-
Grafting, a time-honoured horticultural technique, has revolutionized plant breeding and cultivation by enabling the combination of desirable traits from different plant species or cultivars[1]. The success of grafting relies on various factors, including the strength of the seedlings, proper healing of the grafting junction, and the subsequent recovery of heterografted seedlings[1]. Recent advancements in our understanding of the target of rapamycin (TOR) signalling pathway have shed light on its pivotal role in regulating growth and development in plants[2]. As an evolutionarily conserved protein kinase, TOR integrates diverse signals and coordinates cellular processes related to growth, nutrient signalling, and stress responses. In this perspective paper, we delve into the implications of TOR signalling in grafting, exploring its involvement in pre-grafting growth regulation, graft healing at the junction site, and the possible mechanisms underlying the shoot-root communication in heterografted seedlings. By elucidating the contributions of TOR signalling to grafting processes, we aim to provide valuable insights that can enhance grafting success rates and broaden the scope of agricultural practices and crop improvement strategies.
-
Pre-grafting growth regulation is a crucial aspect of successful grafting, ensuring the development of robust and vigorous seedlings capable of withstanding the grafting process. However, greenhouse conditions can present challenges that compromise the strength and health of seedlings, hindering their suitability for grafting. In the modern greenhouse environment, where controlled conditions prevail, several factors contribute to the occurrence of weak or etiolated seedlings. Insufficient light exposure, improper temperature and humidity control, inadequate nutrient availability, and suboptimal cultural practices often result in elongated and fragile seedlings with reduced photosynthetic capacity. These weakened seedlings not only possess diminished vigour but also display compromised physiological and morphological characteristics that can hinder the success of grafting. Addressing these current problems in the greenhouse is essential for optimizing pre-grafting growth regulation and ensuring the production of resilient seedlings ready for successful grafting procedures.
TOR signalling exhibits high sensitivity to light conditions and effectively integrates light signals with other environmental and hormonal cues to regulate plant growth. During skotomorphogenesis, the activity of TOR is suppressed by COP1, a key component of the light signalling pathway[3]. Upon exposure to light, photosynthetic sugar production and auxin-ROP2 signalling activate TOR[4,5], leading to the stimulation of polysome loading for numerous mRNAs involved in photomorphogenesis[6]. RelA SpoT Homologue 3 (RSH3) was recently identified as a TOR phosphorylation target that regulates chloroplast activity[7]. Notably, recent research has shed light on the role of TOR as a central hub in regulating the rosette habit by orchestrating the ATH1-PIF relay in response to diverse light conditions[8]. This discovery highlights the intricate role of TOR signalling in integrating light-mediated regulatory pathways and shaping plant development and architecture.
It's important to note that the role of TOR in hypocotyl elongation can vary significantly during skotomorphogenesis and after photomorphogenesis. Hypocotyl elongation in the light is primarily regulated by three types of transcription factors: BRASSINAZOLE-RESISTANT (BZR), AUXIN RESPONSE FACTOR (ARF), and PHYTOCHROME INTERACTING FACTOR (PIF)[9], known as the BAP module (Fig. 1a). TOR is activated by auxin signalling and in turn regulates ARF3 and ARF5 at translation level via promoting eIF3h/RISP/eS6-mediated translation reinitiation[5,10,11]. Alongside auxin, brassinosteroid and gibberellin signalling also play significant roles in the BAP transcriptional network. TOR is essential for sucrose-induced hypocotyl elongation in response to darkness by stabilizing BZR1 from autophagy degradation[12]. TOR is also known to participate in the brassinosteroid signalling pathway to regulate hypocotyl elongation through key components such as BES1 and BIN2[13−15]. However, it's worth noting that TOR silencing in the two estradiol-inducible RNAi lines leads to increased hypocotyl length in the absence of sucrose feeding, compared to the same lines without estradiol application[12]. To explain this inconsistency in TOR's effect on hypocotyl elongation under different sucrose conditions, we propose the involvement of another factor.
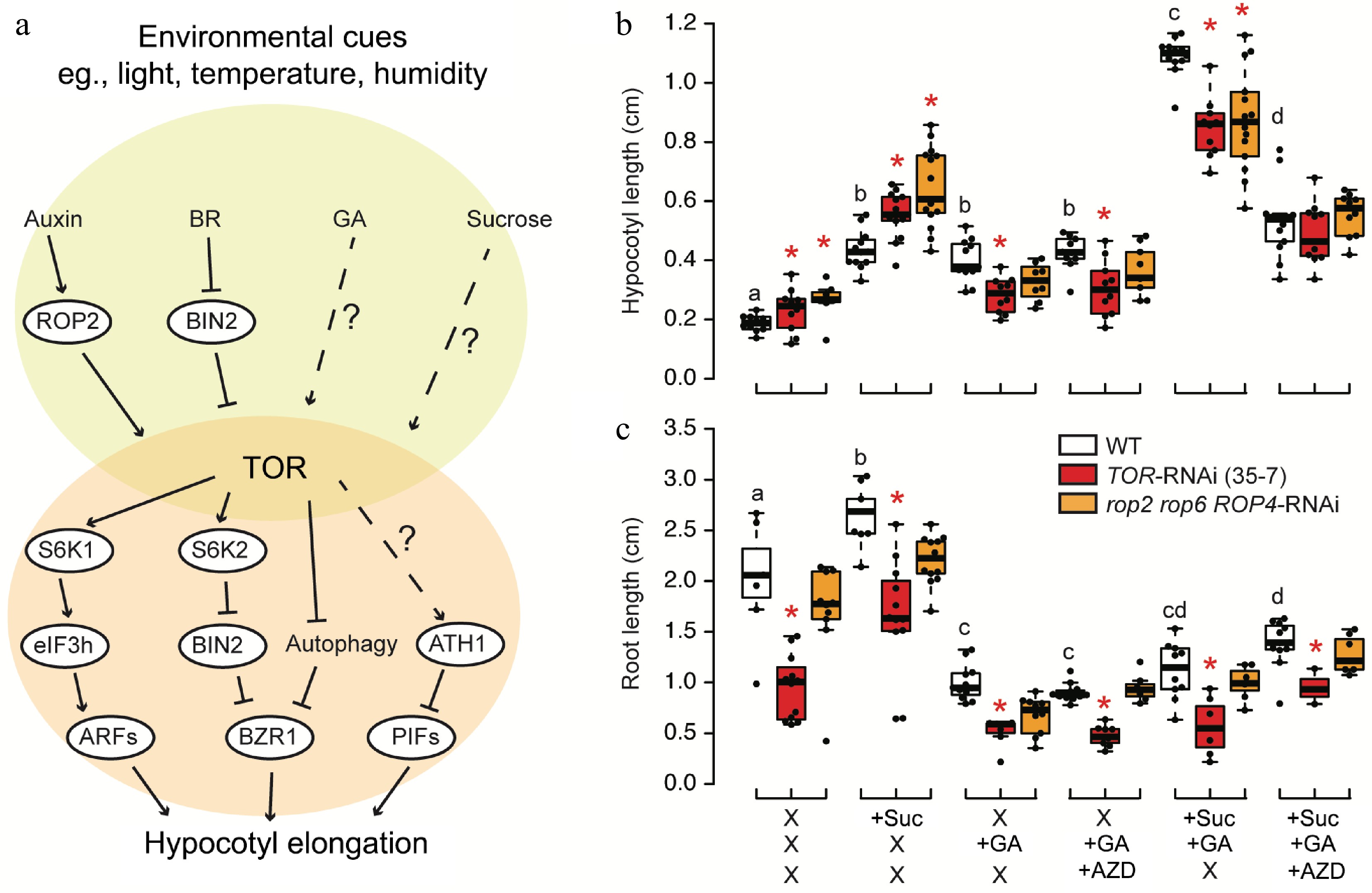
Figure 1.
TOR-mediated regulation of hypocotyl elongation. (a) Proposed model of TOR in the regulation of hypocotyl elongation. TOR serves as a central regulator that integrates upstream phytohormone signaling and sucrose availability to modulate the BAP (BRASSINAZOLE-RESISTANT (BZR), AUXIN RESPONSE FACTOR (ARF), and PHYTOCHROME INTERACTING FACTOR (PIF)) module, which functions in hypocotyl elongation. BR, brassinosteroid; GA, gibberellin; ROP2, RHO-related protein from plants 2; BIN2, BRASSINOSTEROID INSENSITIVE 2; S6K, Ribosomal-protein S6 kinase; eIF3h, eukaryotic translation initiation factor 3h; ATH1, ARABIDOPSIS THALIANA HOMEOBOX 1. (b), (c) Arabidopsis thaliana ecotype Col-0 as wild type (WT), TOR-RNAi (35-7) line and the triple mutant of ROP2/4/6 (rop2 rop6 ROP4-RNAi) were cultivated on 1/2MS medium under long-day conditions (16 h of light) for 7 d, and treated with combinations of 0.5% sucrose, 0.5 µM AZD8055, or 0.5 µM GA3 for another 3 d in darkness. (b) Hypocotyl lengths and (c) root lengths were quantified from seedling grown on two independent plates. Stars or letters indicate the significant difference (n > 10, one-way ANOVA followed by Student's t-test, two-tailed p < 0.05).
Gibberellins (GAs) are crucial hormones that regulate various aspects of plant growth and development, prominently including elongated growth. GAs particularly influence stem and internode elongation, as well as processes like leaf expansion and cell elongation[9]. However, the connection between TOR and GA remains poorly understood. A previous study demonstrated that TOR is activated by GA during wheat seed germination[16]. To investigate the relationship between sucrose, TOR, and GA in hypocotyl elongation, we conducted experiments using TOR RNAi mutant 35-7[17] and upstream activator mutant rop2 rop6 ROP4-RNAi[5]. One-week old seedling were grown for 1 week in the light and treated with combinations of sucrose, GA and TOR inhibitor for another 3 d in the darkness. Both mutants exhibited increased hypocotyl length compared to the wild type in the absence of any stimulator. Both sucrose and GA had comparable effects on hypocotyl elongation in the wild type. Additionally, sucrose and GA showed an additive effect on hypocotyl length in the wild type. In TOR mutants, the absence of sucrose prevented further increase in hypocotyl length mediated by GA, suggesting that TOR represses GA-mediated hypocotyl elongation (Fig. 1b). Collectively, GA-activated hypocotyl elongation relies heavily on TOR, while sucrose can induce hypocotyl elongation in TOR mutants regardless (Fig. 1b). Despite contradicting current knowledge, it is crucial to highlight that most published studies utilized estradiol-inducible TOR RNAi lines, leading to robust TOR silencing[18]. In contrast, the 35-7 line represents a much milder silencing approach[17], and rop2 rop6 ROP4-RNAi specifically signifies the knockout of the auxin-TOR axis[5]. Hence, further efforts are needed to explore the physiological significance of TOR in hypocotyl elongation under low light conditions. In contrast to hypocotyl, GA repressed root growth. In presence of GA, AZD could not further decrease root length and sucrose could hardly rescue root growth, suggesting a role of GA in repressing TOR activity in root. Surprisingly, the root length of ROP triple mutants yielded similar results to the wild type (Fig. 1c), indicating that the auxin-TOR pathway specifically affects shoot architecture rather than the primary root growth, as previously suggested[4].
Besides light conditions, high temperature and humidity often contribute to excessive seedling growth. However, there has been limited research on the regulation of TOR activity under these specific conditions. One mechanism that protects plants from high temperatures is the Spermidine-TOR relay[19]. Furthermore, TOR has been found to regulate heat stress memory through the phosphorylation of E2Fa. Phosphorylated E2Fa, in turn, activates the transcription of ATX1, which establishes H3K4me3 on various target genes involved in thermomemory[20]. Both H3K4me3 and H3K27me3 are well-known markers of bistable chromatin, representing a crucial regulatory layer downstream of TOR signalling in response to stress[21]. However, the precise mechanism by which TOR regulates heat-induced elongation is still not fully understood.
The comprehensive studies of hypocotyl elongation in response to variations in light and temperature has been extensively reviewed by Li et al.[22]. Generally, these environmental factors, along with water availability, have primarily been investigated in the context of stress conditions. However, it is pertinent to discern whether subtle fluctuations in these factors could exert an influence on seedling growth prior to grafting, thereby enabling precise manipulation of growth conditions within greenhouse settings. In the pursuit of this objective, we undertook an investigation to evaluate whether fluctuations in light, temperature, and watering regimes could potentially modulate seedling strength for optimal grafting performance. Our findings, first and foremost, unveil that even a slight deviation of 2 degrees in temperature significantly impacts various growth parameters, consequently influencing the success rate of grafting (Fig. 2a, b). Expanding on this exploration, we ventured to synergize varying durations of light exposure with diverse watering frequencies. After eight days of various treatments, there were slight variations in plant height, while hypocotyl diameters increased under all growth conditions. The seedling shoot index (SSI) is a commonly used measure to assess plant vigour in horticultural practices. Interestingly, SSI significantly decreased after a three-day drought. A milder drought for two days appears to be optimal for enhancing plant vigour under long-day conditions. Our results illuminated the existence of an optimal nexus between daylength and water availability, both of which collectively contribute to fostering enhanced seedling vigour (Fig. 2c, d). These revelations hold significant implications, delineating avenues for augmenting grafting seedling robustness through the judicious manipulation of environmental factors within controlled climate chambers.
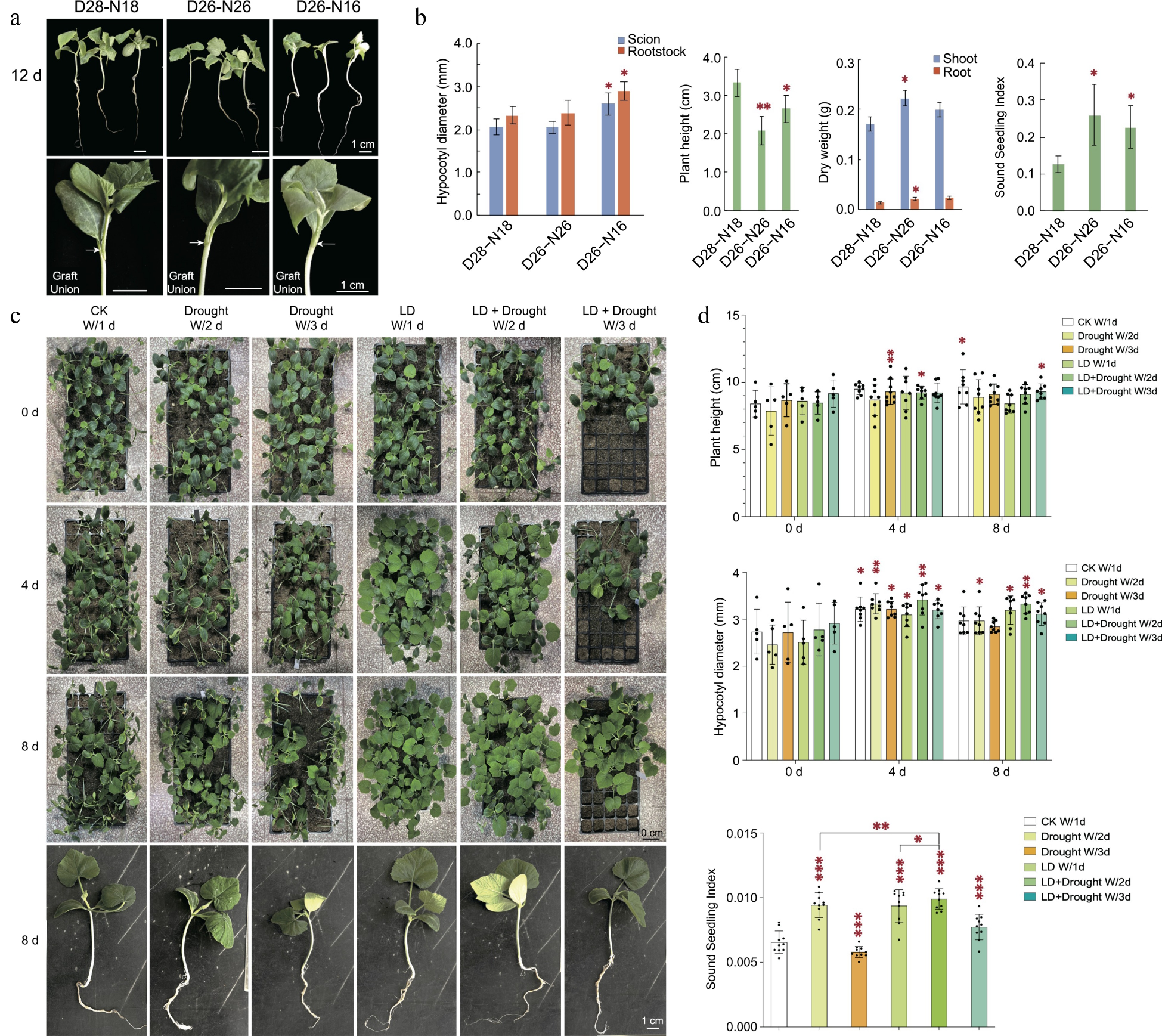
Figure 2.
Impact of environmental factors along with water availability on root-shoot relationship and plant growth. (a) 30−50 successfully grafted cucumber (scion)/pumpkin (rootstock) seedlings were cultivated under three distinct conditions: Day26 °C−Night16 °C (D26−N16), Day26 °C−Night26 °C (D26−N26), and Day28 °C−Night18 °C (D28−N18). Graft unions are indicated by arrows. (b) After 12 d, the grafted cucurbits exhibited varying growth characteristics under different conditions. Specifically, the hypocotyl diameter of grafted cucurbits was highest under D26−N16, the plant height under D28−N18, and the dry weight under D26−N26. The sound seedling index (Hypocotyl diameter (mm) / Plant height (cm) * Total dry weight (g)) for D26−N26 and D26−N16 conditions was significantly higher than for other conditions. Statistical analysis was conducted with three replicate pools, each pool comprising 10 individual grafted seedlings. Statistical significance is denoted by stars when compared to the D28-N18 treatment (One-way ANOVA followed by Student's t-test, two-tailed, *** p < 0.001, ** p < 0.01, * p < 0.05). (c) One-leaf-old spindling pumpkin seedlings (Plant height > 4 cm, Stem diameter < 3 mm) were grown at 28 °C/18 °C (day/night) under six different conditions: CK W/1 d, watered with 200 mL/tray once a day, 16 h/8 h day/night; Drought W/2 d, watered with 200 mL/tray once every two days, 16 h/8 h day/night; Drought W/3 d, watered with 200 mL/tray once every three days, 16 h/8 h day/night; LD W/1 d, watered with 200 mL/tray once a day, 20 h/4 h day/night; LD + Drought W/2 d, watered with 200 mL/tray once every two days, 20 h/4 h day/night; LD + Drought W/3 d, watered with 200 mL/tray once every three days, 20 h/4 h day/night. (d) After 8 d, spindling pumpkin seedlings in the LD + Drought conditions exhibited recovery in plant height and hypocotyl diameter compared to the control group (CK). The sound seedling index (SSI) = (hypocotyl diameter/plant height + root dry weight/shoot dry weight) × seedling dry weight. The SSI analysis demonstrated that environmental factors and drought (Drought W/2 d, LD + Drought W/2 d, and LD + Drought W/3 d) effectively promoted plant vigor. Statistical analysis involved 10 replicates, with each replicate comprising three individual seedlings. Statistical significance is denoted by stars when comparing each treatment to its corresponding 0-day control (mean ± s.d., One-way ANOVA followed by Student's t-test, two-tailed, *** p < 0.001, ** p < 0.01, * p < 0.05).
It's important to note that while TOR signalling is involved in growth regulation in response to light, temperature, and humidity, the specific mechanisms and interactions with other signalling pathways can be complex and context-dependent. Additionally, the extent to which TOR influences growth in relation to these factors may vary among different plant species and environmental conditions. Ongoing research continues to uncover the intricacies of TOR-mediated growth regulation in response to various environmental cues.
-
Grafting is a process that involves the fusion and subsequent healing of two plant tissues. Successful wound healing during grafting relies on the formation of callus cells. TOR plays a crucial role in regulating the activation of various genes and pathways involved in cell proliferation, differentiation, and tissue regeneration. Additionally, TOR signalling has been strongly implicated in wound healing and callus formation in plants[23−25]. Notably, two TOR-downstream pathways have demonstrated significant capability in regulating callus formation and cell differentiation. TOR promotes anabolic processes, such as translation, while inhibiting catabolic processes, like autophagy[26]. Studies involving autophagy mutants have shown that impaired autophagy severely hampers cell differentiation[27]. Moreover, the TOR-Polycomb repressive complex 2 (PRC2) pathway has recently emerged as a core regulator of plant development and stress responses[21,28]. Severe PRC2 mutants, such as fie and clf/swn double mutants, exhibit a callus-like phenotype, indicating the significance of this pathway in regulating cell differentiation during wound healing[29]. However, the specific involvement of TOR in this process remains unclear.
Both sugar and auxin, the most characterized TOR activators, have been studied most in details regarding their systemic roles in tissue regeneration after wounding[30,31]. In the context of grafting, TOR signalling promotes sugar-mediated callus formation at the grafting junction, which is a vital step in successful graft healing[24,30,32]. In the study by Xiong et al., the role of TOR in graft junction healing was initially characterized using rapamycin-sensitive BP12 lines, contrasting with the rapamycin insensitivity observed in wild-type Arabidopsis. Notably, self-grafting of BP12-2 proved unattainable in the presence of rapamycin, underscoring the pivotal role of TOR in graft junction formation[14]. This observation was subsequently corroborated by Miao et al. in the widely utilized cucumber-pumpkin grafting system. An intriguing aspect of their approach involved employing sugar-starved etiolated seedlings for grafting. The introduction of exogenous glucose significantly heightened the grafting success rate. It is noteworthy, however, that xylem reconnection induced by sugar appears to be TOR-independent in cucumber-pumpkin heterografts[32].
During graft formation, auxin is necessary for re-establishing severed vasculature by promoting callus growth and forming new vascular connections between scion and rootstock[33]. While shoot-derived auxin was initially thought to be the primary driver of graft formation[34], recent evidence suggests a more complex scenario. Systemic auxin flow appears not to corelate with vascular regeneration[34], but environmental conditions can influence the process through remote signals, affecting proliferation at the graft site[35]. A systemic increase in auxin levels induced by elevated temperature had a promotive effect on proliferation at the graft site, leading to faster connections of grafted tomatoes[35]. Additionally, cotyledon-derived auxin does not seem to play a major role in vasculature regeneration[36]. However, the role of TOR in auxin-mediated graft healing remains elusive.
-
Following successful grafting, communication between scion and rootstock involves a diverse array of signalling molecules. The majority of these molecules traverse from source to sink organs through the phloem stream, alongside the movement of photosynthates[37]. These molecules include mRNAs, small RNAs, and phytohormones. Notably, sucrose, functioning as an upstream activator of the TOR pathway[18], plays a pivotal role in determining sink strength.
In response to specific abiotic stresses such as sulphur deficiency and drought, plants enact adaptive strategies, often sacrificing shoot growth to conserve energy and resources that can subsequently be directed towards promoting root growth. This growth behaviour entails the inhibition of the TOR pathway in the shoot, triggering autophagy-mediated nutrient recycling[38]. Consequently, sucrose is reallocated from the shoot to the root via specialized sucrose transporters, namely SWEET11/12, which in turn activate the root apical meristem through glucose/sucrose-TOR signalling. In the context of drought conditions, Sucrose Non-Fermenting-1 Related Kinase 2 (SnRK2) come into play, acting in opposition to TOR[39]. These kinases phosphorylate SWEET11/12, an essential modification facilitating sucrose reallocation, particularly crucial under drought stress conditions[40].
In addition to sucrose, we would like also to direct attention towards the role of Translationally Controlled Tumour Protein (TCTP) within the context of the TOR signalling pathway. While TCTP's role in regulating TOR activity has been studied in mammals[41], its involvement in the TOR signalling pathway within plants remains relatively unexplored. In Arabidopsis, two TCTP homologs, TCTP1 and TCTP2, are recognized as vital mobile signals within plant communication networks[42,43]. TCTP1, in particular, has been identified as a crucial regulator of mitosis[44]. Intriguingly, its mRNA exhibits mobility from shoot to root, facilitated by the m5C modification[43]. A previous review also highlighted the possible involvement of TOR in mRNA movement[45]. Notably, Arabidopsis m5C mRNAs are remarkably enriched in the mobile transcripts, encompassing well-characterized targets such as TCTP1 and HSC70.1[43]. Given TCTP1's pivotal role as an upstream activator of TOR[44], a pertinent and unresolved query pertains to the interplay between m5C modification and Glc-TOR signalling specifically in root growth.
To shed light on this intriguing nexus, we subjected the m5C writer mutant[43], dnmt2 nsun2b, to TOR inhibition through AZD8055 (Fig. 3a). Both wild type and dnmt2 nsun2b were grown on ½ MS medium supplemented with or without TOR inhibitor for 7 d in the presence of sucrose. The absence of m5C modification in dnmt2 nsun2b mutant yielded a significant reduction in primary root length, while conspicuously leaving the shoot phenotype unaffected. Strikingly, in comparison to the wild-type, the m5C mutant exhibited augmented resistance to TOR inhibition, suggesting a probable reduction in TOR activity within the root of the dnmt2 nsun2b mutant (Fig. 3b). This intriguing finding raises the prospect of investigating whether the diminished TOR activity in the root of dnmt2nsun2b is attributed to the absence of TCTP1 mRNA transfer from the shoot, and whether the mobility and signalling function of TCTP1 is conserved in the grafted plant species. As such, the dynamic interplay between m5C modification, TCTP1 mRNA mobility, and the intricate Glc-TOR signalling pathway beckons as a promising avenue for future research.
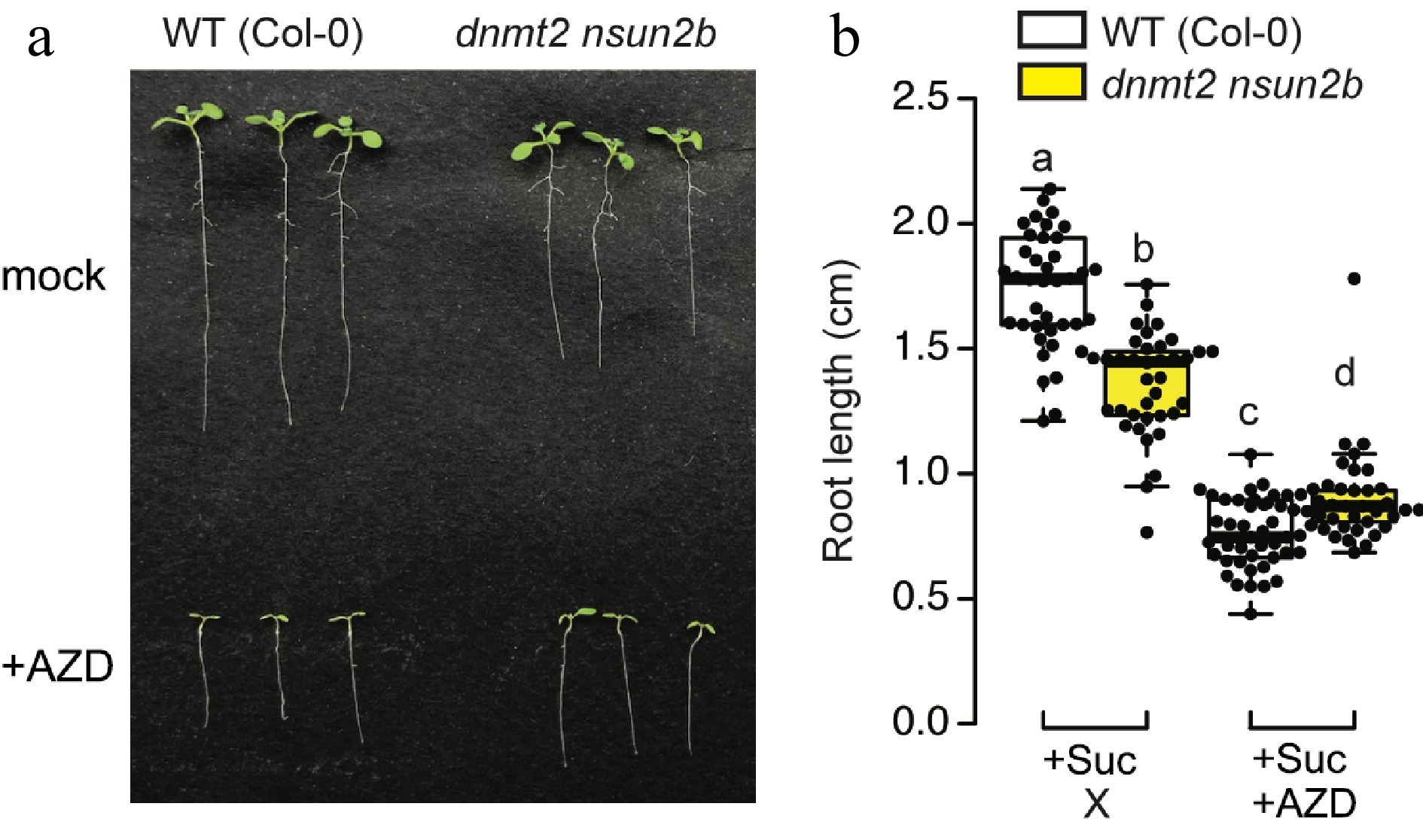
Figure 3.
The interplay between m5C modification and TOR signaling in the context of root growth. (a) Both Arabidopsis thaliana ecotype Col-0 as wild type (WT) and m5C writer mutant (dnmt2 nsun2b) were grown on 1/2MS medium containing 0.5% sucrose under long-day condition (16 h light) for 7 d. 0.5 µM AZD8055 was applied to inhibit TOR activity, while 1 µL DMSO was used in mock. (b) Root length was quantified from more than 40 seedlings grown on two independent plates. Statistical difference is indicated by different letters (One-way ANOVA followed by Student's t-test, two-tailed p < 0.05).
Overall, TOR signalling plays a significant role in shoot-root communication by integrating nutrient signalling, hormonal crosstalk, and carbon metabolism. It allows the shoot and root systems to communicate and coordinate their growth and development in response to changing environmental conditions, nutrient availability, and energy status. Further research is needed to unravel the precise mechanisms and molecular components involved in TOR-mediated shoot-root communication and its impact on grafting biology.
-
TOR serves as a pivotal master regulator orchestrating plant growth and developmental processes by assimilating a multitude of upstream signals and finely modulating crucial downstream cellular mechanisms, encompassing translation, autophagy, and the cell cycle[26]. While the scrutiny of this kinase has been exhaustive within metazoans, its comprehensive exploration within the plant kingdom commenced notably later, specifically around 2017. To this day, a distinct comprehension of TOR's multifaceted functionality remains constrained, primarily attributable to the dearth of diverse mutants and reporter lines, particularly in non-Arabidopsis plant species. This perspective paper seeks to illuminate the latent role of TOR in the grafting process, with the overarching goal of directing heightened research focus towards the TOR signalling pathway within the realm of vegetable crops.
Although the characterization of TOR as a conserved kinase across eukaryotes is well-established, its functional implications might diverge considerably. Notably, TOR signalling pathways undergo a complete reprogramming in photoautotrophic organisms compared to their heterotrophic counterparts[46,47]. Intriguingly, recent research has unveiled pivotal insertions within the TOR protein of salamanders, correlating with their remarkable regenerative capabilities in contrast to mammals[48]. This raises the intriguing possibility of an augmented regenerative role for plant TOR during the course of evolution, considering a low amino acid sequence identity of Arabidopsis TOR, especially at the N-terminus, compared to its animal counterparts[49]. This tantalizing perspective holds significant promise, particularly within vegetable crops subject to multiple annual harvests and in the context of grafting junction healing. Another study in vegetable crops highlights an intriguing future perspective on TOR signalling. The absence of a plant TOR inhibitor posed a significant challenge for TOR research in past decades, given the insensitivity of most studied higher plants to rapamycin due to the non-functional FK506 Binding Protein 12 KD (FKBP12). Intriguingly, in horticultural crops such as tomatoes, SlFKBP12 can facilitate the inhibitory interaction between rapamycin and TOR, mirroring the situation observed in yeast and mammals[50]. The delineation of a comprehensive phylogenetic analysis coupled with an in-depth functional evolution study on TOR and its signalling components emerges as an imperative avenue for future research endeavours.
TOR plays a crucial role in orchestrating efficient shoot-to-root communication by coordinating nutrient and phytohormone signalling. However, research on TOR in vegetable crops, particularly in the context of heterografts, is noticeably limited. This is particularly intriguing given that specific rootstocks have the potential to enhance various agricultural traits in scions including increased stress tolerance and fruit yield[1]. Exploring how the TOR signalling pathway in the scion might be reprogrammed by a specific rootstock becomes an intriguing avenue of inquiry. Furthermore, TOR itself could potentially serve as a conduit for shoot-to-root communication. Notably, within animal systems, the mobility of TOR mRNA has been observed during nerve injury[51]. While such mobility hasn't been observed in Arabidopsis[52], recent advancements in mobile RNA databases, specifically in the context of cucumber (Csa)/pumpkin (Cmo) heterografts, have unveiled CsaTOR (CsGy7G006260) and CmoTOR (CmoCh19G005520) as mobile shoot-to-root transcripts. Remarkably, CmoTOR has been identified as being modified by m5C in the vascular extrude[53]. This intriguing discovery raises the tantalizing prospect that TOR mRNA mobility might indeed be a phenomenon that varies across diverse plant species.
To gain comprehensive insights into this dynamic, it becomes imperative to delve into the regulation of TOR mRNA transcription, a facet that presently remains strikingly unexplored. During tomato fruit ripening, TOR is silenced transcriptionally to promote ethylene production[54,55]. However, the mechanism remains obscure. The only known instance of a well-studied transcription factor from the jasmonic acid (JA) signalling pathway, MYC2, binding to the promoter region of TOR has been identified in tomatoes[56]. Unravelling the intricacies of this transcriptional control is pivotal in comprehending the potential mobility of TOR mRNA within heterografts. By bridging this gap in knowledge, we can not only expand our understanding of TOR's communication capabilities but also pave the way for unlocking the underlying mechanisms governing shoot-to-root signalling in the intricate context of plant grafting.
-
The authors confirm contribution to the paper as follows: Zhang W, Han L, Dong Y initiated the project and designed the experiments; Zhang W, Huang Y, He J, Dong Y performed experiments; Huang Y, He J were supervised by Zhang W. Zhang W, Dong Y made figures and wrote the manuscript; Han L, He F revised the manuscript. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analyzed during this study are included in this published article.
We thank Dr. C. Meyer (INRAE, France) for providing the 35-7 line, Dr. Y. Zhou for providing the dnmt2 nsun2b mutant, the ikann-editorial team for language editing (ikann-editing@outlook.com), the 2020 Marie-Curie fellowship 885864 TOR in acTIon MSCA-IF-EF-ST to Y. Dong, and the 2115 Talent Development Program of China Agricultural University to W. Zhang.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Zhang W, Han L, Huang Y, He J, He F, et al. 2024. Unveiling the significance of Target of Rapamycin (TOR) signalling in grafting. Vegetable Research 4: e004 doi: 10.48130/vegres-0024-0003
Unveiling the significance of Target of Rapamycin (TOR) signalling in grafting
- Received: 11 September 2023
- Revised: 11 December 2023
- Accepted: 03 January 2024
- Published online: 31 January 2024
Abstract: Grafting is a widely employed horticultural technique that enables the combination of desirable traits from different plant species or cultivars. The success of grafting depends on various factors, including the strength of the seedlings, proper healing of the grafting junction, and the subsequent recovery of heterografted seedlings. In recent years, the target of rapamycin (TOR), a conserved protein kinase, has emerged as a key regulator of growth and development in plants. This perspective paper delves into the implications of TOR signalling in the grafting processes, including the role of the TOR signalling pathway in the regulation of seedling vigour prior to grafting, healing of graft junction and shoot-to-root communications. Particularly, we highlight the role of gibberellin and m5C modification in the regulatory network of TOR. However, further research is needed to unravel the precise molecular mechanisms underlying TOR's involvement in grafting and to optimize its application for improved grafting outcomes.
-
Key words:
- Grafting /
- Target of rapamycin (TOR) /
- Environmental cues /
- Graft junction /
- Shoot-root communication












