HTML
-
The current study was designed to search for the occurrence of protozoa and fungi in the Egyptian habitat. These mycobiomes include varieties of microorganisms such as plasmodial slime molds and ascomycetes which are usually associated with soil and dead woody trunks of trees, respectively.
Myxomycetes live in various environments, such as soil, where they are distinguished by their macroscopic fructification. According to the classification of Ainsworth[1], true slime molds, also known as plasmodial slime molds, were placed in the protozoa group, commonly known as Mycetozoa. The characterized fructification of these fungal groups permitted several studies on their ecology and distribution for over 200 years[2]. The protoplasm of these extinct microbes, known as eucaryotes, moves in a shuttle-like manner to produce multinucleate, coenocytic, saprobic plasmodium. The distinctive peridium and capillitium of the sporangium, as well as the occasional presence of calcium carbonate (sometimes known as 'lime') on the exterior surface, are the major characteristics that can be used to identify these sporocarps. Furthermore, the ameboflagellate and plasmodial vegetative stages of their life cycles are alternate[3]. They inhabit different localities such as deserts, forests, water and snowy mountains[4].
Lepidoderma de Bary, a plasmodiocarpous genus, was published for the first time by Rostafiski[5] and described in detail by Martin & Alexopoulos[6] It was systematically placed in the order Physarales, family Didymiaceae. Moreover, it is a nivicolous organism that mostly thrives in snow. Lepidoderma crustaceum is an uncommon species since it is found in very few locations worldwide, particularly the nivicolous situations in Switzerland, USA[7], Italia[8] and western Crete[9].
The ascomycete fungus Arthrinium Gustav Kunze belongs to the family Apiosporaceae, order Xylariales, and the class Sordariomycetes. The type species of this widespread saprophytic ascomycete, known as Arthrinium caricicola, has been around for more than 200 years and has been discovered in association with numerous plants[10,11]. As plant pathogenic fungi, some Arthrinium species, such as A. arundinis, have been identified, which cause barley kernel blight. The cause of wheat damping-off is A. sacchari[12, 13] which has also been recorded as an endophyte in the tissues of some plants, lichens, and marine algae[14−17]. A. bambusicola species has oval to rounded, brown, multi-guttulate to roughen, granular conidia with delicately pale slits in the outer margins are diagnostic of the Nov., which is described and shown in this study[18].
The current work is characterizing the protozoan, L. carestianum and the ascomycetous A. bambusicola for the first time in Egypt based on the morphological characteristics of both species, in addition to the sequencing of COI and ITS genes. Recording of these species is considered an addition to the Egyptian myco-biota taxa.
-
Lepidoderma carestianum – Figs 1 & 2

Figure 1.
Sporocarp of L. carestianum. (a) On the soil surface. (b) & (c) Lime scales covered the peridium of sporocarp (3X). (d) Hypothallus (2X).
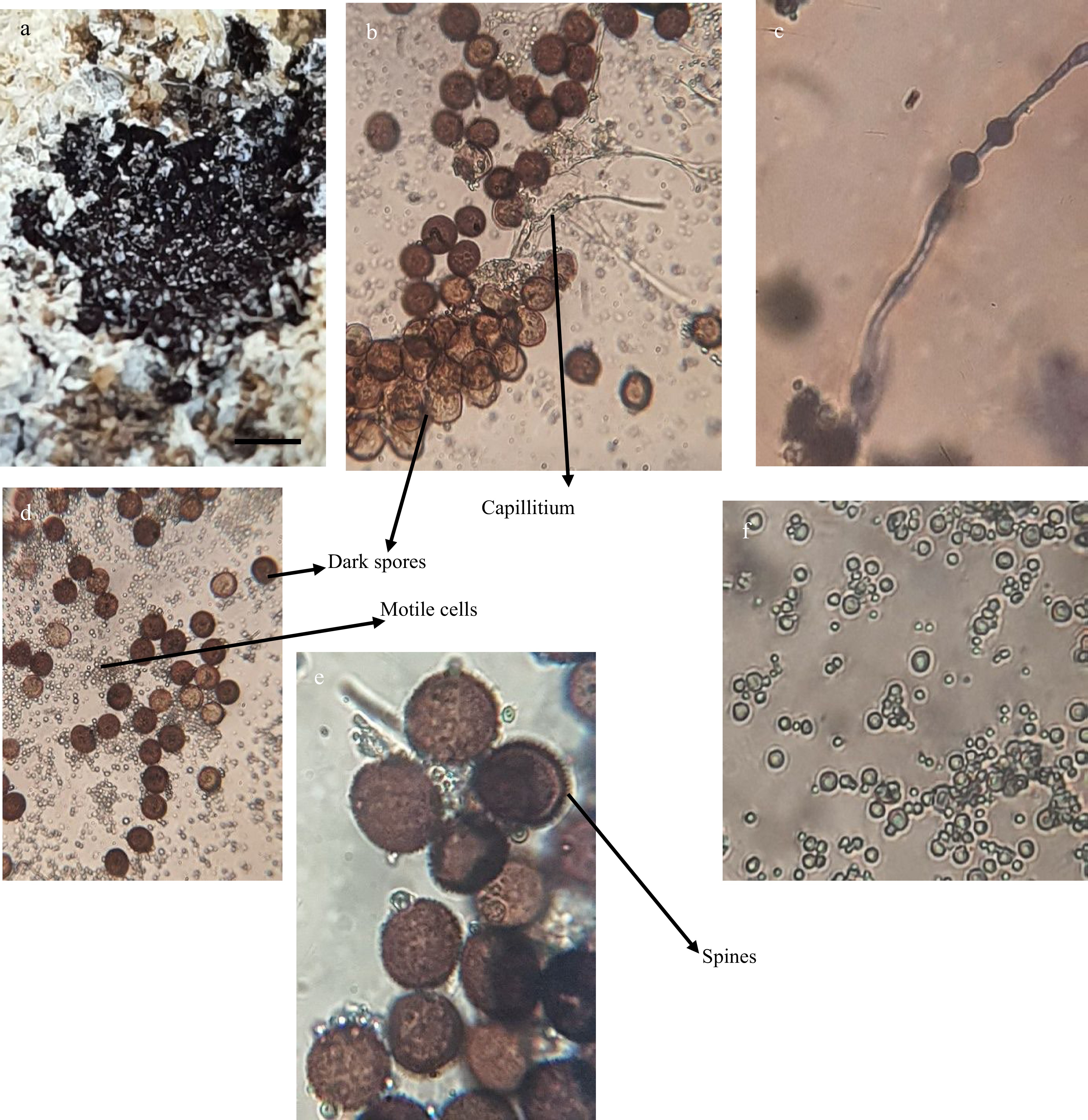
Figure 2.
A black powder inside sporocarp of L. carestianum (2X). (b) Dark spores. (c) Capillitium thread with swellings. (d) Dark spores and masses of motile cells. (e) Enlarged spores with spinose walls. (f) Enlarged motile cells. (b, c, d and f: 400X. e: 1,000X)
GenBank accession number: OM630523
The examined protozoan's sporocarp was discovered on the soil in Tanta University's Science Faculty garden during a chilly November in 2021 when the temperature ranged between 8 and 20 ± 2 °C.
-
Irregularly branched, flattened and sessile sporocarp measured 30−45 × 60−65 mm diameter (Fig. 1a). Peridium that covered the entire surface of the sporocarp is distinguished as thin, membranous, or sub-cartilaginous with the presence of white creamy lime scales (Fig. 1b & c). Thin, continuous, creamy white hypothallus was found under the sporocarp body in contact with the plant debris (Fig. 1d).
Brown spiny globose to subglobose spores of this tested slime mold were found after breaking the lime scales as masses of black powder (Fig. 2). The size of these spores was recorded as 10−10.2 × 10−10.5 μ. Also, a lot of motile, different sizes and shaped cells were present (Fig. 2f). Thread like capillitium with some globular swellings appeared among the spores and cells (Fig. 2c).
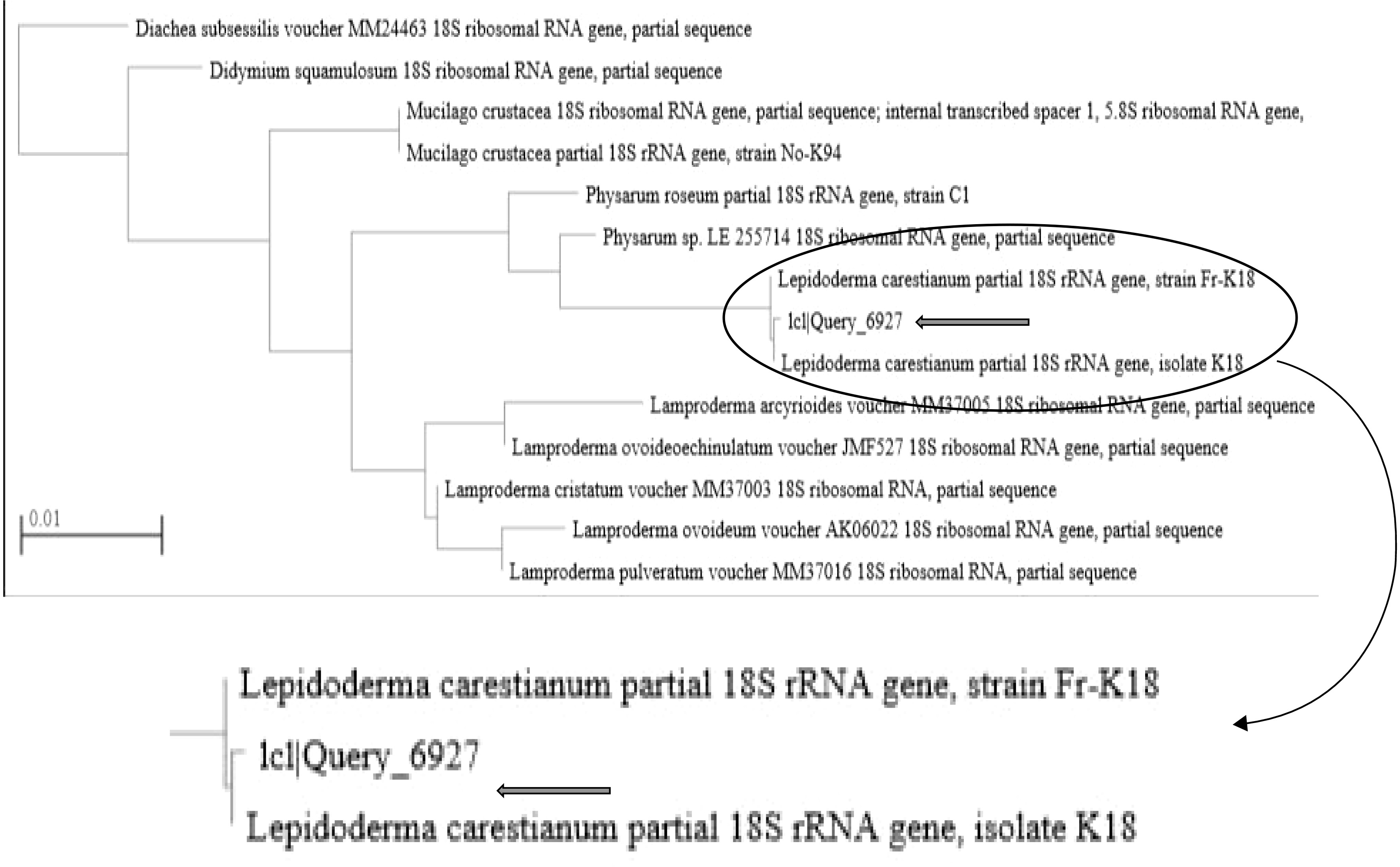
The sequences producing significant alignments with the sequence of the tested myxomycete (1c1|Query_6927) is indicating 93.95% identity to two isolates of L. carestianum (HE614609.1 and AM231296.1) with query cover 94% for each (Table 1). Even though the tested strain had a higher percentage identity with the strains of Physarum, Diderma, and Mucilago than it did with Lepidoderma, it was nonetheless classified as L. carestianum. This choice is based on every aspect of the isolate's morphological and microscopic characteristics noted. The query also returns Lepidoderma with a cover percentage. The value for L. carestianum strain was the highest (94%). Additionally, the tested isolate was clustered with L. carestianum, as shown by the phylogenetic tree (Fig. 3).
Table 1. Molecular data of (1c1|Query_6927) strain with other previously reported myxomycetous strains.
Tested strain Name Accession No. Query
cover
(%)Percent identity
(%)1c1|Query_6927 Lepidoderma carestianum HE614609.1 94 93.95 Lepidoderma carestianum AM231296.1 94 93.95 Physarum vernum KC759102.1 49 95.33 Physarum vernum KC759101.1 49 95.33 Physarum nivale DQ903680.2 49 95.33 Diderma crustaceum JQ277927.1 49 95.02 Mucilago crustacea MH348907.1 50 94.12 Arthrinium bambusicola – Fig. 4
GenBank accession number: ON076927
Demonstrative examination:
Colonies with elevated mycelia, colorless reverse with ability to grow fast were observed. Conidiomata was discovered embedded in the colony as dark, atypically shaped objects that were surrounded by numerous brown septate hyphae. Smooth, oval conidia are hyaline when young and then turn brown by age, occasionally characterized with tapered ends (Fig. 4).

Figure 4.
Morphology and microscopic examination of A. bambusicola: (a) White colony. (b) Reverse with black spots due to conidiomata (2X). (c) & (d) Conidiomata surrounded by septate hyphae (100X & 400X). (e) & (f) Conidia. (400X & 1,000X).
Using available ITS sequence data retrieved from GenBank, a phylogenetic analysis of the tested strain and other Arthrinium isolates was performed. The tested isolate (Arth) shared a 95% similarity with the previously known strain Arthrinium according to the BLAST search results and the phylogenetic tree of the ITS sections of the acquired sequences. Figure 5 depicts A. bambusicola (MFLU 20-0528 ITS).
-
In this study, L. carestianum was identified for the first time in the Egyptian environment. The recent changes in the Egyptian environment, which has become exceptionally cold in the winter with minimum temperatures reaching 6 °C, may have something to do with this form of fungi's emergence there. Therefore, we predict that this cold environment encourages the appearance of this myxomycete fungus in Egypt. This discussion is based on the previous records of Lepidoderma in extremely cold localities around the world[19]. The specific features of the sporocarp of the examined myxomycete are in agreement with the extensive morphological description provided by previous researchers[20]. Furthermore, the microscopic structures of myxomycete being studied match those analyzed and published in earlier investigations[6, 21]. However, neither research investigations, nor reviews by other researchers, made note of the obvious motile cells discovered in the tested isolate. The characteristic features listed by the description of A. bambusicola are in accordance with those cited in other literature[17]. The resulting data from molecular and phylogenetic analyses supported and confirmed the morphological identification. Recently, DNA sequences of different genes like ITS, TEF, and TUB, were employed to delimit and recognize closely related Arthrinium species where species identification based on morphological features is problematic[22].
-
This study recorded the first occurrence of L. carestianum and A. bambusicola in an Egyptian context. While A. bambusicola was saprophytically associated with Melia azedarach, whereas the sporocarp of L. carestianum was found on the soil surface in a terrestrial habitat. Morphological keys, and molecular analysis of each fungus using the indicated DNA sequences, showed high levels of identity and query coverage with other isolates of the same species previously recorded in GenBank, that was used to confirm and validate each fungus' identification.
-
During a field experiment, a mature sporocarp of L. carestianum was discovered on the surface of a soil mixture made up of clay, manure, and 10% sand and growing alongside Vicia faba plants. Dimensions (mm), colour, surface roughness, and appearance along with microscopic examination were all measured precisely and reported.

Figure 6.
Melia azedarach tree. (a) Healthy tree. (b) Olivaceous green growth of A. bambusicola on the dead branch (2X).
On a dead branch of a Melia azedarach tree, masses of olivaceous green velutinous spores belonging to A.bambusicola were seen (Fig. 6). The examined branch was taken to the lab, where A. bambusicola was isolated using PDA plates that were provided by the antibiotic chloramphenicol as an antibacterial agent. It was then distinguished based on colour, orientation, and texture concurrently with the microscopic investigation of different fungal structures according to previous studies[18, 23−25].
Genetic analysis
-
The examined species were identified molecularly and recorded in the National Center for Biotechnology Information (NCBI). The whole DNA genome of the myxomycete specimen's ground-up spores was used for genetic analysis, and DNA was extracted in accordance with the manufacturer's instructions for the E.Z.N.A.® Fungal DNA Mini Kit (D3390-01, Omega BIO-TEK,USA). While A. bambusicola's cellular genomic DNA was extracted in accordance with the manufacturer's instructions using the DNeasy Plant Mini Kit (provided by QIAGEN). Following that, each piece of genomic DNA was individually amplified using the PCR method[26].
The COI gene was chosen for sequencing because it can be used for barcoding and phylogenetic reconstruction in dark-spored myxomycetes[27] and Micron-Corp Company, (Korea) assistance was used to complete this phase. While ITS sequencing study proved that A. bambusicola was the correct species. The ITS sequence was examined using the primers ITS4: R-(5'-GCTGCGTTCTTCATCGATGC-3′) and ITS1: F-(5′-CTTGGTCATTTAGGAAGTAA-3′). By performing distance matrix analysis with the neighbor joining method, a phylogenetic tree was created. Neighbor-joining (NJ) trees were created in MEGA x[28].
Diagnosis and identification of the tested fungi
-
The authors declare that they have no conflict of interest.
- Copyright: © 2022 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
| El-Debaiky SA, Mahmoud YAG. 2022. First record of Lepidoderma carestianum and Arthrinium bambusicola from Egypt. Studies in Fungi 7:10 doi: 10.48130/SIF-2022-0010 |