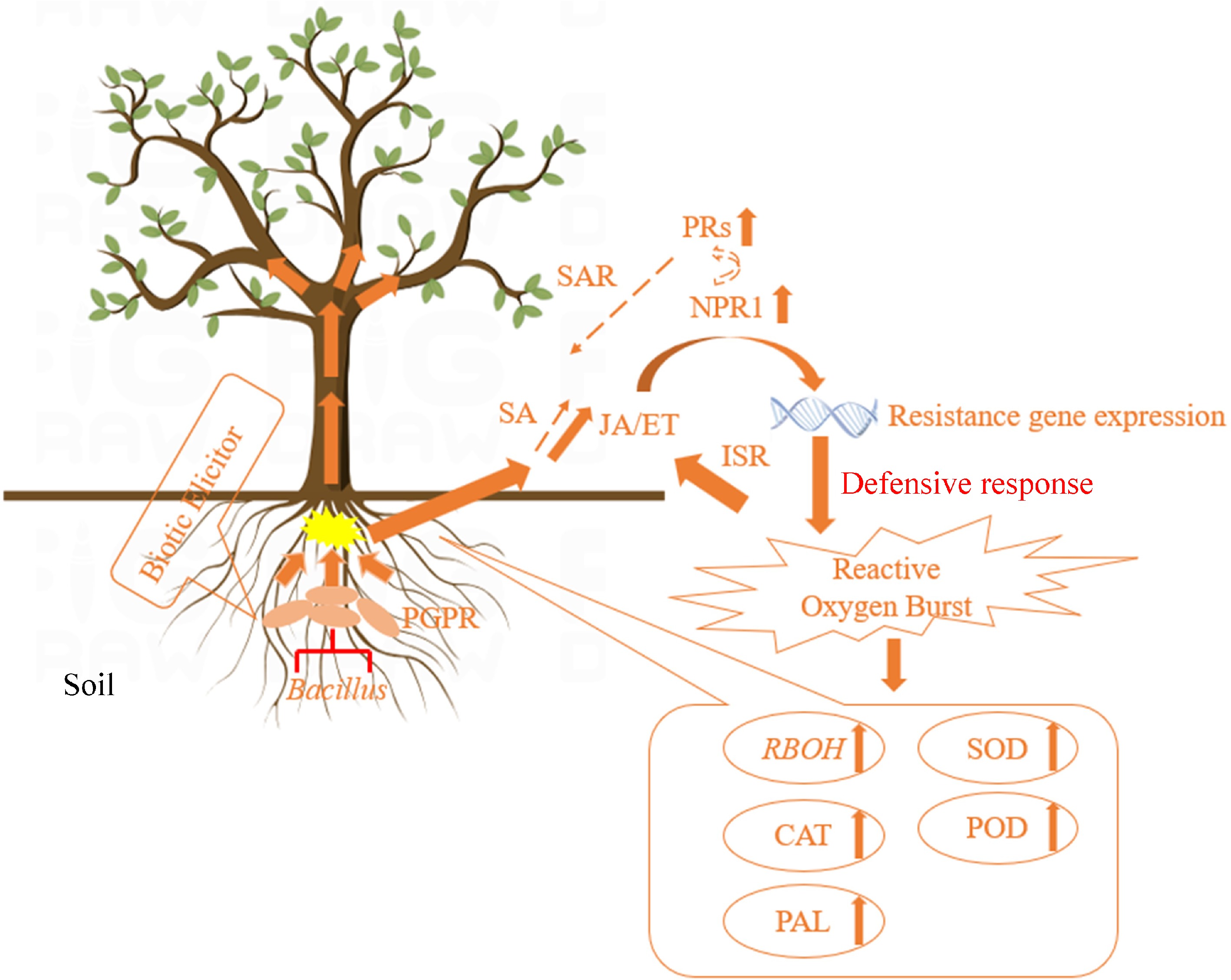
Figure 9.
Pattern diagram of Bacillus-induced plant resistance[191].
-
Bacterial small RNA (also known as sRNA) is a class of transcribed RNAs that do not normally encode proteins and participate in signaling by interacting with mRNA and proteins. sRNAs are 50–500 nt in length, they are mostly encoded by spacer regions in the genome, with some of them being encoded by the complementary strand of the gene coding regions[192]. sRNAs in the spacer regions of genes affect the translation or stability of target mRNAs, thus regulating different target genes[193]. sRNAs are involved in the regulation of various physiological processes, such as iron ion metabolism[194], carbon metabolism[195], amino acid metabolism[196], bacterial virulence[197], acid stress[198], oxidative stress[199], and protease activity[200].
Small non-coding RNA has been found in many bacteria, including E. coli and B. subtilis, but sRNA functions in E. coli have been more intensively studied[201]. sRNAs are important post-transcriptional regulators, and in B. subtilis, a total of 108 sRNAs have been predicted and characterized[202]. FsrA, the first trans-encoded RNA discovered in B. subtilis, targets mRNAs related to iron metabolism and requires the involvement of the chaperone proteins FbpA, FbpB, and FbpC[203]. SR1 was the first bifunctional sRNA identified in B. subtilis. SR1 inhibits translation by base-pairing with mRNAs that encode arginine catabolism manipulator rocABC and transcriptional activator of rocDEF, ahrC[196]. In addition, SR1 encodes a small peptide, SR1P, which interacts with GapA[204], and its mutation accelerates the formation of B. subtilis spores but also leads to their poor quality[205]. SR1P forms a GapA/SR1P complex with the glycolytic enzyme GapA and interacts with RNase J1 to promote the degradation of RNase J1 targets[204]. Hfq is a conserved protein first discovered as a housekeeping factor required for replication of RNA phage Qβ in E. coli[206]. Whole genome sequencing has shown that almost half of gram-positive and gram-negative bacteria contain Hfq proteins encoded by homologous genes[207]. Deep sequencing of B. subtilis Hfq-associated RNA HfqBS showed that it binds to sRNAs, antisense RNAs, and guide sequences within mRNAs[208], suggesting that it may play a role in post-transcriptional regulation (Fig. 10).
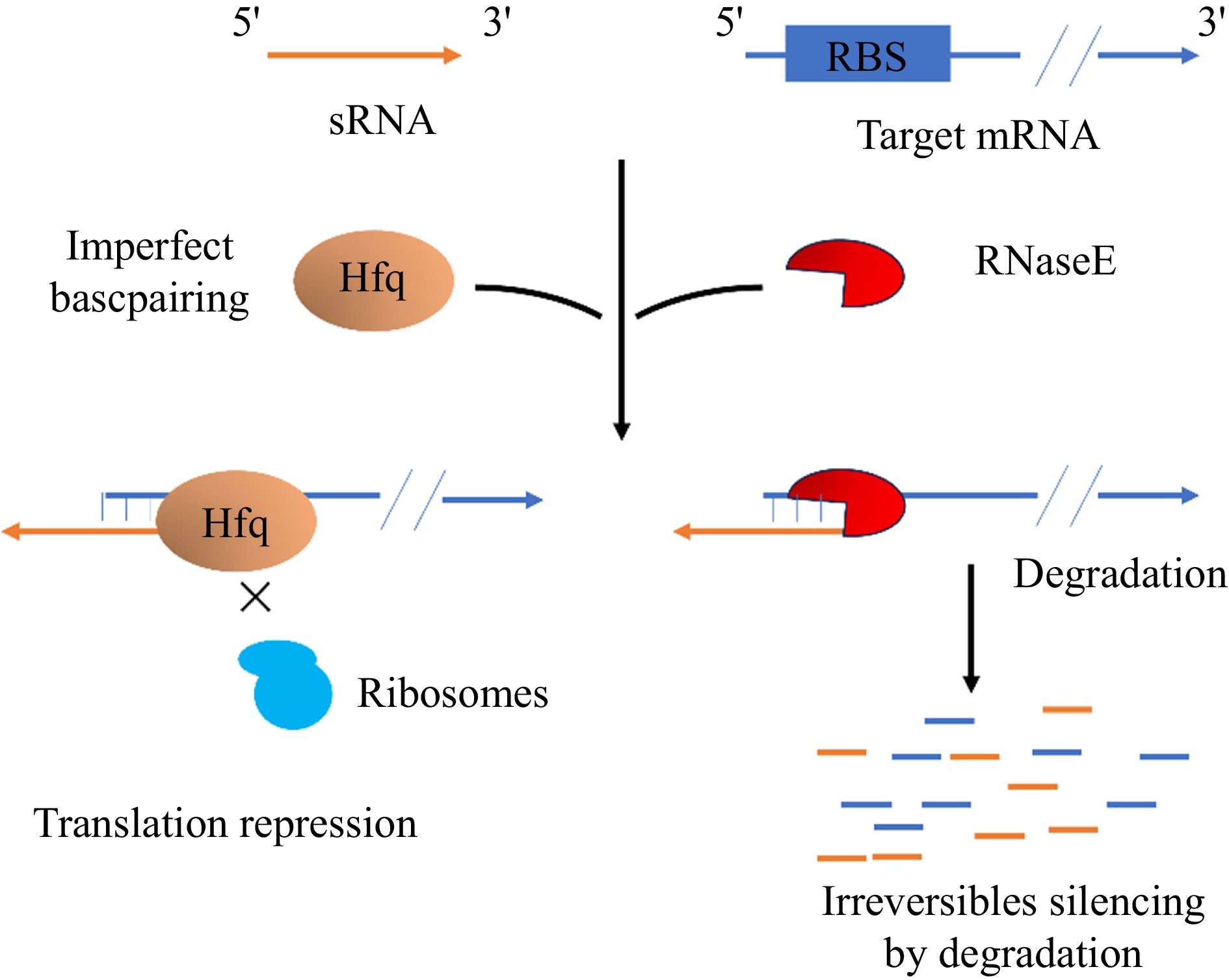
Figure 10.
A class of trans-encoded sRNAs imperfectly base-pair with target mRNAs to repress translation and speed up the degradation[209].
In addition, Jagtap et al. showed that Hfq positively regulates the expression of chemotaxis- and flagellum-related genes in B. subtilis[210], implying its novel function in B. subtilis. CsrA (carbon storage regulator A) is a widely conserved and abundant sRNA binding protein (~60 aa) involved in the regulation of carbon metabolism, biofilm formation, and virulence[211]. CsrA binds to 2−6 single-stranded GGA motif-containing sites around target mRNA SD sequences[212], inhibiting[213] or activating translation[214], affecting RNA processing[215], and altering transcript elongation[216]. In B. subtilis, CsrA regulates only one target, hag mRNA, and is sequestered by protein FliW rather than by sRNA[217]. FliW interacts with the C-terminal extension of CsrA and antagonizes it in a non-competitive manner, preventing it from binding to hag mRNA[218].
-
Agriculture is the foundation of the Chinese national economy, pesticides are an important means of production to ensure agricultural production, but also need special means of production that requires scientific use and special management. According to the Food and Agriculture Organization of the United Nations, (FAO), the amount of agricultural pesticides used in China in 2021 will be 244,800 tonnes, of which 66,100 tonnes of pesticide, 64,100 tonnes of fungicides, 101,500 tonnes of herbicides, and fewer than 0.01 tonnes of rodenticides. From the perspective of pesticide registration, there are more than 90 kinds of biopesticide active ingredients registered in China, and more than 3,000 registered products, accounting for about 11% to 13% of the total number of registered pesticides in China. Among them, in 2021, there will be 21 new pesticides registered in China, and the number of biopesticides is the largest, accounting for 71.4% of new pesticides, and microbial pesticides account for 42.8%, showing that biopesticides are accelerating the pace of development. The development of pesticides is closely related to the green development of agriculture, and green pesticides have become an indispensable part of modern agricultural production and agricultural economic development. The creation of highly effective, low-toxicity, and environmentally friendly green pesticides is an indispensable step in promoting green agriculture and increasing the efficiency of pesticides (Fig. 11).
As an important biocontrol resource, Bacillus has prospects of broad application. The use of Bacillus to control plant diseases has been widely reported at home and abroad[113,147]. Because of its complex bacteriostasis mechanism and broad spectrum bacteriostasis, Bacillus has been widely used in agricultural production. For example, the most widely known B. thuringiensis, the endotoxin produced by it, and the cryptocrystalline protein have been developed as mature biopesticides, and have good biological control effects on the Grapholita molesta, the cocoa pod borella, and the larva of rhinocerodon copra (Coleoptera: Chelonidae)[219].
Bacillus also has a very good control effect on pathogenic fungi. Djaenuddin et al.[220] found that the B. subtilis preparation could effectively inhibit the occurrence of corn downy mildew caused by Peronosclerospora philippinensis and improve the growth of corn plants, and the subtilis treatment group often had higher chlorophyll content of corn plants than the control group. Fajaruddin et al.[221] showed that the mixture of liquid silicon fertilizer and B. preparations had significant effects on the growth and tolerance of fungi Pyricularia oryzae Cav. The formula of 12 mL liquid silica and 6 gm/ basin Bacillus can effectively increase the plant height and tiller number of rice, and significantly reduce the average damage percentage of rice. Klein et al.[222] found that C. acutatum can cause postbloom fruit drop of citrus, and the application of B. subtilis ACB-69 strain can effectively control it. In their paper, they developed and optimized a preparation of B. subtilis with potential for large-scale application, and evaluated its in vitro and in vivo effects against C. acutatum. The results showed that the formula containing B. subtilis talc + urea (0.02%) effectively inhibited the growth of mycelia and spore germination, and had a high potential to inhibit post-bloom fruit drop of citrus. Since 1991, the field control efficiency of rice sheath blight has been stabled at 60%−81%, and the cumulative application area has reached 8 million hm2. The biological control bacterium B. subtilis B3 (trade name Maifengning) was screened by Nanjing Agricultural University and the field control efficiency of wheat sheath blight was 50%~80%. The microbial pesticide 'Baikang' (1 billion spores/g B. subtilis wettable powder) jointly developed by Yunnan Agricultural University and China Agricultural University has been registered by the Ministry of Agriculture, with a cumulative promotion area of about 4,667 hm2, mainly preventing and controlling rice dry blight, Notoginseng root rot, and tobacco black tibia. B. subtilis GB03, trade name Kodiak, was developed by Gustafson Company, mainly used in soil-borne fungal disease control, especially cotton and soybean seed treatment, sales in the United States market are good. B. subtilis MBI600, trade name Subtilex, was developed by Microbio Ltd., mainly through root use or seed dressing to control root diseases caused by fusarium, aspergillus, etc., beans, cotton, and peanuts. B. subtilis QST713, marketed as Serenade, was developed by Agra Quest to combat leaf diseases caused by both fungi and bacteria in vegetables and fruits. The combination of B. amyloliquefaciens FZB42 and B. subtilis GB122 makes Bio Yield, a fungicide developed by Gustafson for the control of powdery mildew, downy mildew, blight, and gray mold in vegetables, cherries, grapes, gourds, and walnuts. In Germany, B. amyloliquefaciens FZB42 was used as a microbial fertilizer and B. amyloliquefaciens CECT 5940 was used as a feed additive. B. subtilis A-13 developed in Australia has good control and an increased effect on wheat and carrot blight and other soil-borne diseases. The wettable powder of B. subtilis is the most registered biological pesticide, which is mainly used to control powdery mildew, gray mold, blight, wilt, bacterial wilt, downy mildew, soft rot, and other diseases of cucumber, tomato, Chinese cabbage, pepper, and other vegetables. The company introduced Provilar, a biocide formulated from the strains of B. velezensis RTI301 and B. subtilis RTI477. Provilar is an environmentally friendly biopesticide that helps combat leaf spots and white mildew. Its liquid formulation contains endospores of bacteria and their metabolites, which can prevent and sustain leaf diseases.
Compared with other Bacillus bacteria, B. velezensis has a wide distribution, rapid growth, good genetic stability, environmental friendliness, and is relatively safe. B. velezensis secondary metabolites also have broad-spectrum antibacterial activity. They play important roles in feed, plant protection, food industry, forestry, and sewage treatment. In recent years, research on B. velezensis has mainly focused on the isolation, identification, and application of the strains, mechanisms of its antibacterial activity, antibacterial substance isolation, gene cluster identification, optimization of fermentation conditions, and formulation development. B. velezensis is widely used to prevent and treat plant fungal and bacterial diseases. Lee et al.[223] found that B. velezensis strain YP2 effectively inhibited the occurrence of gray mold and sclerotinia in leafy vegetables, and the control effect of this strain after spraying diluted 10 times on the powdery mildew of red mustard and Chang mustard was 91.8% and 80.9%, respectively. In addition, B. velezensis YP2 promoted seed germination and plant growth in mustard. Liu et al.[224] found that B. velezensis strain Bv-303 has good application potential for the biological control of bacterial blight disease in rice. In vitro, B. velezensis strain Bv-303 strain inhibited the growth of Xanthomonas oryzae pv. oryzae by 85.7%–88.0%, and this strain was resistant to heat, acid, alkali, and ultraviolet light. In rice leaves, the fermentation solution, fermentation supernatant, and bacterial suspension of strain Bv-303 improved the resistance of rice plants to bacterial blight disease. The fermentation solution had the best effect and increased the resistance by 62.7% without any adverse effects on rice seed germination or seedling growth. Liu et al.[225] found that the B. velezensis strain S161 effectively controlled the postharvest green mold (Penicillium digitatum) of citrus. The cell-free supernatant of strain S161 strongly inhibited mycelial growth, spore germination, and bud tube elongation in vitro. The cell-free supernatant of strain S161 had a good protective effect on citrus fruits, and the control effect on citrus green mold was 95.6%. Further studies showed that the cell-free supernatant of strain S161 degraded and ruptured mycelial membranes and spores, induced excessive accumulation of ROS in mycelial cells, and caused oxidative damage to pathogenic fungal cells. In addition, treatment with the cell-free supernatant of strain S161 downregulated genes related to spore germination, growth, reproduction, and virulence. Husna et al.[226] showed that B. velezensis has a significant inhibitory effect on soft rot, a postharvest bacterial disease of eggplant fruits caused by Pectobacterium carotovorum subsp. carotovorum. The cell-free supernatant of B. velezensis significantly inhibited the growth of P. carvotorum in vitro and in vivo. The methanol-soluble precipitate obtained from the cell-free supernatant of B. velezensis also significantly inhibited P. carvotorum, and the main antibacterial substance was identified as the surfactant subtype. In addition, B. velezensis alleviated oxidative damage caused by ROS by enhancing the activity of ROS-scavenging enzymes, thereby improving the disease resistance of eggplant fruits. Dr. Lu's team from the Beijing Plant Protection Station screened 5,000 strains, isolated the first domestic B. velezensis strain CGMCC No. 14384 in a tropical salt-tolerant and high-temperature environment, and obtained a nationally authorized invention patent in 2019. Simultaneously, numerous indoor and field experiments and demonstrations were performed, the fermentation process and bactericide formulation were optimized, and a new type of bactericidal B. velezensis was created. Biological pesticides have a wide spectrum of control, high antibacterial activity, and strong inhibitory effect on over 20 species of plant pathogenic fungi. The field test results showed that the control effect over strawberry powdery mildew on leaves and fruits was 86.6% and 71.1%, respectively, which is equivalent to or even higher than that of chemical fungicides. It also had a significant control effect over tobacco powdery mildew and wolfberry powdery mildew. Formex recently launched Ataplan, a new biopesticide for the Brazilian market. This product contains B. velezensis CEPA RTI301 and B. subtilis CEPA RTI477, which are used against major soil-borne fungal pathogens, such as fusarium, trichotonia, and anthrax. These products contain endospores, have high flexibility, and are compatible with chemicals. Thus, Ataplan can be used as a supplement to chemical fungicides. Owing to its mechanism of action and ability to colonize the roots, the seedling protection period is extended, providing a more effective defense for the crop. Unlike other fungus-derived products, Ataplan can be used under all weather conditions, and its use does not affect existing field management. Therefore, this product is recommended as a seed-and-furrow treatment agent.
The colonization ability of Bacillus biocontrol strains in the field is the key to its biocontrol effect. At present, under laboratory conditions, a large number of Bacillus strains with strong antagonistic activity against plant pathogens can be screened out. However, when these antagonistic strains are applied as biological control factors to control plant diseases and insect pests, can they be stably colonized in the field, or can they maintain a certain population number and successfully colonize in the leaves and root encircling of plants under the condition of field application of pesticides or residues? The formation of dominant populations is one of the key factors for the large-scale development and commercialization of living biopesticides in agricultural production. At present, most biocontrol preparations for Bacillus are live preparations. When promoting and applying in production, whether enough live preparations can be sprayed in the critical period of plant disease occurrence directly affects the biocontrol effect. In addition, the field application of live bacteria preparations is often affected by external factors such as temperature, humidity, soil, and pH value, so the control effect is not very stable.
-
Bacillus species are the most common biocontrol bacteria that are widely used for plant defense. Unlike chemical control, biological control has attracted considerable attention because of its high efficiency, low toxicity, and environmental friendliness. A large number of biocontrol bacteria have been reported to be able to control anthracnose, among which Bacillus and Actinobacteria have been the most extensively studied. Currently, the biocontrol effects of LP compounds produced by Bacillus are actively studied, however, most of the research is still quite limited, and other biocontrol mechanisms remain unclear. In addition, the number of biologically active substances produced by Bacillus is relatively low; therefore, it is necessary to expand the production and improve the fermentation process. The development of new Bacillus-based biocontrol formulations is of practical significance for agriculture.
This work was supported by grants from Hainan Provincial Natural Science Foundation-the Scientific Research Foundation for Advanced Talents (Grant No. 322RC591, 324RC455), the National Natural Science Foundation of China (Grant No. 31960552, 32260698), Hainan Province Science and Technology Talent Innovation Project (Grant No. KJRC2023B14), the earmarked fund for Tropical High-efficiency Agricultural Industry Technology System of Hainan University (Grant No. THAITS-3). The authors would like to thank the Key Laboratory of Green Prevention and Control of Tropical Plant Diseases and Pests, College of Tropical Agriculture and Forestry, Hainan University, for the support of the experimental site and equipment.
-
The authors confirm contribution to the paper as follows: conceptualization: Miao W, Jin P; funding acquisition: Jin P; supervision: Miao W, Jin P, Liu W; validation: Liu W; writing – original draft preparation: Chu L, Xuan Z, Lin Z, Fang Y, Pan X, Wang J. writing – review & editing: Jin P, Xie Q, Chu L. All authors have read and agreed to the published version of the manuscript.
-
All data generated or analyzed during this study are included in this published article.
-
The authors declare that they have no conflict of interest.
-
Received 30 July 2024; Accepted 15 October 2024; Published online 16 January 2025
-
# Authors contributed equally: Pengfei Jin, Linglong Chu, Zhe Xuan
- Copyright: © 2025 by the author(s). Published by Maximum Academic Press on behalf of Hainan University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Jin P, Chu L, Xuan Z, Lin Z, Fang Y, et al. 2025. Bacillus velezensis, a new valuable source of bioactive molecules within plant microbiomes and natural weapons for the biocontrol of plant pathogens. Tropical Plants 4: e001 doi: 10.48130/tp-0024-0044
Bacillus velezensis, a new valuable source of bioactive molecules within plant microbiomes and natural weapons for the biocontrol of plant pathogens
- Received: 30 July 2024
- Revised: 02 October 2024
- Accepted: 15 October 2024
- Published online: 16 January 2025
Abstract: Bacillus spp. are widely used as a biological agent in the management of crop diseases. B. velezensis is a novel species of the genus Bacillus that produces various secondary metabolites, shows a broad spectrum of antimicrobial activities, and promotes plant growth. In addition, it is widely used for the biocontrol of plant diseases. Here, the discovery, classification, antimicrobial substances and related gene clusters, effect on microbiota, quorum sensing system, antagonistic mechanism, and application of B. velezensis are reviewed. This review summarizes the current knowledge, highlights existing problems regarding applications of B. velezensis for plant protection, and discusses future directions for research on this bacterial species. The present review will provide a useful reference for studies on B. velezensis and speed up the development of new formulations for its practical applications.