HTML
-
Polyurethane (PU) is a synthetic polymer produced by reacting polyols and polyisocyanates that is widely utilized in medical, agricultural, automotive and industrial fields (Howard 2002, Tokiwa & Calabia 2009, Krasowska et al. 2012, Magnin et al. 2019, Tan & Ohwada 2019). In particular, polyurethane-based binder systems are extensively used in composite solid propellants, usually consisting of PU and its derivatives like alkyne terminated polybutadiene with urethane segments (PUPB), ammonium perchlorate (AP), aluminum powder (Al) solids, binders and other additives like plasticizers (Bunyan et al. 1993, Haska et al. 1997, Libardi et al. 2010). Solid propellants are widely used in space and tactical systems as energetic materials (Davenas 2003, Libardi et al. 2010). An increasing amount of solid propellants are produced every year, all of which need disposal owing to their deterioration or obsolescence. In the past, solid propellant materials have been disposed via ocean dumping, open area burning or detonation in a safe zone (Gautam et al. 2007, Mahajan & Gupta 2015); however, when solid propellants are released into the biosphere, energetics are xenobiotic contaminants that threaten ecosystems, humans and other biota with toxic waste materials (Pichtel 2012). These conventional means of processing propellants or other polymer materials can worsen land and water pollution while exacerbating safety concerns. Therefore, in order to address environmental pollution problems caused by propellant waste processing, sustainable solutions are urgently needed for biodegrading the PU and PUPB that comprise the backbone structures of propellant materials. Out of all currently available methods, microbial degradation has been accepted as the most environmentally friendly method for the disposal of these polymer materials (Sangale et al. 2019, Sarkhel et al. 2019).
Among all the major matrices of solid propellants, no microorganism capable of directly degrading the PUPB matrix has been reported. However, recent advances in our understanding of how other types of PU biodegrade suggest how to biologically process and degrade PUPB in the future (Mathur & Prasad 2012, Khan et al. 2017). Some studies have assessed the potential and characteristics of fungi associated with PU degradation, while other studies have evaluated the extracellular enzymes used by fungi to utilize PU as a carbon source (Álvarez-Barragán et al. 2016, El-Morsy et al. 2017, Pathak & Navneet 2017, Khan et al. 2020). For example, El-Morsy et al. (2017) isolated Monascus sp., which can degrade PU from plastic-contaminated soils in Egypt. Khan et al. (2020) also reported that Aspergillus flavus isolated from the intestines of crickets can degrade PU with a mean mass loss of 1.9% per week.
In this study, we aimed to screen different fungal strains from explosive materials-contaminated soils and test their ability to degrade standard PU film and PUPB patches. Initial identification analysis was also conducted to identify fungal strains with the potential to degrade PU and PUPB. Here, we report Fusarium solani capable of degrading standard PU films with the potential ability to degrade PUPB patches in laboratory conditions as well as its taxonomic identification and enzyme activity.
-
Soil samples contaminated by explosive materials were collected from central China. Soil collection was carried out using a sterilized auger. From each site, five soil sub samples were collected while maintaining 5 cm soil depth from the surface layer. Collected soil samples were placed in sterilized bags and thoroughly mixed. Samples were transported to the laboratory and stored at 4℃ for further analyses.
Preparation of PU film and the PUPB patch
-
The standard PU used in the present study was a type of polyester polyurethane, (Poly ]4, 4' -methylenebis (phenyl isocyanate) -alt-1, 4-butanediol/di (propylene glycol)/polycaprolactone] (PU/PCL), Sigma-Aldrich Corporation, CAS: 68084-39-9, USA). PU beads (22 g) were dissolved in 600 ml of tetrahydrofuran (Aladdin Industrial Corporation, China) and oscillated in a reciprocating multi-amplitude orbital shaker for 1 day at 30℃ (ZWF-200, Zhicheng, China). The PU solution was then poured onto petri dishes (10 mL/per dish) and allowed to solidify for 48 h in a desiccator at room temperature. Dried PU films were removed from the petri dishes and stored at room temperature. PUPB materials were prepared using 2, 4-toluene diisocyanate (TDI), propargyl (3-isocyanato-4-methylphenyl), carbamate (PTI) and HTPB, all of which were provided by the Aerospace Chemical Power Laboratory of Hubei Institute of Aerospace Chemotechnology, China. PUPB patches were made by cutting PUPB material into 1.5 × 2.0 cm rectangles 0.2 cm thick.
Isolation testing of fungi from contaminated soil
-
The serial dilution plating method was used to dilute soil samples as described by Waksman (1922) with the purpose of minimizing fungi in the soil of each dilution. Soil sample dilution was conducted in two replicates, and each replicate was diluted four times and labeled 10-1 to 10-4. At each level of dilution, soil extracts were obtained by shaking 1 g of soil in 9 mL of sterilized water for 1 hour and next centrifuged for 10 min at 2000 rpm. Supernatants were serially diluted under aseptic operating conditions and 1 mL of 10-1 diluted fungal solution was placed into the centrifuge tube containing 9 mL sterile water, which was then shook and mixed evenly to obtain a 10-2 concentration of fungal solution. Finally, 20 μL of each concentration of diluent was placed onto a Potato Dextrose Agar (PDA) petri dish with a pipette and spread evenly across the surface of the petri dish using a sterile glass coating rod.
Upside-down culture dishes were incubated under dark conditions at 28℃ for 3-5 days. Post-incubation fungal colonies were transferred to new PDA plates. Pure fungal cultures were obtained by sub-culturing each of the different colonies onto new PDA plates. Finally, 29 strains were obtained.
Degradative testing of strains
-
A sterilized razor blade was used to divide the colony of each strain into 2 mm sections, after which one piece of each purified culture was transferred to Malt Extract Agar (MEA), and a sterilized PU film with a diameter of 80 mm was laid over the surface of each medium (three replicates). Simultaneously, a purified culture section from each colony was transferred to MEA, and a sterilized PUPB patch was laid over the surface of each MEA medium (three replicates). Cultures were incubated at room temperature (26 ± 2℃) for 90 days in a sterile culture room, and decomposition of PU and PUPB was observed.
Sample collection
-
The abilities of 29 fungal strains to degrade PU and PUPB were tested. Fungal degradative abilities were analyzed by determining mass loss after 90 days of observation. After 90 days of incubation, PU films and PUPB patches were both collected, washed thoroughly with distilled water, shade-dried and weighed. According to collected data, mass loss of PU films and PUPB patches were calculated using the following formula (Mathur & Prasad 2012).
$ \mathrm{M}(\%)=\frac{(\mathrm{M} 1-\mathrm{M} 2) \times 100}{\mathrm{M} 1} $ In the formula, M (%) is the percentage of mass loss, M1 (g) is the initial mass before degradation and M2 (g) is the final mass after degradation.
Macro- and micro-morphological photography
-
Fungal colonies were incubated at 25℃ for four weeks on PDA. Micro-morphological structures were photographed using a Nikon compound microscope (Nikon ECLIPSE Ni) fitted with a Canon (EOS 600 D) digital camera. Micro-morphological changes in PU and PUPB structures after fungal degradation were observed via Scanning Electron Microscope (SEM, Sigma 300). Measurements were taken using the Tarosoft (R) Image Frame Work program. Images used for figures were processed with Adobe Photoshop CS6.
DNA extraction, PCR amplification, sequencing and phylogenetic analyses
-
Genomic DNA was extracted from the mycelium grown on PDA at 25℃ for four weeks using Biospin Fungus Genomic DNA Extraction Kit (BioFlux® Hangzhou, China). Three genes were used in our study, viz. internal transcribed spacer region (ITS) using primer pair ITS5/ITS4 (White et al. 1990), the large subunit nuclear ribosomal (LSU) using primer pair LR0R/LR5 (Vilgalys & Hester 1990), the translation elongation factor 1-alpha gene (tef1-α) using primer pair EF1/EF2 (O'Donnell et al. 1998) and the second largest subunit of RNA polymerase Ⅱ (rpb2) using primer pair 5f2/11ar (Liu et al. 1999, Reeb et al. 2004). Amplification reactions were performed in a total volume of 25 μL of PCR mixtures containing 8.5 μL ddH2O, 12.5 μL 2X PCR MasterMix (TIANGEN Co., China), 2 μL DNA template and 1 μL of each primer. The PCR thermal cycling program for LSU, ITS and tef1-α were set as described in Wang et al. (2019) PCR products were sent for sequencing at Qingke Company, Kunming City, Yunnan Province, China. Sequences were deposited in GenBank (Table 1).
Table 1. Taxa names, strain numbers, host information, locations and corresponding GenBank accession numbers of the sequences used for the phylogenetic analyses
Taxon name Strain number Isolate habitat/host Location GenBank accession numbers LSU ITS tef1-α rpb2 Geejayeesia atrofusca NRRL 22316 Staphylea trifolia USA AF178392 AF178423 AF178361 JX171609 Fusarium catenata NRRL 54992 Zebra shark multiple tissues USA MG189913 KC808255 KC808213 KC808354 Fusarium catenata NRRL 54993T Zebra shark multiple tissues USA MG189914 KC808256 KC808214 KC808355 Fusarium croci CBS 115659 Potato Germany JX435206 JX435206 JX435156 JX435256 Fusarium croci CBS 142423T Citrus sinensis Italy LT746264 LT746264 LT746216 LT746329 Fusarium croci CPC 27187 Citrus sinensis Italy LT746265 LT746265 LT746217 LT746330 Fusarium cyanescens CBS 518.82T = NRRL 37625 Human foot The Netherlands EU329684 EU329684 FJ240353 EU329637 Fusarium falciformis CBS 318.73 = NRRL 22660 Trichosanthes dioica India JX435208 JX435208 JX435158 JX435258 Fusarium falciformis CBS 475.67T Human Puerto Rico MG189915 MG189935 LT906669 LT960558 Fusarium gamsii CBS 217.53 = NRRL 22655 Plywood Nigeria MG189916 MG189936 DQ247637 LT960559 Fusarium gamsii CBS 143207T = NRRL 32323 Human bronchoalveolar lavage fluid USA DQ236462 DQ094420 DQ246951 EU329576 Fusarium haematococa CBS 119600ET Dying tree Sri Lanka KM231664 KM231797 DQ247510 LT960561 Fusarium illudens NRRL 22090 Beilschmiedia tawa New Zealand AF178362 AF178393 AF178326 JX171601 Fusarium keratoplastica NRRL 43373 Contact lens Malaysia EF453072 EF453072 EF452920 EF469959 Fusarium lichenicola NRRL 28030 Human Thailand DQ236397 DQ094355 DQ246877 EF470146 Fusarium lichenicola NRRL 34123 Human eye India DQ236687 DQ094645 DQ247192 EU329635 Fusarium macrospora CBS 142424T Citrus sinensis Italy LT746281 LT746266 LT746218 LT746331 Fusarium macrospora CPC 28192 Citrus sinensis Italy LT746282 LT746267 LT746219 LT746332 Fusarium mahasenii CBS 119594T Dead branch of live tree Sri Lanka JF433045 JF433045 DQ247513 LT960563 Fusarium metavorans CBS 130400 = NRRL 43489 Human cornea USA DQ790528 DQ790528 DQ790484 DQ790572 Fusarium metavorans CBS 143194 = NRRL 22782 Human corneal ulcer Spain EU329670 EU329670 DQ246850 EU329528 Fusarium petroliphila NRRL 46706 = FMR 8340 Human blood Qatar EU329715 EU329715 NA EU329664 Fusarium plagianthi NRRL 22632 Hoheria glabrata New Zealand AF178386 AF178417 AF178354 JX171614 Fusarium pseudensiformis CBS 241.93 = NRRL 53635 Human Suriname JX435198 JX435198 JX435148 JX435248 Fusarium pseudensiformis CBS 125729T Unknown dead tree Sri Lanka KC691584 KC691584 DQ247512 NA Fusarium solani CBS 140079ET = NRRL 66304 = FRC S-2364 Solanum tuberosum Slovenia KT313633 KT313633 KT313611 KT313623 Fusarium solani NRRL 32484 = FRC S-1242 Human USA DQ236491 DQ094449 DQ246982 EU329583 Fusarium solani NRRL 43474 Human eye USA EF453097 EF453097 EF452945 EF469984 Fusarium solani KUMCC 20- 0230 Soil China JX435189 MW393522 MW460712 MW460711 Fusarium suttoniana CBS 124892 Human nail Gabon DQ236659 JX435189 DQ247163 JX435239 Fusarium suttoniana CBS 143214T = NRRL 32858 Human wound USA MG189926 DQ094617 LT906672 EU329630 Fusarium tonkinensis CBS 115.40T = NRRL 53586 = IMI 113868 Musa sapientum Vietnam MG189927 MG189941 LT906673 LT960564 Fusarium tonkinensis CBS 143038 Human cornea The Netherlands EF453092 MG189942 EF452940 LT960565 Fusarium vasinfecta CBS 130182 = NRRL 43467 Human USA AF178392 EF453092 AF178361 EF469979 Our strain sequence is indicated in bold. "NA" sequences are unavailable. Ex-type strains are indicated with superscript "T". Ex-epitype strains are indicated with superscript "ET". Sequences of representative taxa were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/), and accession numbers are listed in Table 1. Newly generated sequences in this study were assembled using BioEdit 7.0.9.0 (Hall 1999). Individual gene regions were separately aligned in the MAFFT v.7.110 web server (http://mafft.cbrc.jp/alignment/server/) (Katoh et al. 2019). Gene alignments were improved by manually deleting ambiguous regions plus gaps and combined using BioEdit 7.2.3. Final alignments containing LSU, ITS, tef1-α and rpb2 were converted to NEXUS format (.nxs), employing CLUSTAL X (2.0) (Thompson et al. 1997). The FASTA format was translated into PHYLIP format via Alignment Transformation Environment (ALTER) online program (http://www.sing-group.org/ALTER/) and used for maximum likelihood analysis. Maximum likelihood analysis (ML) was carried out in CIPRES Science Gateway v.3.3 (http://www.phylo.org/portal2/; Miller et al. 2010) under RAxML-HPC2 on XSEDE (8.2.12) (Stamatakis 2014) with the GTR+GAMMA substitution model and 1, 000 bootstrap iterations. Bayesian analyses of six simultaneous Markov chains were run for 2, 000, 000 generations, and trees were sampled every 100th generations. Phylogenetic trees were visualized in FigTree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/, Rambaut 2012). The constructed tree was edited using Microsoft PowerPoint and saved as a PDF format.
Enzyme activity determination
-
For detecting lipase, esterase and protease activities, cultured fungal mycelia were transferred to a peptone agar medium (1% peptone, 0.5% NaCl, 0.01% CaCl2·H2O, 1.5% agar) (w/v), PDA medium (1% peptone, 0.1% yeast extract, 0.005% CaCl2, 1.5% agar) (w/v) and basal medium (2 % sucrose, 0.5% yeast extract, 2% KCl and 1.5% agar) (w/v), respectively (Sierra 1956, Castro et al. 1992, Vermelho et al. 1996, El-Morsy et al. 2017). Peptone agar medium was supplemented with 1% of autoclaved tween 80, PDA medium was supplemented with 1% tween 20 (El-Morsy et al. 2017). Plates were incubated at 28℃ for 7 days. Protease production was detected by staining the medium with 0.25% Coomassie brilliant blue (methanol-acetic acid-water 5:1:4 (v/v/v); Beijing Solarbio Science and Technology Co., China) (Vermelho et al. 1996). An opaque halo could be easily observed around the colonies, indicating tested micro-organisms experienced lipolytic activity on the peptone agar medium (Sierra 1956). Esterase production by the fungi strain was indicated by a white precipitate of calcium salt around colonies on the PDA (Castro et al. 1992). Protease production by the fungal isolates was indicated by the formation of clear zones around colonies on the basal medium (Vermelho et al. 1996). Results were evaluated by calculating an index of relative enzyme activity (RA) (Bradner et al. 1999). RA was calculated using the following equation, and diameters were measured in cm.
$ \mathrm{RA}=\frac{(\mathrm { clear\ zone\ diameter }-\mathrm { colony\ diameter) }}{\mathrm { clear\ zone\ diameter }} $
Selection testing of degradative strains via mass loss of degraded PU film and PUPB patch
-
Biodegradative capacity was monitored by measuring the mass loss of PU films and PUPB patches before and after incubation with isolated fungus (Ibrahim et al. 2011). Out of the 29 monitored fungal strains, the fungal isolate strain H14 reduced the mass of PU films and PUPB patches to a total loss of 25.8% and 1.3%, respectively, after 3 months (values were arrived at by averaging of three replicates, Table 2). Results showed that fungal strain H14 has the greatest ability for PU degradation compared to other fungal strains. Mass loss values also showed H14 has the ability to degrade PUPB patches (Table 2).
Table 2. PU film and PUPB patch mass loss resulting from inoculated Fusarium solani H14
Polymer type No. M1 (g) M2 (g) Difference (g) M (%) Average M (%) PU 1 0.4026 0.3020 0.1006 24.98 2 0.4002 0.2895 0.1105 27.66 25.8 3 0.3985 0.3001 0.9840 24.69 PUPB 1 0.3791 0.3741 0.0050 1.32 2 0.3375 0.3334 0.0041 1.21 1.3 3 0.5142 0.5073 0.0069 1.34
Screening and isolation of PU- and PUPB-degrading fungi
-
Fusarium solani (Mart.) Sacc., Michelia 2(no. 7): 296 (1881). Fig. 1
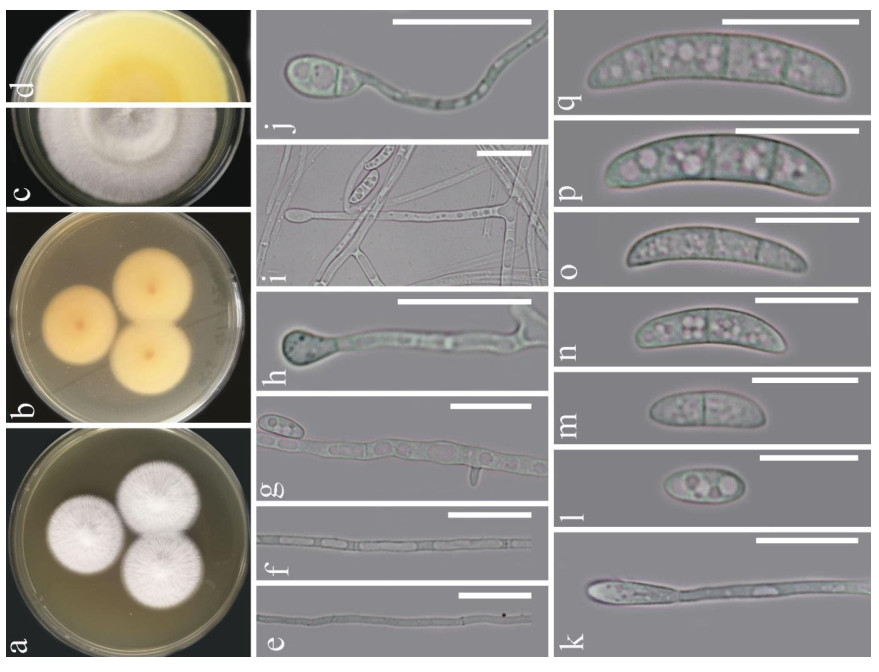
Figure 1. Fusarium solani (KUMCC 20-0230). a-d Sporulated culture. e-g Mycelium. h-k Conidiogenous cell and conidia. l-q Conidia. Scale bars: e-k = 20 μm, l-q = 15 μm.
Index Fungorum number: IF190352; Facesoffungi number: FoF01873
= Neocosmospora solani (Mart.) L. Lombard & Crous, Stud. Mycol. 80: 228 (2015)
Morphology description - Colonies on PDA (Fig. 1), reaching 20-25 mm diam., after four weeks at 25℃, mycelium white, floccose, radiate, with abundant aerial mycelium, circular, umbonate at the center, margins entire, texture velvety. Reverse luteous. Conidiophores borne on aerial mycelium, 50-100 μm long, slightly tapering upward, micronematous, mononematous, erect, simple, straight or slightly flexuous, smooth-walled, thin-walled, hyaline, sometimes reduced to conidiogenous cells. Conidiogenous cells monoblastic, integrated, determinate, terminal, hyaline. Conidia 14-25 × 3.5-6 (x = 18.6 × 5.1, n = 30) μm, solitary, acrogenous, simple, smooth-walled, 2-3-septate, straight to curved, hyaline, ellipsoidal, reniform.
Material examined - central China, from soils contaminated with explosive materials, 21 December 2019, Heng Gui, HKAS 112165, living culture KUMCC 20-0230.
Phylogenetic analyses
-
The phylogenetic analysis was conducted with 34 taxa in Fusarium and one outgroup taxon, Geejayeesia atrofusca (NRRL 22316). The aligned sequence matrix comprised tree gene regions including gaps (LSU: 833 bp, ITS: 498 bp, tef1-α: 712 bp and rpb2: 854 bp) for a total of 2897 characters. The RAxML analysis of the combined dataset yielded a best scoring tree with a final ML optimization likelihood value of (-10227.648965). Estimated base frequencies were as follows: A = 0.240280, C = 0.278119, G = 0.257975, T = 0.223626; substitution rates AC = 2.083626, AG = 4.760418, AT = 2.617861, CG = 1.090249, CT = 10.654651, GT = 1.00; and gamma distribution shape parameter α = 0.166032. In Bayesian posterior analysis, GTR+I+G model was used for LSU and ITS, GTR+G model was used for tef1-α and SYM+I+G model was used for rpb2. In the phylogenetic tree obtained from ML and BI analysis (Fig. 2), our strain was positioned among the group of F. solani with high bootstrap support (100% ML and 1.00 BYPP, Fig. 2), indicating our strain is F. solani.

Figure 2. Phylogram generated from Bayesian Inference analysis based on combined LSU, ITS, tef1- α and rpb2 dataset. Bootstrap support values for maximum likelihood (ML) equal to or higher than 80 % and Bayesian posterior probabilities (BYPP) equal to or greater than 0.95 are indicated above the nodes. The new isolate from this study is indicated in red bold. The tree is rooted with Geejayeesia atrofusca (NRRL 22316).
Degradation of PU films and PUPB patches by Fusarium solani H14
-
After 3 months of heavy fungal colonization by Fusarium solani H14, the surface of the PU film was discolored, its color changed from white to yellowish-brown and surface aberrations, damage and yellow spots appeared after being thoroughly washed. There were conspicuous changes on the PU film surface (Fig. 3). At the same time, visible mycelium could be seen attached to the surface of the PUPB patch (Fig. 4).
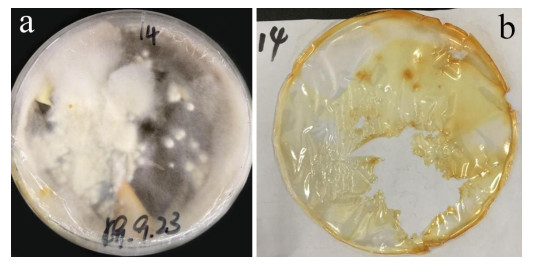
Figure 3. Photographs of the PU film on medium with fungal growth before and after washing. a Mycelia growth on the PU surface film 90 days after incubation. b PU film degraded by Fusarium solani H14.
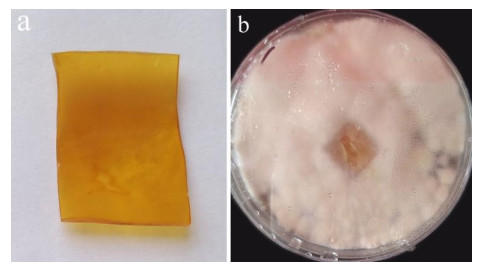
Figure 4. Photographs after degradation of the PUPB patch by Fusarium solani H14. a PUPB-sterilized patch. b Mycelia growth on the PUPB-sterilized patch 90 days after incubation.
Scanning electron microscopy
-
Scanning electron microscopy was performed on PU films and PUPB patches 90 days after degradation, and images showed that the surface of the flat and smooth PU film formed holes, underwent folding and experienced cracking and irregular fissuring alongside the appearance of an extensive network of fungal hypha (Fig. 5). Hypha covered the surface of the PUPB patch, and pores in the surface of the PUPB patch were clearly visible (Fig. 6).
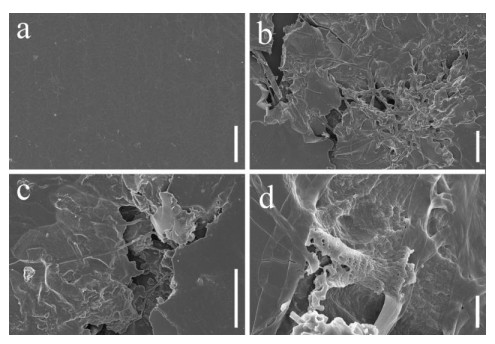
Figure 5. Scanning electron microscopy micrographs of PU film ultrastructure after Fusarium solani H14 growth. a Control. b-d Fusarium solani H14 with PU film 90 days after the biodegradation experiment. Scale bars: a = 20 μm, b, c = 40 μm, d = 4 μm.
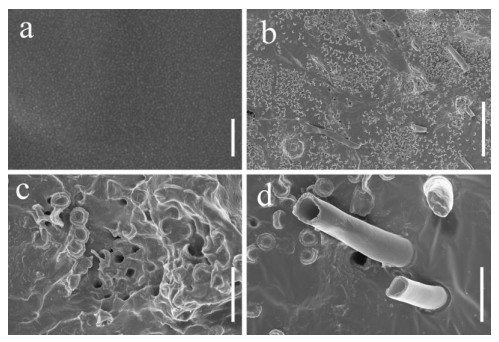
Figure 6. Scanning electron microscopy micrographs of the PUPB patch ultrastructure after Fusarium solani H14 growth. a Control. b-d Fusarium solani H14 with the PUPB patch 90 days after the biodegradation experiment. Scale bars: a = 100 nm, b = 100 μm, c, d = 10 μm.
Enzyme activity by Fusarium solani H14
-
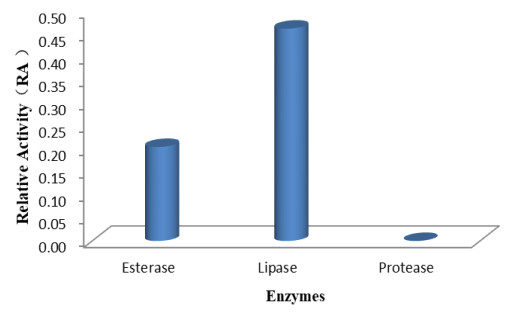
Our findings revealed that the isolate Fusarium solani H14 could produce esterase and lipase, and RA is 0.46 and 0.21, respectively (Figs 7, 8). However, in the protease production experiment, no formation of transparent haloes around the cultures could be found.
Taxonomy
-
Biodegradation as an ecologically friendly means of mitigating the accumulation of polymer types and reducing environmental pollution has attracted increasing attention in recent years. The number of polymer-degrading microorganisms isolated from different types of soils has also increased, including actinomycetes, fungi and bacteria (Zheng et al. 2005, Bhardwaj et al. 2013, Kale et al. 2015, Nakei et al. 2015, Brunner et al. 2018, Magnin et al. 2019). Fungal biodegradation of polymers remains a relatively unexplored field of study compared with bacteria and their associated enzymes (Loredo-Treviño et al. 2011). PU is susceptible to microbial attacks in acidic and neutral soils, and predominant degrading microbes are fungi under a wide range of soil (Barratt 2003, Cosgrove et al. 2007). PU degradation involves the degradation of polyester bonds and polyether bonds (Nakajima-Kambe et al. 1999). PU biodegradation results from hydrolysis of ester bonds, as the hydrolysable ester bonds are more biodegradable than polyether ones (Howard et al. 1999, Magnin et al. 2019). Studies have shown that the degradation of solid PU may be affected by the adsorption of external enzymes from clay. Therefore, in order to screen out fungi with PU- and PUPB-degrading potential, we directly isolated soil fungi to determine their degradative ability (Ibrahim et al. 2011).
In this study, Fusarium solani H14 was screened from soil contaminated with explosive rocket propellant material. It was found to be capable of degrading standard PU film. Results from the degradative testing test method indicated a 25.8% mass loss for PU by F. solani after 90 days, and there was a notable change on the surface of the PU film. SEM micrographs revealed that standard PU films experienced folding, cracking, and erosion along with irregular fissuring. This result is in line with previous studies (Crabbe et al. 1994, Ibrahim et al. 2011, Zafar et al. 2013). Ibrahim et al. (2011) reported F. solani caused significant mass loss in the PS-PUR blocks in the shaken cultures and petri dish test method, respectively, and Zafar et al. (2013) also reported F. solani caused significant physical deterioration. In contrast, mass loss for PUPB was relatively low (approximately 1.3%) after 90 days of incubation with F. solani H14; however, extensive mycelia colonization and numerous holes were found on the surface of PUPB under SEM examination suggesting that PUPB degradation might be a slower process.
Effective degradation of polymer by microorganisms is directly related to polymer structure (such as molecular orientation, crystallinity, cross-linking and chemical groups present in the molecular chains) (Howard 2002) and microbial enzymes (such as ureases, proteases and esterase) (Mathur & Prasad 2012). Other studies have reported polymer degradation involving the binding of microbial cells to polymer with subsequent floc formation, followed by degradation of the substrate; more microbial cells coated with PU has led to greater PU degradation (Howard et al. 1999, Howard 2012, El-Morsy et al. 2017, Iram et al. 2019). Furthermore, additives, antioxidants and stabilizers used in the manufacturing of polymer decelerate the rate of degradation and could also be toxic to microorganisms (Arutchelvi et al. 2008, Kale et al. 2015).
Various enzymes play essential roles in the biodegradation of polymers; these enzymes include laccase, cutinase, hydrolase, esterase, protease and urease (Barratt 2003, Loredo-Treviño et al. 2011, Bhardwaj et al. 2013). Fungi feature higher levels of enzyme biodegradation activity compared to bacteria, and enzymes are specific in their actions on substrates (Bhardwaj et al. 2013, Banerjee et al. 2014). Factors affecting the production of enzymes include PH, medium composition and temperature (Vermelho et al. 1996). Enzymes involved in polymer degradation are extracellular and membrane bound (Mathur & Prasad 2012). In the process of degradation, extracellular enzymes function as key players and are actively involved in the biodegradation of polymers by cleaving ester bonds to degrade the PU substrate (Ibrahim et al. 2011, Raaman et al. 2012, Ma & Wong 2013, El-Morsy et al. 2017). The biodegradation of PU begins with surface erosion initiated by microbial enzymes. The chemical process that occurs during biodegradation is usually divided into the assimilatory process and dissimilatory processes (Tan & Ohwada 2019). In the present study, we carried out experiments on enzymes produced by F. solani H14, selected esterase, protease and lipase for the enzyme production experiment and found F. solani H14 had a high ability to produce lipase and esterase and no ability to produce protease. Lipase and esterase could play an important role in the ability of F. solani H14 to degrade PU films; however, whether the holes formed on the PUPB surface are related to lipase and esterase or other enzymes needs further study.
It is also necessary to optimize treatment conditions to improve the biodegradation capacity of F. solani H14 on PUPB by extending the degradation time, improving characteristics of the medium (PH, composition of medium, temperature) as well as analyzing the structure of the propellant materials and enzyme production factors. These must be carried out in order to increase hyphae colonization on PUPB (Ali et al. 2014). Our test provides a pathway for further PUPB-related degradation experiments. Future experiments aimed at enhancing the ability of F. solani H14 to degrade PUBP could include a series of hybridization trials, hybridizing strain H14 and screening resultant strains for increased rates of PUBP biodegradation PUBP.
PU-degrading fungi are generally isolated from contaminated soils, sand, wall paint, plastic waste, dumping areas, compost and plastic debris floating near lakeshores (Loredo-Treviño et al. 2011, Zafar et al. 2013). However, we report on the first F. solani isolation from soils contaminated with explosive rocket propellant materials in China. We also are the first to report F. solani degradation of a propellant material prepared from an elastomer-based PUPB. Further in-depth research on the mechanisms behind PUPB biodegradation is required to solve the issue of degrading rocket propellant materials.
-
In this study, Fusarium solani H14 was isolated from soil samples (contaminated with explosive materials) and illustrated with morphological evidence and phylogenetic analyses. The results of PU- and PUPB-degrading ability of this species can be summarized as follows: mass loss analyses revealed reductions in mass of standard PU film and PUPB patches; and scanning electron microscopy images showed that the surface of the standard PU film and PUPB patches formed holes, underwent folding and experienced damage and irregular fissuring from the erosion of fungal hypha. Two possible degradation enzymes, lipase and esterase, were produced by F. solani H14. The above findings confirm the degradation effect of F. solani H14 on standard PU film and PUPB patches. This is a preliminary study that provides a potential roadmap for solving problems associated with environmental pollution, particularly related to disposing rocket propellant waste materials via microbial degradation in the future.
-
This work was financed by Open Research Fund Program of Science and Technology on Aerospace Chemical Power Laboratory (STACPL320181B04). We also would like to thank the support from the National Natural Science Foundation of China (NSFC21975066, NSFC21875061).
- Copyright: © 2021 by the author(s). This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
| GC Ren, AM Pang, Y Gao, SX Wu, ZQ Ge, TF Zhang, DN Wanasinghe, S Khan, PE Mortimer, JC Xu, H Gui. 2021. Polyurethane-degrading fungi from soils contaminated with rocket propellant and their ability to decompose alkyne terminated polybutadiene with urethane. Studies in Fungi 6(1):224−239 doi: 10.5943/sif/6/1/15 |