-
The WRKY family is one of the largest transcription factor families in plants, and is an important part of the signal network that modulate many plants processes[1]. WRKY family transcription factors are not only involved in the regulation of growth and development, but also in the regulation of plant response to biotic and abiotic stresses.
WRKY transcription factors contain a highly conserved WRKYGQK amino acid sequence and zinc finger motif (Cys(2)-His(2) or Cys(2)-His-Cys), and bind to the W box cis-acting element (TTGAC(C/T)) in the promoter of their target genes[2]. Some WRKY transcription factors can also bind to other cis-acting elements. For example, SUSIBA2 (HvWRKY46) binds to both W-box element and SURE element[3], and OsWRKY13 binds to both W-box element and PRE4 element[4]. The WRKY domain and zinc finger motif of WRKY transcription factor play an indispensable role in the protein-DNA recognition processes[5,6]. The WRKY domain contains a conserved heptapeptide (WRKYGQK), whose amino acid substitutions could affect the binding properties of WRKY domain to W-box[7]. WRKYGEK and WRKYGKK are the most common variants[8]. In a few WRKY proteins, WRKY motif is also replaced by WIKY, WRMC, WSKY, WKRK, WVKY or WKKY[9]. The W-box core sequence is necessary for WRKY transcription factor proteins binding, and its adjacent sequence also affect the binding preference of WRKY protein[5].
The WRKY proteins are generally divided into three groups according to the number of WRKY domains and the type of zinc finger motifs[2]. Group I contains two WRKY domains, and Group II and III contain one WRKY domain. Group II and Group III are distinguished according to their C-terminal zinc finger motifs, as Group II includes a C2H2 zinc finger motif, while Group III includes a C2HC zinc finger motif. Group II members can be further divided into five subgroups (IIa, IIb, IIc, IId and IIe) based on their amino acid sequences. Besides the above three groups, some atypical proteins with WRKY domain and incomplete zinc finger motif are classified into Group IV[10].
Grapes are one of the most economically important fruit crops in the world. Sustainable production of high-quality fruits in an ever-changing environment is the major goal of the grape industry. How to cope with biotic and abiotic stresses is the main focus of the grape biotechnology research community. WRKY transcription factors are important transcriptional regulators involved in plant growth, development and stress response. Their expression is affected by environment and growth status, which helps to find clues to their functions by transcription detection. At present, the functions of nearly 20 members of the grape WRKY transcription factor family have been identified. Here we attempt to summarize the research progress on biological functions, regulating networks and potential applications of the grape WRKY family transcription factors.
-
Grape WRKY family members have been identified and analyzed based on whole genome sequencing[11−15]. In two independent studies, a total of 59 putative WRKY family members were identified from the 12× V. vinifera cv. Pinot Noir (PN40024) genome sequences[11,12]. In another study, Wang et al.[13] also identified a total of 59 putative WRKY family members from three public databases (12× PN40024 grape genome sequence, NCBI GenBank database, and 8× coverage of the grape genome), but there are differences from the former two studies in the functional annotation of two candidate genes (VIT_02s0154g00210 and XM_002277347.4). Zhang & Feng[14] obtained 80 putative WRKY family genes from the NCBI GenBank database, which may have resulted from the repeated calculation of genes in a heterozygous locus. Romero et al.[15] identified 61 putative WRKY family proteins based on the latest V. vinifera reference genome (PN40024 12×.v2, vcost. v3 annotation). It is noticeable that two putative WRKY family members, VIT_02s0154g00210 and XP_002277383.1, varied among these studies. VIT_02s0154g00210 only has a partial zinc finger motif (CX4C) at the C-terminus, which is similar to OsWRKY33 and OsWRKY38 in rice[10]. XM_002277347.4, which encodes the XP_002277383.1 protein in the NCBI GenBank database, is located on chromosome 1 (2348150 to 2350401), and is annotated as unknown protein (GSVIVT01020752001) due to different predictions of its translation starting site in the PN40024 grape genome (12× PN40024, v1 annotation). After removing the redundancy, a total of 61 putative WRKY family members were identified from the PN40024 grape genome (PN40024 12×.v2, vcost. v3 annotation) and NCBI database (Vitis vinifera Annotation Release 102) (Table 1).
Table 1. Putative grape WRKY genes identified in the grape genome.
Gene name Gene number Location Gene number Former names VvWRKY1 GSVIVT01012196001 chr1:628,681..633,595 VIT_01s0011g00720 VvWRKY2 GSVIVT01020060001 chr1:10,977,206..10,982,627 VIT_01s0026g01730 VvWRKY3 GSVIVT01010525001 chr1:21,460,152..21,461,423 VIT_01s0010g03930 VlWRKY3[41], VvWRKY8[51], VvWRKY3[49] VvWRKY4 GSVIVT01019419001 chr2:512,179..513,535 VIT_02s0025g00420 VvWRKY5 GSVIVT01019511001 chr2:1,228,115..1,229,951 VIT_02s0025g01280 VvWRKY6 GSVIVT01035426001 chr4:1,210,142..1,211,252 VIT_04s0008g01470 VvWRKY7 GSVIVT01035884001 chr4:5,247,432..5,248,815 VIT_04s0008g05750 VvWRKY8 GSVIVT01035885001 chr4:5,265,887..5,267,902 VIT_04s0008g05760 VpWRKY3[59] VvWRKY9 GSVIVT01035965001 chr4:6,569,931..6,576,445 VIT_04s0008g06600 VvWRKY10 GSVIVT01033188001 chr4:9,363,044..9,365,026 VIT_04s0069g00920 VvWRKY11[17] VvWRKY11 GSVIVT01033194001 chr4:9,399,822..9,400,803 VIT_04s0069g00970 VvWRKY12 GSVIVT01033195001 chr4:9,409,805..9,411,286 VIT_04s0069g00980 VvWRKY13 GSVIVT01019109001 chr4:16,664,476..16,666,517 VIT_04s0023g00470 VvWRKY14 GSVIVT01034968001 chr5:530,097..531,696 VIT_05s0077g00730 VvWRKY15 GSVIVT01025491001 chr6:364,396..365,481 VIT_06s0004g00230 VvWRKY16 GSVIVT01024624001 chr6:8,290,089..8,292,826 VIT_06s0004g07500 VaWRKY33[57] VvWRKY17 GSVIVT01000752001 chr7:381,054..383,675 VIT_07s0141g00680 VvWRKY18 GSVIVT01028129001 chr7:4,044,185..4,045,807 VIT_07s0005g01520 VvWRKY19 GSVIVT01028147001 chr7:4,200,461..4,202,183 VIT_07s0005g01710 VvWRKY20 GSVIVT01028244001 chr7:4,899,940..4,903,113 VIT_07s0005g02570 VvWRKY21 GSVIVT01022067001 chr7:16,322,554..16,324,062 VIT_07s0031g00080 VvWRKY22 GSVIVT01022245001 chr7:17,794,621..17,797,164 VIT_07s0031g01710 VvWRKY23 GSVIVT01022259001 chr7:17,958,339..17,960,800 VIT_07s0031g01840 VvWRKY13[28,33,52,53] VvWRKY24 GSVIVT01030258001 chr8:9,796,097..9,798,804 VIT_08s0058g00690 VvWRKY33[29], VvWRKY24[49] VvWRKY25 GSVIVT01030174001 chr8:10,843,800..10,846,082 VIT_08s0058g01390 VpWRKY1[16] VvWRKY26 GSVIVT01025562001 chr8:14,033,449..14,039,296 VIT_08s0040g03070 VvWRKY26[34] VvWRKY27 GSVIVT01034148001 chr8:14,828,036..14,830,056 VIT_08s0007g00570 VvWRKY28 GSVIVT01015952001 chr9:16,094,245..16,096,198 VIT_09s0018g00240 VaWRKY14[55], VvWRKY40[30] VvWRKY29 GSVIVT01012682001 chr10:618,665..621,093 VIT_10s0116g01200 VaWRKY12[20], VpWRKY31[45] VvWRKY30 GSVIVT01021252001 chr10:3,008,699..3,010,258 VIT_10s0003g01600 VvWRKY31 GSVIVT01021397001 chr10:4,894,494..4,896,041 VIT_10s0003g02810 VvWRKY32 GSVIVT01021765001 chr10:10,756,001..10,759,241 VIT_10s0003g05740 VvWRKY33 GSVIVT01023600001 chr11:7,838,041..7,845,641 VIT_11s0037g00150 VpWRKY2[16] VvWRKY34 GSVIVT01029265001 chr11:17,821,900..17,823,218 VIT_11s0052g00450 VvWRKY35 GSVIVT01020864001 chr12:878,978..881,517 VIT_12s0028g00270 VvWRKY36 chr12:2,348,148..2,350,455 XP_002277383.1 VvWRKY37 GSVIVT01030453001 chr12:5,678,804..5,681,082 VIT_12s0059g00880 VvWRKY38 GSVIVT01030046001 chr12:9,116,720..9,122,740 VIT_12s0057g00550 VvWRKY39 GSVIVT01029688001 chr12:13,065,135..13,067,116 VIT_12s0055g00340 VvWRKY40 GSVIVT01032662001 chr13:1,716,836..1,718,877 VIT_13s0067g03130 VvWRKY41 GSVIVT01032661001 chr13:1,719,393..1,720,803 VIT_13s0067g03140 VvWRKY42 GSVIVT01036223001 chr14:8,753,748..8,755,749 VIT_14s0081g00560 VvWRKY43 GSVIVT01033063001 chr14:25,479,103..25,481,917 VIT_14s0068g01770
VvWRKY43[49]VvWRKY44 GSVIVT01011356001 chr14:28,924,779..28,926,499 VIT_14s0108g00120 VvWRKY45 GSVIVT01011472001 chr14:29,916,401..29,920,389 VIT_14s0108g01280 VvWRKY46 GSVIVT01018300001 chr15:11,499,221..11,503,179 VIT_15s0021g01310 VvWRKY47 GSVIVT01027069001
chr15:18,191,021..18,193,470VIT_15s0046g01140 MrWRKY30[31], VlWRKY48[42], VvWRKY46[19,44] VvWRKY48 GSVIVT01026969001 chr15:18,940,954..18,942,146 VIT_15s0046g02150 VvWRKY49 GSVIVT01026965001 chr15:18,957,235..18,958,812 VIT_15s0046g02190
VvWRKY22[26]VvWRKY50 GSVIVT01028823001 chr16:18,360,079..18,360,711 VIT_16s0050g01480 VvWRKY51 GSVIVT01028718001
chr16:19,477,129..19,479,547VIT_16s0050g02510 VqWRKY52[43], VvWRKY52[44], VvWRKY30[21], VdWRKY53[56] VvWRKY52 GSVIVT01008553001 chr17:922,644..925,087 VIT_17s0000g01280 VvWRKY1[18,40] VvWRKY53 GSVIVT01008046001 chr17:6,316,168..6,320,034 VIT_17s0000g05810 VvWRKY53[49] VvWRKY54 GSVIVT01009441001 chr18:8,392,053..8,393,721 VIT_18s0001g10030 VvWRKY55 GSVIVT01037686001 chr19:6,882,399..6,884,987 VIT_19s0090g00840 VvWRKY56 GSVIVT01037775001 chr19:7,760,183..7,767,013 VIT_19s0090g01720 VvWRKY57 GSVIVT01014854001 chr19:10,665,189..10,669,055 VIT_19s0015g01870 VvWRKY58 GSVIVT01001332001 chr1_random:297,588..311,938 VIT_01s0011g00220
VvWRKY2[37,54,60]VvWRKY59 GSVIVT01007006001 chrUn:29,694,084..29,699,952 VIT_00s0463g00010 VvWRKY60 GSVIVT01001286001 chr2:4,989,461..4,989,778 VIT_02s0154g00210 VvWRKY61 GSVIVT00037762001 chr16:18360079..18360711 VIT_16s0050g01480 -
The grape WRKY family gene members are divided into different groups[11-14]. Group I and Group III have obvious structural features, and the number of their members are 12 and six, respectively. Two members with incomplete zinc finger structure belong to Group IV. The remaining 41 members are assigned to Group II and subdivided into five subgroups (IIa, IIb, IIc, IId, and IIe) using the phylogenetic tree based on their amino acid sequences. Among them, five subgroups of Group II are slight differences among different studies.
In the grape WRKY family proteins, 56 members contain the conserved WRKYGQK sequence, and the other five members have variants in the heptapeptides, of which four members are replaced by WRKYGKK, and one member is WKKYGQK. Several grape WRKY transcription factors containing conserved WRKYGQK sequence can bind to the W-box element in the promoter of target genes[15−21]. It is worth noting that four out of five heptapeptide variants belonged to the subgroup IIc. Amino acid substitution of conserved hexapeptides may affects the binding activities of WRKY proteins to the W-box. For instance, tobacco NtWRKY12 containing WRKYGKK sequence could specifically bind to WK-box (TTTTCCAC) but not to W-box. When WRKYGKK of NtWRKY12 was replaced by WRKYGQK or WRKYGEK, the binding to WK box was abolished[7]. The five grape WRKY members that have various amino acid compositions in the heptapeptide might recognize and bind to other cis-acting elements in the promoters of target genes.
Since almost all grape WRKY transcription factors possess W-box elements on their own promoters, they could bind to themselves or to the promoters of other WRKY transcription factors to regulate self-expression or other WRKY transcription factors.
-
In higher plants, the WRKY transcription factor family usually contains a dozen to a hundred members. A uniform nomenclature with consecutive numbering may help avoid confusion and facilitate scientific communication. Different nomenclature systems have been used among gene family members. For example, the nomenclature of WRKY family members in Solanum lycopersicum[22], S. tuberosum[23], and Arabidopsis thaliana[2] are based on their phylogenetic similarity to AtWRKY, grouping on the evolutionary tree, and direct assignment, respectively. It will be much simpler if a common method, such as naming family member genes according to their chromosome order or group order[24,25] is used. However, the reality is probably too complicated to have a uniform naming system. Even in the most studied plant genomes of Arabidopsis and rice, naming of family member genes does not always follow a unique rationale, and in most cases, a unique number for each member was assigned based on the knowledge of the researchers[2,26,27].
Due to the differences in putative members and naming methods, a WRKY number used for a grape WRKY member in a publication does not uniquely represent a certain protein[11−15]. In previous studies[11−13], although the most common naming system for the grape WRKY family genes is based on their order in the V. vinifera genom, there are differences in predicted family members and the naming of members with uncertain positions, resulting in confusion in naming. For example, the grape WRKY family gene VIT_04s0008g01470 located on chromosome 4 has been named VvWRKY8[11], VvWRKY6[13], VvWRKY53[12], and VvWRKY50[14] in different studies. When researchers identify and study a new WRKY family gene for a gene function purpose, this gene is often labeled with a number based on its homologous gene number identified in model plants[17,20,28−31]. For example, the grape WRKY family gene VIT_15s0046g01140 , homologous to AtWRKY30, was named VvWRKY30 in a previous study[31].
Since multiple naming systems are used for grape WRKY family genes, the same gene may be represented by different VvWRKY numbers, and vice versa. For example, the grape WRKY family gene VIT_01s0011g00220 are named VvWRKY4[11], VvWRKY58[13], and VvWRKY3[12] in different studies. Therefore, a uniform naming system for grape WRKY family genes is urgently needed for the grape research community. With this in mind, we here recommend that each WRKY member is re-assigned with a unique number on a continuous basis. The future discoveries of new VvWRKY members will be subsequently assigned a new number. In order to avoid confusion caused by the new numbering system, we recommend the use of the numbering system based on the study of Wang et al.[13]. All WRKY family members in this article are numbered in Table 1. We encourage the use of the names in Table 1 in future studies to avoid the confusion which often arises when multiple names are assigned to a given WRKY gene family member.
-
The expression of WRKY transcription factors in various tissues and in response to different treatments had been extensively studied, in which the functions of about 20 grape WRKY family genes have been revealed. Due to the limitation of grape transgenic technology and growth cycle, most of the research on the functions of grape WRKY transcription factors were carried out in Arabidopsis or tobacco.
The expression of WRKY members in different environments provided important clues for studying their functions. Moreover, the functions of homologous proteins in model plants also shed light on the functional study of grape WRKY members.
-
Plant WRKY proteins have been reported to participate in several plant growth and development processes, such as seed development, dormancy and germination, leaf senescence and trichome development[32]. Expression of grape WRKY family genes at different developmental stages and in different tissues have been examined[11−13]. The functions of some grape WRKY members in grape growth and development have been identified.
VvWRKY13 regulates the synthesis of Et (ethylene) and ABA, affecting the growth and development of grapevines. VvWRKY13, cloned from a grape cv. ‘Zuoyouhong’, was found to be ubiquitously expressed in various grapevine tissues[28]. Ectopic expression of the VvWRKY13 gene in Arabidopsis could also promote the synthesis of Et and ABA, showing constitutive triple responses, delayed seed germination, smaller stomatal aperture size, and several other phenotypes[28,33].
Grape WRKYs also play important roles in regulating fruit development. VvWRKY26 might regulate grape fruit quality by participating in vacuolar transport and acidification, as well as flavonoid pathways[34]. VvWRKY26, which had the closest homology to petunia PhPH3 and Arabidopsis AtTTG2 (AtWRKY44), could fulfill the PH3 function in the regulation of vacuolar pH and restored the wild type pigmentation phenotype by up-regulating the expression level of the pH structural genes and the anthocyanin-related genes in the petunia ph3 mutant[34]. In the early stage of berry development, VvWRKY26 was highly expressed in the inner integument of the seed coat and might be involved in the regulation of the expression of many proanthocyanidin related genes[34]. It was supported that VvWRKY26 function as a proanthocyanidin regulator and might also affect seed development and plant fertility in grapevine as its Arabidopsis homolog AtTTG2[35].
VvWRKY22, cloned from V. vinifera ‘Cabernet Sauvignon’, was involved in the sugar metabolism of grape berries[36]. VvWRKY22 was co-expressed with 16 sugar-related genes in grape berries, and the expression of VvWRKY22 in grape suspension cells was inhibited by sucrose, but induced by fructose and ABA. Overexpression of VvWRKY22 in grape suspension cells reduced the content of sucrose, glucose, and fructose, while the silencing of VvWRKY22 gene in grape suspension cells increased the content of these sugars. The role of VvWRKY22 on sugar metabolism might be accomplished by regulating the expression of sugar and ABA-related genes, and the protein activity of VvWRKY22 might be directly regulated by VvSnRK1.1 (Sucrose non-fermenting-1-related protein kinase 1.1) and VvSnRK1.2.
VvWRKY2 played a role in lignin biosynthesis and xylem development[37]. VvWRKY2 was specifically and highly expressed in the lignified tissues of grapevine stems (sclerenchyma and xylem cells). Ectopic expression of VvWRKY2 in tobacco delayed xylem formation and changed lignin composition in stems and petioles by regulating the expression of genes involved in lignin biosynthesis pathway and cell wall formation[37]. VvWRKY2 activated the expression of VvCH4 involved in the lignin biosynthesis pathway.
-
The expression profiles of grape WRKY family genes in response to biotic and abiotic stresses, or plant hormones have been studied to a certain extent. The transcription of most WRKY protein members can be altered by at least one stress treatment, which suggests that they are widely involved in plant response to biotic or abiotic stress[11−15].
Plant defense responses to phytopathogen attack are regulated through a complex network of signaling pathways that involve SA (Salicylic acid), JA (Jasmonic acid), Et, and ABA[38,39]. JA and Et work synergistically in the inducing of defense-related genes against necrotrophs. SA works antagonistically to JA/Et, enhancing susceptibility to necrotrophic pathogens while promoting resistance to hemibiotrophs and biotrophs. ABA plays a positive or negative role in the plant defense response to necrotrophs, depending on the specific plant-pathogen interaction[39]. WRKY genes are often induced by these hormones and also involved in regulating downstream responses mediated by these hormones. Expression of some marker genes is commonly used to indicate the activation of these hormone signal transductions.
The functions of some grape WRKY members under biotic stress have been studied (listed in Table 2 and Fig. 1). However, most of their molecular mechanisms leading to the functions are still poorly understood. Besides VvWRKY40 and VlWRKY46, these grape WRKY members generally prompted resistance to biotrophs or hemibiotrophs (Fig. 1). VvWRKY1, VdWRKY53, and VvWRKY33 have been found to promote plant resistance to necrotrophs, and biotrophs or hemibiotrophs. While VlWRKY3 and VqWRKY52 can increase plant resistance to hemibiotrophs and semi-biotrophs, but increase susceptibility to necrotrophs. These results suggest that these WRKY transcription factors in grape manipulate various responses to pathogen attacks.
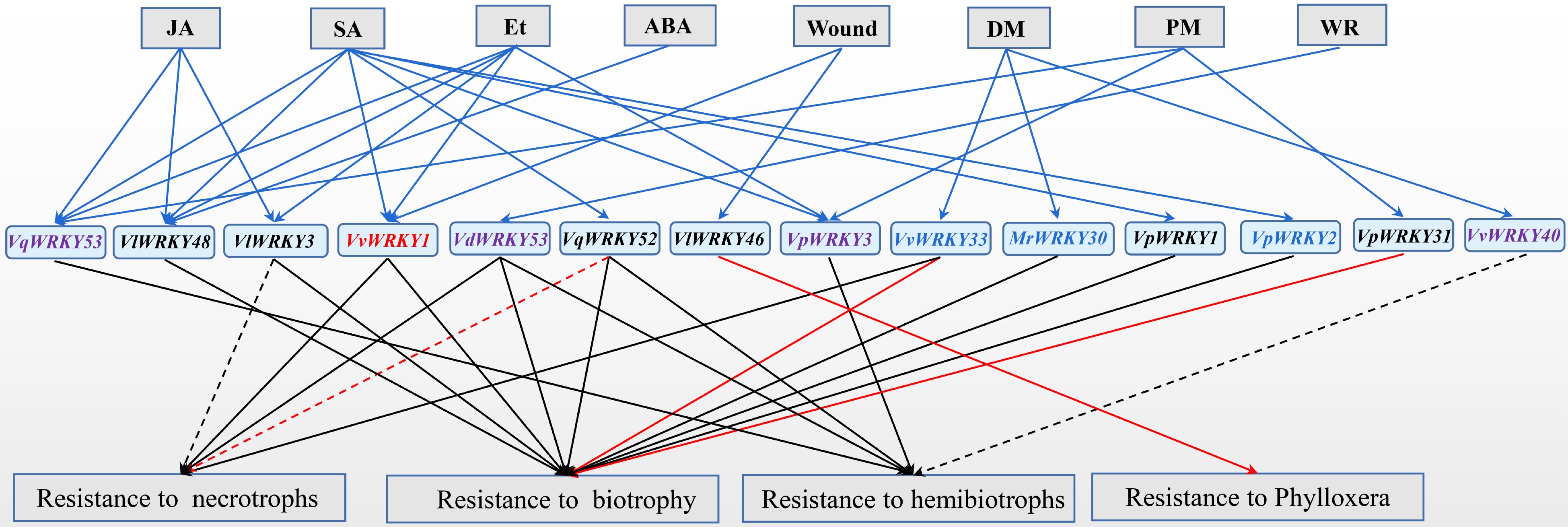
Figure 1.
Functions of grape WRKY transcription factors in biotic stresses. The expression of grape WRKY members were induced (solid blue arrows) by pathogen attack (DM, Plasmopara viticola; PM, Erysiph ecichoraceaum; WR, Coniella diplodiella), hormone (JA, Methyl jasmonate; SA, Salicylic acid; Et, Ethylene), and wound. Grape WRKY members enhanced (solid arrows) or repressed (dotted lines) plant resistance to different types of pathogens. Grape WRKY members in red font were involved in regulating the expression of JA signaling pathway-related denfense genes. Grape WRKY members in black font were involved in regulating the expression of SA signaling pathway-related defence genes. Grape WRKY members involved in regulating the expression of JA signaling pathway-related defence genes (red font), SA signaling pathway-related defence genes (black font), both SA and JA signaling pathway-related defence genes (blue font), or unknown (purple font). The function of WRKY members were identified in grapevines (red arrows) or other plants (black arrows).
Table 2. Functional analysis of grape WRKY proteins under biotic stresses.
Former name Gene function VlWRKY3[41] Enhanced resistance to Golovinomyces cichoracearum, but increased susceptibility to Botrytis cinerea in Arabidopsis VpWRKY3[59] Enhanced tolerance to Ralstonia solanacearum in tobacco VvWRKY33[29] Enhanced resistance to Plasmopara viticola in grapevine; participate in resistance to B. cinerea in Arabidopsis VpWRKY1[16] Enhanced resistance to Erysiphe necator in Arabidopsis VvWRKY40[30] Increased susceptible to Phytophthora capsica in tobacco VpWRKY31[45] Enhanced resistance to E. necator in susceptible V. vinifera VpWRKY2[16] Enhanced resistance to E. necator in Arabidopsis MrWRKY30[31] Enhanced resistance to Hyaloperonospora parasitica in Arabidopsis VlWRKY48[42] Enhanced resistance to G. cichoracearum in Arabidopsis VlWRKY46[19] Attenuated phylloxera attack and delayed nymph development in susceptible grape cultivars VqWRKY52[43,44] Enhanced resistance to G. cichoracearum and Pseudomonas syringae pv. tomato DC3000, but increased susceptibility to B. cinerea in Arabidopsis. Enhanced resistance to B. cinerea in gene-edited grape VdWRKY53[56] Enhanced resistance to Coniella diplodiella, Pst DC3000 and G. cichoracearum in Arabidopsis VvWRKY1[18,40] Enhanced resistance to Pythium, Peronospora tabacina, and E. cichoracearum in tobacco. Enhanced resistance to P. viticola in grapevine VqWRKY53[50] Enhanced resistance to Pst DC3000 in Arabidopsis The expression of these grape WRKY transcription factors is affected by pathogen attack and plant hormones, and they also act as transcriptional regulators to regulate plant hormone-mediated defense response. The functions of these transcription factors on biotic stress is mostly attributed to the regulation of plant hormone-mediated defense response. VpWRKY2[16], VvWRKY1[18,40], VvWRKY33[29], and MrWRKY30[31] are involved in regulating the expression of JA signaling pathway-related genes, while VlWRKY3[41], VvWRKY33[29], VpWRKY1[16], VpWRKY2[16], MrWRKY30[31], VlWRKY48[42], VlWRKY46[19], and VqWRKY52[43,44], and VpWRKY31[45] are involved in regulating the expression of SA signaling pathway-related genes. VpWRKY2 is also involved in regulating the expression of Et signaling pathway-related genes[16].
To study the molecular functions and regulating networks, target genes of grape WRKY members have been identified. For example, VvWRKY52 enhanced grape resistance to P. viticola by activating the expression of JA pathway-related defense genes, including JAZ1.1, JAZ1.2 and LOX[40]. VvWRKY33 specifically activated the expression of VvPR10.1 gene, prompting the resistance to P. viticola in grapevine[29]. VlWRKY46 played a role in SA-mediated defense-regulatory network by directly binding to the downstream structural gene VvCHIB, and attenuated phylloxera attack and delayed nymph development in susceptible cultivars[19].
Stilbene also plays an important role in protecting plants from pests, pathogens and abiotic stresses[46,47]. There was a high correlation between VvWRKY family genes and stilbene synthase genes (STS) expression in the microarray and RNA-Seq data[48−50]. Some grape WRKYs could directly bind to the promoters of stilbene synthase genes, and regulate their expression. Four VvWRKY genes, VvWRKY3, VvWRKY24, VvWRKY43, and VvWRKY53, were involved in regulating stilbene biosynthetic pathway in different hierarchies[49]. In particular, VvWRKY24 played a role in regulating the expression of VvSTS29 gene alone, while VvWRKY3 and VvWRKY53 worked through the combined effect with VvMYB14. The VqWRKY53 involved in positively regulating the expression of stilbene synthase genes VqSTS32 and VqSTS41 to promote plant disease resistance[50]. VvWRKY8 interacted with VvMYB14 and prevented it from binding to the promoters of VvSTS15 and VvSTS21, thereby reducing the resveratrol accumulation[51]. VqWRKY31 could directly bind to the promoters of VvSTS1 and VvSTS2, and promote their expression[45]. Over-expression of VqWRKY31 conferred powdery mildew resistance in susceptible V. vinifera plants by prompting salicylic acid signaling, and specific stilbenoids and flavonoid synthesis[45].
Grape WRKY transcription factors, as transcriptional activators or inhibitors, play roles in many processes of plant responses to abiotic stress (Table 3 and Fig. 2). Except for VvWRKY13 and MrWRKY30, other WRKY members enhance plant resistance to one or more abiotic stresses. VvWRKY13 negatively regulates plant resistance to drought and salt stress, while MrWRKY30 promotes plant resistance to salt stress but negatively regulates plant resistance to cold stress.
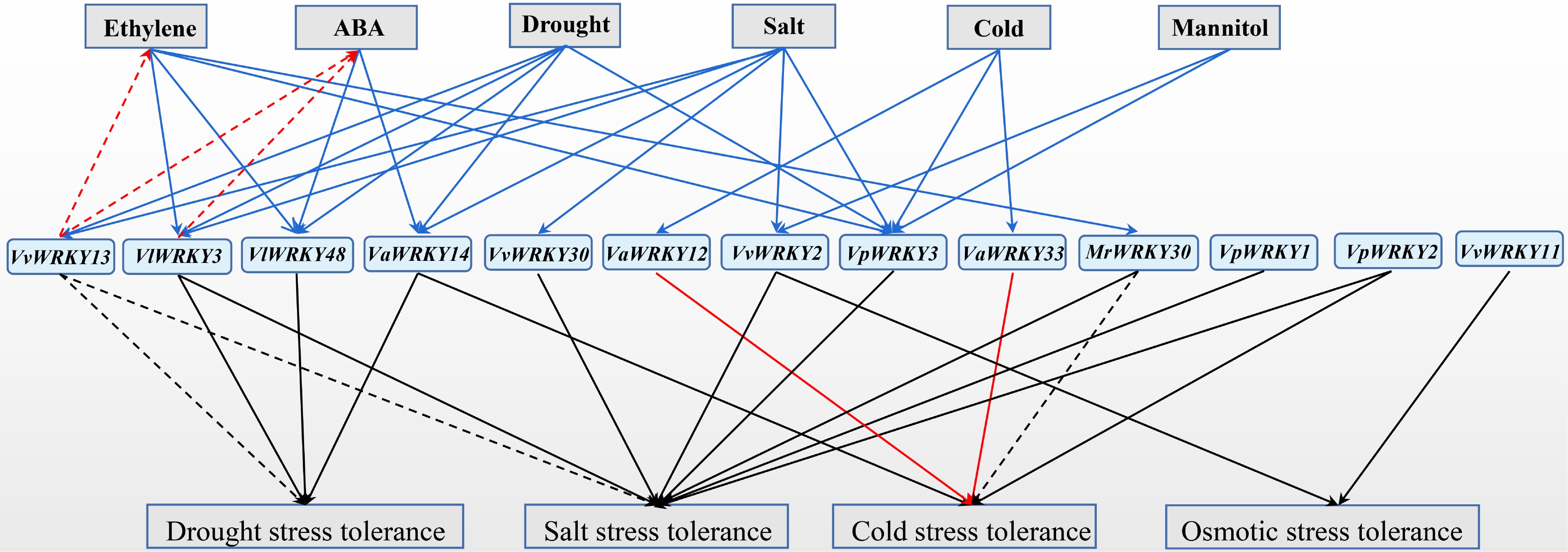
Figure 2.
Functions of grape WRKY transcription factors in abiotic stresses. The expression of grape WRKY members were induced (solid blue arrows) by abiotic stress, and hormones (ABA, Abscisic acid; Et, Ethylene). Grape WRKY members were involved in plant hormone synthesis (red dotted arrows). Grape WRKY members enhanced (solid arrows) or repressed (dotted arrows) plant abiotic stresses tolerance.
Table 3. Functional analysis of grape WRKY proteins under abiotic stresses.
Gene name Gene function VlWRKY3[41] Enhanced tolerance to the salt and drought stress in Arabidopsis VpWRKY3[59] Enhanced tolerance to the salt stress in tobacco VvWRKY11[17] Enhanced tolerance to the osmotic stress in Arabidopsis VaWRKY33[57] Enhanced tolerance to the cold stress in Arabidopsis and grape calli VvWRKY13[52,53] Negatively regulated drought and salt stress tolerance of Arabidopsis VpWRKY1[16] Enhanced tolerance to the salt stress in Arabidopsis VaWRKY14[55] Enhanced tolerance to the drought stress in Arabidopsis VaWRKY12[20] Enhanced tolerance to the cold stress in Arabidopsis and grape calli VpWRKY2[16] Enhanced tolerance to the salt and cold stress in Arabidopsis VlWRKY48[42] Enhanced tolerance to the drought stress in Arabidopsis MrWRKY30[31] Enhanced tolerance to the cold stress in Arabidopsis, and negatively regulated salt stress tolerance of Arabidopsis VvWRKY30[21] Enhanced tolerance to the salt stress in Arabidopsis VvWRKY2[54] Enhanced tolerance to the salt stress and osmotic stress in tobacco Under abiotic stress, grapevine WRKY family genes stimulate the synthesis of plant hormones ABA and Et, the accumulation of cell osmotic substances, the removal of ROS, and the expression of stress-related genes, etc.
Ectopic expression of VvWRKY13 in Arabidopsis promoted constitutive ABA and Et production, and impaired plant tolerance to drought and salt stress by decreasing the accumulation of cell osmotic substance, increasing the ROS level, and down-regulating the expression of stress-related genes[28,33,52,53]. VlWRKY3 promoted stress-induced ABA production by promoting the expression of ABA synthesis genes, thereby enhancing the resistance of Arabidopsis to drought and salt stress[41].
VvWRKY13[52,53], VvWRKY30[21], and VvWRKY2[54] are found in regulating the accumulation of osmotic substances, and VlWRKY3[41], VaWRKY14[55], VvWRKY23[52,53], VaWRKY12[56], VlWRKY48[42], VvWRKY30[21], MrWRKY30[31] involved in regulating the level of ROS, affecting plant resistance to abiotic stress.
The expression of stress-responsive genes in transgenic materials reveal that these grape WRKY members have different functional mechanisms in response to abiotic stresses. VaWRKY14 could enhance the expression of stress-related genes including COR15A, COR15B, COR413, KIN2 and RD29A in Arabidopsis under drought stress[55]. VaWRKY12 prompted the expression of genes encoding antioxidant enzymes including peroxidases and glutathione S-transferases in Arabidopsis under drought stress[56]. Ectopic expression of MrWRKY30 in Arabidopsis enhances cold stress tolerance by activating the AtCBF-mediated signaling pathway to induce the downstream AtCOR47 gene, but impairs salt stress resistance by suppressing the expression of antioxidant genes[31].
-
Only a few proteins involved in regulating the functions of grapes WRKY members have been identified. A cold-responsive ethylene response factor VvERF92 directly regulated the response of VaWRKY33 to low temperature and ethylene[57]. VvSnRK1.1 and VvSnRK1.2 interacted directly with VvWRKY22, and might be involved in regulating the role of VvWRKY22 in sugar metabolism[36]. An E3 ubiquitin ligase EIRP1 (Erysiphe necator-induced RING finger protein 1), which played a positive regulatory role in plant immunity, directly interacted with VpWRKY11, and promoted degradation of VpWRKY11 by 26S proteasome[58]. The cytoplasmic effector protein PvRXLR111 secreted by P. viticola interacted directly with VvWRKY40 and increased the stability of VvWRKY40 protein, thereby inhibiting plant immunity[30].
The external environment affects not only the expression of WRKY members, but also the subcellular localization of the grape WRKY members. For instance, VaWRKY12 was located in the nucleus and cytoplasm under normal temperature, but it localized only in the nucleus at low temperature[20].
-
In this article, we summarized the research progress regarding functions of grape WRKY family members, and suggested future naming systems to avoid confusion in sharing scientific findings of grape WRKY family members.
Grape WRKY family transcription factors play important regulatory roles, not only in grapevine growth and development, but also in their responses to biotic and abiotic stress. Although functions of some WRKY proteins have already been identified, there are still many unanswered questions that require future investigation. Currently, most functional studies and regulating network of VvWRKY family genes were conducted using model plants (tobacco and Arabidopsis) or in vitro grape systems. Although some abiotic and biotic stresses can be well simulated in model plants, the growth characteristics of grape, as a perennial vine, are markedly different from the model plants, which may negatively impact the functional analysis of grape WRKY members in model plants. In addition, many grape pathogens are unable to infect these model plants, and some Arabidopsis pathogens are also unable to infect grapes. Therefore, it is necessary to verify the function of grape WRKY transcription factors in an in vivo system of grapevines such as a stable grape transformation system.
This work was supported by National Key R & D Program of China (2019YFD1002501 to S.R. Song), Shanghai Municipal Commission for Science and Technology (2021-02-08-00-12-F00751 to J. Lu), Yunnan Province Science and Technology Department (202005AF150023 to J. Lu), and China Postdoctoral Science Foundation (18Z102060107 to W. Wu).
-
The authors declare that they have no conflict of interest.
- Copyright: 2022 by the author(s). Exclusive Licensee Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Wu W, Fu P, Lu J. 2022. Grapevine WRKY transcription factors. Fruit Research 2:10 doi: 10.48130/FruRes-2022-0010
Grapevine WRKY transcription factors
- Received: 28 March 2022
- Accepted: 19 June 2022
- Published online: 25 July 2022
Abstract: Grape is one of the most economically important fruits and is cultivated worldwide, but the viticulture faces challenges of various biotic and abiotic stresses. WRKY transcription factors play important roles in regulating plant responses to these stresses, in addition to their roles in plant growth. Genome-wide identification and functional analysis of grape WRKY family genes have been conducted in recent years. However, different approaches were found in naming the grape WRKY family gene members among these reports, which causes a great deal of confusion and has become a barrier in the sharing of research findings in the research community. Here we attempt to comprehensively review the research progress on grape WRKY family transcription factors, and attempt to assign unified names for them.
-
Key words:
- WRKY transcriptional factor /
- Plant growth and development /
- Abiotic stress /
- Biotic stress /
- Grapevine












