HTML
-
Many ancient literature indicate that synergistic collaboration among fungi and plants have taken place more than 400 million years ago (Krings et al. 2007). Morphologically fungi exist as both microscopic and macroscopic classes. As a result of its existence at the microscopic level, only a few numbers of fungi are recognized and depicted till now. The appellation "Endophytes" has been used in an extensive perception as per its description to comprise bacteria (Kobayashi & Palumbo 2000), fungi (Petrini 1991, Stone et al. 2000), actinomycetes (Bills et al. 2004, Stone et al. 2000, Tan et al. 2006), algae (Peters 1991) and some insects (Feller 1995) residing within the plant tissues deprived of any ostensible symptoms of the disease. Nearly all type of vascular plants are well-known to harbor endophytic microorganisms (Arnold et al. 2000, Sturz 2000). The ecological condition of the host also influences the endophytic scattering in a population (Hata et al. 1998). Endophytic fungi can enter the plant through its root and colonize different plant parts including flowers and fruits as well, but through stomata or wounds in the plant many phyllosphere endophytes can also entre as those wounds or stomata acts as passage to such microorganisms (McCully 2001, Danhorn & Fuqua 2007, Schulz & Boyle 2006).
Nepenthes is the tropical carnivorous plant, has been known to have evolutionarily modified leaves as the peripheral digestive organs via leaf epiascidiation process (folding of the leaf towards the interior side with the ventral or upper surface flattering the inside of the pitcher) in order to receive nutrients from prey trapped inside the pitcher cups. For the digestion of the prey, Nepenthes reduces the pH of its fluid to facilitate the enzymatic reactions and to regulate the bacterial population present in the micro-habitat (Chan et al. 2016).
The present study aimed to isolate endophytic fungi from leaves, stems, roots and pitcher cups of an endemic plant of Meghalaya, N. khasiana. It is a carnivorous IUCN listed - endangered plant. N. khasiana is the only representative of the family Nepenthaceae from India and is considered to harbor vast range of medicinal properties used in traditional medicine by local people. Thus, the aspect of assessing the diversity of endophytic fungi will help to understand the association of fungal endophytes with the host plant.
-
For the isolation of endophytic fungi, N. khasiana was selected as the host plant. The plants were randomly collected for a period of 1 year (2015-2016) at monthly intervals from the West Jaintia Hills district of Meghalaya. The plants were collected aseptically in sterilized polythene bags which were taken to the laboratory and processed within 24 hours of collection.
Surface sterilization
-
The samples were surface sterilized following the slightly modified protocol of Bayman et al. 1997. The plant parts were vigorously washed under running tap water to remove dust and soil particles. The protocol given by Suryanarayanan et al. 1998 (modified) was used for the isolation of endophytic fungi from the sterilized plant samples. Different plant parts (leaves, stems, roots and pitcher cups) of N. khasiana were chopped into small fragments of 0.5 cm diameters. These segments were then surface sterilized by immersing them in 70% ethanol for 1-3 minute, followed by treatment in 2% (v/v) sodium hypochlorite (NaOCl) for 3 minutes, then again immersing them in 70% ethanol for 30 seconds and then lastly rinsed with sterile distilled water to remove any traces of surface sterilants. The excess moisture adhering to the treated segments were eliminated by blotting with sterilized Whatman No.1 filter paper.
Efficiency of surface sterilization
-
The effectiveness of surface sterilization was performed by washing the surface sterilized samples with sterile water few times followed by transferring the samples in 5 ml of sterile water. The sterilized sample-water mixtures thus obtained were then stirred for 1 min. An aliquot of 0.5 ml water obtained above (after removing the plant parts) was then inoculated on Potato Dextrose Agar (PDA) medium and incubated at 27℃ for 7 days. Plates which show negative result considered as successfully surface disinfected and the procedure thus used for the isolation of endophytic fungi (Schulz et al. 1993).
Isolation of endophytic fungi
-
For the isolation of endophytic fungi, samples were dried under laminar airflow to avoid contamination. Prepared samples were inoculated on to a Petri dish containing PDA (Potato Dextrose Agar) medium amended with Streptomycin (200mg/l) to suppress any bacterial growth. The Petri dishes were sealed using ParafilmTM and incubated at 25 ± 2℃ in an incubator for 7 to 15 days. Further to the development of colonies on culture plates were studied by isolating them and again sub-cultured as pure culture on PDA and Czapeck Dox Agar (CDA) media. For characterization of the morphology of fungal isolates, slides were prepared and stained with lacto-phenol blue reagent and examined under a bright field microscope. Identification was done by referring standard monographs (Ellis 1976, Domsch et al. 1980, Barnett & Hunter 1972, Subramanian 1971) and with the help of their morphological characteristics such as growth pattern, hyphal characters, colour of colonies on the medium, surface texture, margin character, aerial mycelium, mechanism of spore production and characteristics of the spores. The isolated fungal endophytes have been deposited to the Microbial Repository Centre (MRC), IBSD, Manipur, India. Culture samples that did not produce any spores were categorized separately and given a code based on colour of mycelium and morphological feature of colony. Molecular characterization (ITS sequence of rDNA) was done to confirm the identification of such sterile fungal strains and the nucleotide sequences obtained in this study were deposited to the NCBI- GenBank database with following accession numbers - MH458928.1, MH458932.1, MH510306.1, MH458934.1 and MH458933.1.Confirmation of the presence of endophytic fungal spores and elongation of new hyphae in different tissue parts of the host plant was performed with the help of Scanning Electron Microscopy (SEM) (Fig. 2).
Data analysis
-
With the intention to estimate the diversity of endophytic fungi, the colonization frequency (%CF) of endophytic fungi was calculated and determined by using the formula given by Hata & Futai (1995). Dominant endophytes were calculated as percentage of colony frequency of an endophyte divided by the sum of the percentage of colony frequency of all endophytes x100 (Tayung & Jha 2006).
$ {\rm{CF}}\left(\% \right) = \left({{{\rm{N}}_{{\rm{Col}}}}/{N_t}} \right) \times 100 $ Where, NCol = Number of segments of plant tissue colonized by each fungus and Nt = Total number of segments of plant tissue studied.
The fungal diversity of endophytic population was estimated with the following diversity indices. The reason for using these diversity indices was to take advantage of the strength of each index and to predict the complete structure of the population. All the statistical analyses were achieved with the software package PAST3 and MS Excel (Hammer et al. 2001) following diversity indices were calculated: (a) Simpson's Index. (b) Simpson's index of diversity. (c) Specie richness. (d) Shannon-Weaver diversity index. (e) Evenness was calculated.
(a) Simpson's index (D) was calculated by following formula (Simpson 1951): D =∑ (n/N) 2 Where, n = Total number of isolates of a particular species and N = Total number of isolates of all species.
(b) Simpson's index of diversity = 1-D (D is Simpson's index)
Species richness is a measure of the number of species found in a sample. This particular measure of species richness is known the Menhinick's index: (c) Species richness = S/√N Where, S = Total number of species.
Index of general diversity (H') or Shannon & Weaver (1949) diversity (Shannon & Weaver 1949): (d) Shannon Index (H') = -∑ pi ln pi
Where, pi = n/N, n is the total number of isolates of a particular species, N = Total number of isolates of all species and ln = Natural Log.
Pielou's evenness J' (Pielou 1995), which is expressed by the Shannon information scaled by the maximum information, to measure species evenness for each community: (e) J' = H'/ ln (S) Where, H'= Observed value of Shannon index and S = Total number of species observed.
Sample collection
-
During the study period, a total of 576 plant segments (leaves, stems, roots and pitcher cups) VCwere plated for the isolation of endophytic fungi. 4 segments per plates were inoculated on PDA medium plates with three replicate each (Fig. 1) and to maintain pure culture, Czapeck dox agar (CDA) medium plates have been used. Highest numbers of fungal endophytes were isolated from the leaf of N. khasiana followed by pitcher cup, stem and root for the study period. SEM images also confirm the presence of fungal endophytes within the different plant tissues of the host plant N. khasiana (Fig. 2). The fungal isolates were mainly composed of Ascomycota (25 genera; 35 species), followed by Zygomycota (3 genera; 6 species) and Oomycota (3 genera; 5 species).
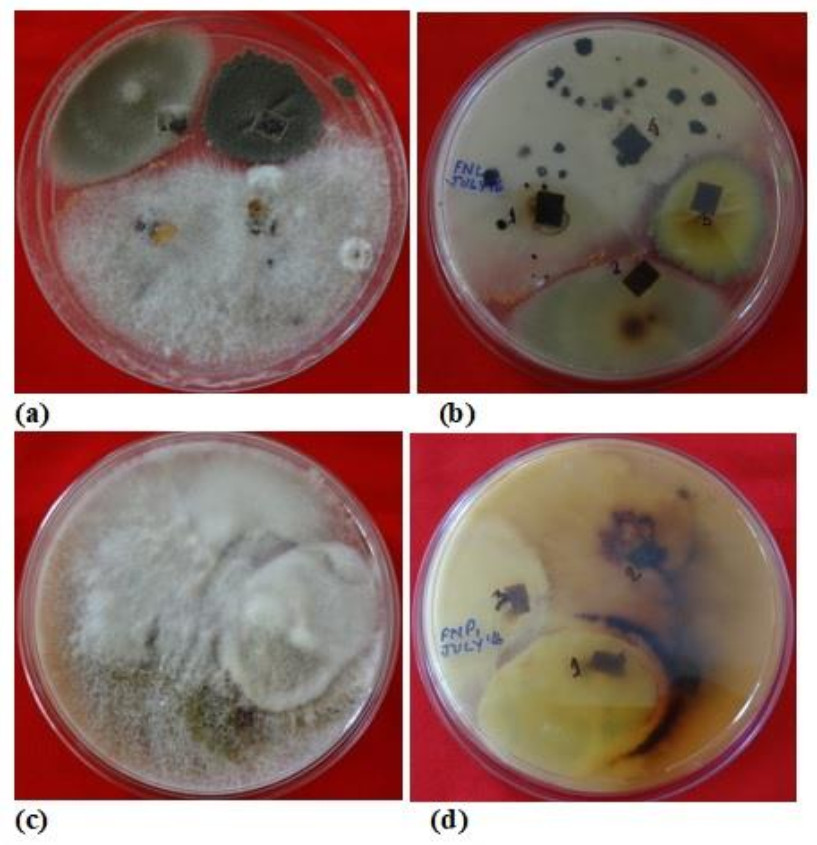
Figure 1. Plates showing mix cultures of fungal endophytes isolated from different plant parts of N. khasiana.
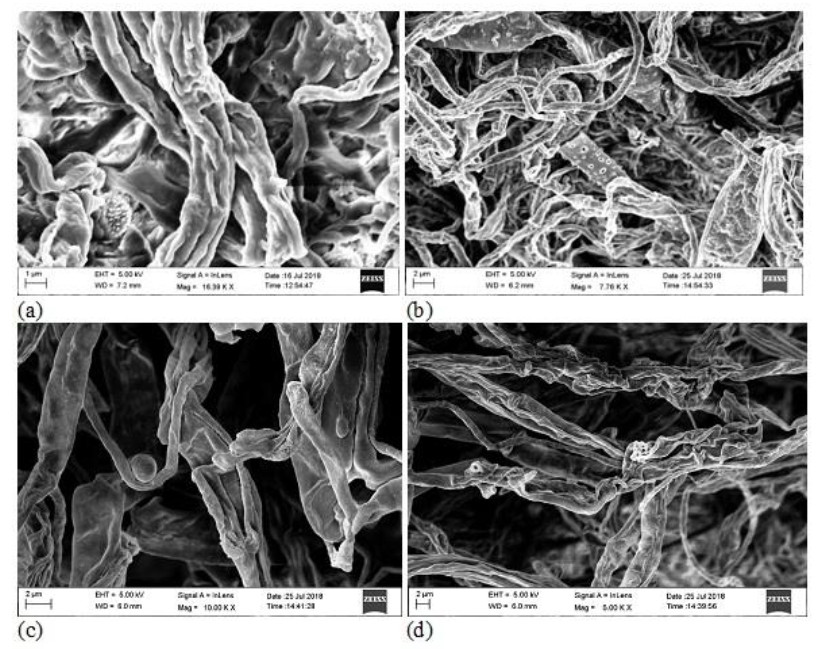
Figure 2. SEM micrograph showing the presence of endophytic fungal spore in (a) leaf and (b) stem; the elongation of new hyphae from mycelia of fungal endophytes in (c) root and (d) pitcher cup.
A total of 39 endophytic fungi were isolated and identifies from the different plant parts of N. khasiana during the one year of study period. Table 1 depicts the list of endophytic fungi isolated from N. khasiana. Among these isolates, it was observed that the phylum Ascomycota dominated the endophytic assemblage within the host plant. In our study we detected that 61.53% of isolates belonged to Ascomycota followed by Oomycota, Zygomycota and mycelia sterilia with 12.82 % each. The fungal assemblage was found to be dominated by the class Sordariomycetes (25.64%) followed by Eurotiomycetes (20.51%), Dothideomycetes (10.25%), and Incetraesedis and Mycelia sterilia (12.82% respectively), Mucoromycetes (7.69%), Mortierellomycetes (5.12%), Pezizomycetes and Leotiomycetes (2.56% each). Endophytic fungi with maximum average percentage of colonization frequency (%CF) were considered to be dominant. During the sampling period, Juxtiphoma eupyrena and Talaromyces ruber exhibited highest dominance in the leaf with 15% each followed by Globisporangium irregulare with 8.33%. The endophytic assemblage in the stem was dominated by Mycelia sterilia white (16.39%), J. eupyrena (13.66%) and T. ruber with 12.29%. The dominant root endophytes were MS brown (22.78%) and T. ruber (10%). Whereas, in pitcher cup it was dominant by MS white (17.5%) followed by Colletotrichum gloeosporioides and Gliomastix cerealis with 10% each.
Table 1. List of fungal endophytes isolated from different plant parts (leaf, stem, root and pitcher cup) of N. khasiana during the study period of 2015-2016.
Sl. Endophytic fungal isolates L S R PC No. Phylum: Ascomycota- 17 Genera, 24 Species Class- Sordariomycetes Order- Hypocereales 1 Acremonium murorum (Corda) W. Gams 1971 + - - + 2 Cosmospora butyri (J.F.H. Beyma) Gräfenhan, Seifert & Schroers 2011 + - - - 3 Fusarium proliferatum (Matsush.) Nirenberg ex Gerlach & Nirenberg 1982 + - - - 4 F. redolens Wollenw. 1913 + + - - 5 Gliomastix cerealis (P. Karst.) C.H. Dickinson 1968 + + + + 6 Rectifusarium ventricosum (Appel & Wollenw.) L. Lombard & Crous 2015 - + + - Order- Sordariales 1 Humicola fuscoatra Traaen 1914 - + + - 2 Trichocladium griseum (Traaen) X. Wei Wang & Houbraken 2018 - + + + Order- Incertaesedis 1 Arthrinium arundinis (Corda) Dyko & B. Sutton 1979 + - - + Order- Glomerellales 1 Colletotrichum gloeosporioides (Penz.) Penz. & Sacc. 1884 + - - + Class- Dothiodeomycetes Order- Pleosporales 1 Alternaria alternate (Fr.) Keissl. 1912 - + + - 2 Juxtiphoma eupyrena (Sacc.) Valenz.-Lopez, Crous, Stchigel, Guarro & + + + + Cano 2017 Order- Capnodiales 1 Cladosporium cladosporioides (Fresen.) G.A. de Vries 1952 + + + + Order- Tubeufiales 1 Tubeufia cerea (Berk. & M.A. Curtis) Hohn. 1919 + - + - Class- Eurotiomycetes Order- Eurotiales 1 Penicillium brevicompactum Dierckx 1901 + + + + 2 P. glabrum (Wehmer) Westling 1911 + - + + 3 P. simplicissimum (Oudem.) Thom 1930 + + + - 4 P. jensenii K.W. Zaleski 1927 - - - + 5 Talaromyces islandicus (Sopp) Samson, N. Yilmaz, Frisvad & Seifert 2011 - - - + 6 T. purpureogenus Samson, N. Yilmaz, Houbraken, Spierenb., Seifert, + + + + Peterson, Varga & Frisvad 2011 7 T. ruber (Stoll) N. Yilmaz, Houbraken, Frisvad & Samson 2012 + + + + 8 T. rugulosus (Thom) Samson, N. Yilmaz, Frisvad & Seifert 2011 + + + + Class- Pezizomycetes Order- Pezizales 1 Phymatotrichopsis omnivore (Duggar) Hennebert 1973 - + - - Class- Leotiomycetes Order- Incertaesides 1 Scytalidium lignicola Pesante 1957 - - + - Phylum: Oomycota- 3 Genera, 5 Species Class- Incertaesides Order- Peronosporales 1 Globisporangium intermedium (de Bary) Uzuhashi, Tojo & Kakish. 2010 - - + - 2 G. irregular (Buisman) Uzuhashi, Tojo & Kakish. 2010 + + + + 3 Phytophthora cactorum (Lebert & Cohn) J. Schrot. 1889 - - - + 4 P. cinnamomi Rands 1922 + + - + 5 Pythium aphanidermatum (Edson) Fitzp. 1923 - - + - Phylum: Zygomycota- 3 Genera, 5 Species Class- Mortierellomycetes Order- Mortierellales 1 Mortierella sp. 1 + - - - 2 Mortierella sp. 2 - - - + Class- Mucoromycetes Class- Mucorales 1 Gongronella butleri (Lendn.) Peyronel & Dal Vesco 1955 + - - - 2 Rhizopus microspores Tiegh. 1875 - - + - 3 Rhizopus sp. - + - - Mycelia sterilia (MS)- 5 isolates 1 MS (Black) - + - - 2 MS(Brown) - + + + 3 MS (Red) - + - - 4 MS (White) + + + + 5 MS (Yellow) + - + - Total- 23 Genera, 34 Species and 5 Mycelia sterilia L = leaf, S = stem, R = root, PC = pitcher cup, '+' = present and '-' = absent We found that Alternaria alternata, Fusarium proliferatum, Gongronella butleri and Mortierella sp.1 were isolated only from the leafs, Periconia macrospinosa, Phymatotrichopsis omnivore, Mycelia sterilia (black) and Mycelia sterilia (Red) were specific only to the stem, in root Globisporangium intermedium, Pythium aphanidermatum, Rhizopus microspores, Rhizopus sp. and Scytalidium lignicola were restricted to the root tissues and Cosmospora butyri, Mortierella gamsii, Mortierella sp. 2, Penicillium jensenii and Phytophthora cactorum were isolated only from the pitcher cups. However, Cladosporium cladosporioides, Gliomastix cerealis, Juxtiphoma eupyrena, Penicillium brevicompactum, Talaromyces purpureogenus, T. ruber, T. rugulosus, MS (brown) and Mycelia sterilia (white) were found to be present in all the plant parts of the sampling plant.
In which, Globisporangium irregulare (83.33%), Penicillium glabrum (66.66%) and MS white (58.33%) showed highest percentage of colonization frequency in leaf (Fig. 3). It was found that endophyte J. eupyrena (83.33%) and Fusarium redolens (33.33%) showed maximum %CF in stem (Fig. 4), Talaromyces ruber (66.66%), T. purpureogenus and MS (white) with %CF 58.33% was maximum in root (Fig. 5). However, C. gloeosporioides and G. cerealis showed %CF 66.66 in pitcher cup (Fig. 6), where the highest %CF was recorded by MS (white) with 116.66%.
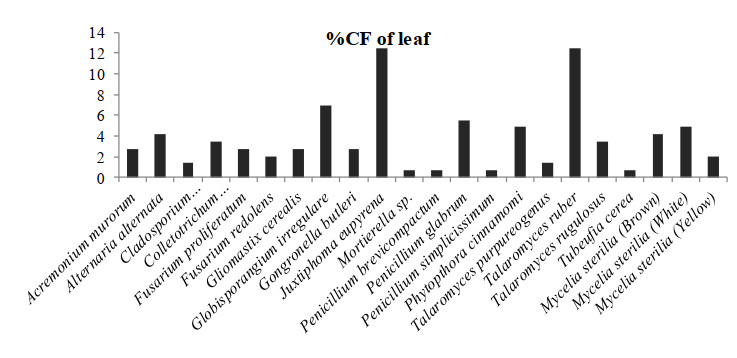
Figure 3. Percentage colonization frequency of endophytic fungi isolated from the leafs of N. khasiana during the study period of 2015-2016
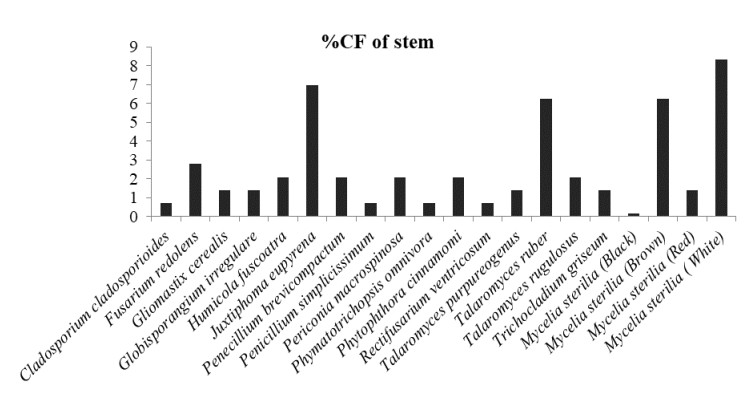
Figure 4. Percentage colonization frequency of endophytic fungi isolated from the stem of N. khasiana during the study period of 2015-2016
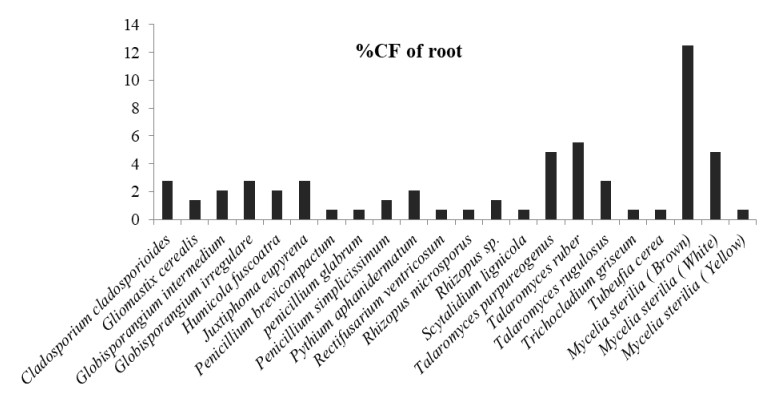
Figure 5. Percentage colonization frequency of endophytic fungi isolated from the root of N. khasiana during the study period of 2015-2016
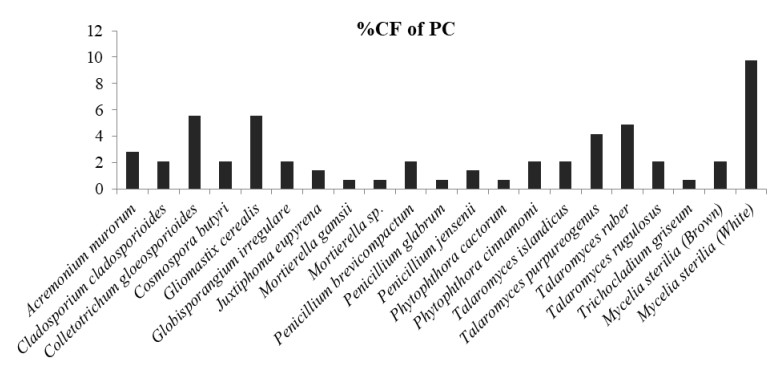
Figure 6. Percentage colonization frequency of endophytic fungi isolated from the pitcher cup (PC) of N. khasiana during the study period of 2015-2016.
Shannon index (H') was found to be highest in the leaf with the value of 2.78 followed by pitcher cup with 2.76 whereas it was recorded to be the least in root with value of 2.68, which indicates the vise-versa result for Simpson's diversity index (1-D). Simpson's index of dominance (D) was found to be highest in root with value of 0.09 and d index was least in leaf (0.07). Species richness (S/√N) was recorded to be highest in leaf with the value of 2.34 and minimum in leaf with value of 1.64. As like other indices, Species evenness (E) was also recorded and it was found to be highest in leaf with value of 0.96 whereas, it was recorded to be least in root with value of 0.89 (Table 2).
Table 2. Diversity indices of fungal endophytes isolated from N. khasiana during the study period of 2015-2016.
Plant tissue Shannon index (H') Simpson's index of Dominance (D) Simpson, s diversity index (1-D) Species richness (S/√N) Species evenness (E) Leaf 2.78 0.07 0.92 1.64 0.96 Stem 2.68 0.08 0.91 2.07 0.92 Root 2.68 0.09 0.90 2.26 0.89 Pitcher cup 2.76 0.07 0.90 2.34 0.90
-
In the present investigation, the endophytic fungi isolated from N. khasiana were mainly composed of Ascomycota (25 genera; 35 species), followed by Zygomycota (3 genera; 6 species) and Oomycota (3 genera; 5 species). However, high percentage of ascomycota as endophytic fungi were also reported by the work of Goveas et al. (2011) from Coscinium fenestratum- a red list endangered medicinal plant, it could be due to the ability of Ascomycota to produces ascospores which helps them to strive against other microorganisms through the harsh environmental circumstances. A total of 5 mycelia sterilia were also isolated. The group of mycelia sterilia consists of several morphological fungal varieties, but then again not forming spores under laboratory conditions. This group of fungi is significantly predominant in endophytic studies (Lacap et al. 2003). Other studies on endophytic fungi are also reported the presence of sterile forms in their survey (Suryanarayanan et al. 1998, Rajagopal 1999, Maheswari 2011). Several phylloplane fungi such as Alternaria, Aureobasidium, Cladosporium, Epicoccum belonged to Hyphomycetes were isolated as endophytes from a wide range of plant species growing in different habitats (Bills 1996, Peláez et al. 1996). These phylloplane fungi are proficient of penetrating the outer layers of the leaf or may be confined in the substomatal cavities (Cabral et al. 1993). In our study we also have witnessed similar finding where some of the phylloplane fungi viz. Alternaria, Cladosporium etc. were isolated as fungal endophytes.
In the case of the present investigated plant the endophytic fungi isolated were Gliomastic cerealis, Juxtiphoma eupyrena, Talaromyces purpureogenus, MS (Brown) and MS (white) showed highest colonization ferequency (%CF) as compared to other isolated of the plant. And among all, J. eupyrena was observed as commonly occurring endophytes in all the plant parts of N. khasiana. Similar occurrence of J. eupyrena was observed in the tissues of Artemisia thuscula by Cosoveanu et al. (2018). It was observed that in selected plant for present investigation, the Shannon diversity index (H') was higher in the aerial part of the plant i.e. in leaf followed by the underground part (root) and least was observed in stem of the plant, this is the opposite of the previous finding by Jin et al. (2013). However, Simpson's index of dominance (D) showed the vise-versa result. Species richness was also reported to be higher in leaf and lower in roots during the two year of the study period. A smilar finding was reported by various researchers viz. Kumar & Hyde (2004), Raviraja (2005), Huang et al. (2008), Sun et al. (2008), Xing et al. (2010), Chaeprasert et al. (2010), Thalavaipandian et al. (2011), Siqueira et al. (2011) and Suwannarach et al. (2011). This could be due to the fact that N. khasiana is a carnivorous plant that grows in the soil where the nutritional value and pH of the soil is low and therefore, they adjust their requirements through the captured and digested prey. Moreover, the surface area of leaf is supplementary as compared to the roots of pitcher plants. The time of leaf exposure may also have accounted for the better density of endophytic fungi (Frohlich et al. 2000, Toofanee & Dulymamode 2002, Arnold & Herre 2003). Huang et al. (2008) also pointed out the tissue specific trait of endophytic fungi although most of the endophytes only exhibited tissue preference this partially supports our finding where a higher number of endophytes has been isolated from a specific tissue as compared to other tissues of the host plant.
-
From the present investigation, it can be concluded that the carnivorous plant Nepenthes khasiana, which is one of the endemic plants of the north-eastern state of Meghalaya, India, harbor a great diversity of endophytic fungi. The colonization frequency of the isolates from different tissues of the host plant significantly differed between leaf, stem, root and pitcher cup. Hence, this entire study can be considered as an earnest attempt in exploring the diversity of fungal endophyte and their association with the pitcher plant, and therefore, the further study to explore the facts that how these fungal endophytes are helping the plant in the digestion of prey, synthesis of several secondary metabolites, endurance towards the harsh environment, etc. is much needed. Additionally, isolated endophytic fungal strains may also be evaluated for their antagonistic activity against plant pathogen of the host plant.
-
The authors appreciatively acknowledge the Head of the Department, Centre for Advanced Studies in Botany, North- Eastern Hill University, Shillong for providing necessary laboratory facilities and DBT- infrastructure and CAS for providing necessary instrument facilities. The corresponding author is thankful to University Grants Commission for providing financial assistance in the form of Maulana Azad National Fellowship. Authors would like to appreciate the Head and technical staff of Institute of Advanced Study in Science and Technology (IASST), Guwahati for their help in taking SEM images.
-
The authors declare no conflict of interest.
- Copyright: © 2021 by the author(s). This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
| F Naseem, H Kayang. 2021. Endophytic fungal diversity of endemic carnivorous plant Nepenthes khasiana in Meghalaya, India. Studies in Fungi 6(1):138−150 doi: 10.5943/sif/6/1/7 |












