-
Cassava is one of the most important crops for food security and income generation for small-scale producers in the tropics[1], and it has increasing significance due to its climate resilience and productivity[2]. Despite its importance, the cassava research community has historically been relatively small, and innovation in variety development has lagged[3]. Recently, a substantial improvement has been made with the NextGen Cassava breeding investment, where genomics-assisted breeding is a primary component[4−9].
However, a substantial challenge persists in the necessity to cross heterozygous parents in cassava breeding. When using heterozygous parents, fixing essential traits, and introgressing new traits through backcrossing have been challenging due to inbreeding depression caused by high genetic loads[5,10−14]. Consequently, this constraint limits the efficiency of breeding programs and delays the development of cassava varieties with specific traits preferred by farmers and consumers[15].
In response to this challenge, an innovative initiative is proposed – inbred-parent-based hybrid cassava breeding. This pioneering initiative aims to transform cassava breeding from a heterozygous-parent-based mutation-covering mode into an inbred-parent-based mutation-purging one. A similar transformation in maize, over a century ago, laid the foundation for continuous yield increase[16,17], and this method has been proven successful in various crops such as sorghum, rice, canola, sunflower, and more recently, diploid potato[15]. Ongoing discussions on hybrid breeding in clonally propagated root and tuber crops underscore the potential of this transformative approach[4,18].
The transformation to inbred-parent-based hybrid cassava breeding will significantly improve the predictive ability of breeders, enabling more accurate trait selection and informed parent crossing. Improved well-known cassava cultivars with specific desired traits can be quickly adopted by small-scale producers, thereby ensuring increased cassava productivity, and ultimately strengthening food security.
-
In the tropics, cassava has significant advantages over corn in staple carbohydrate production. It can survive through long dry seasons and produce edible roots for small-scale producers. In terms of dry matter yield, cassava also outperforms corn in tropical regions on a country-wide scale. For example, when considering 30% dry matter content in cassava and 87% in corn, in Nigeria, cassava yields 2.36 tons/ha of dry matter, compared with 1.68 tons/ha in corn on average from 2017−2021. Similarly in Vietnam, cassava achieves a dry matter yield of 5.90 tons/ha, while corn yields 4.17 tons/ha[19]. These advantages highlight the crucial roles of cassava in ensuring food security under climate change.
Initiating inbred-parent-based hybrid breeding represents a transformative shift in cassava breeding strategies. Hybrid breeding will not only enhance the efficiency of the breeding programs but also increase breeding programs' adaptability to emerging biotic and abiotic stresses induced by climate change. Instead of waiting for 6−10 years required for improvement via recurrent selection, by introgressing essential traits using backcrossing, cassava breeding programs can respond quickly to the dynamic needs of ever-changing markets and environmental conditions. Although simulations suggest that inbred-hybrid breeding may not always outperform outbred recurrent selection for quantitative traits[18], it is crucial to recognize the benefits of backcrossing for traits controlled by major genes or loci, for instance, cassava mosaic disease (CMD) resistance, waxy starch, and high beta-carotene content.
Furthermore, inbred-parent-based hybrid breeding will also lead to substantial cost savings for breeding programs to develop multiple products for the diverse market demands. Instead of managing multiple large, separate breeding pipelines and populations for diverse products, breeding programs can concentrate on the improvement of a foundation population. By combining the foundation population improvement and line conversion techniques, breeders can efficiently develop varieties for specific market segments (Fig. 1). This foundation improvement and line conversion simplifies the development process, saving both time and resources. All the product development is built on the achievements of the foundation population, requiring much less effort and resources than managing separate pipelines.
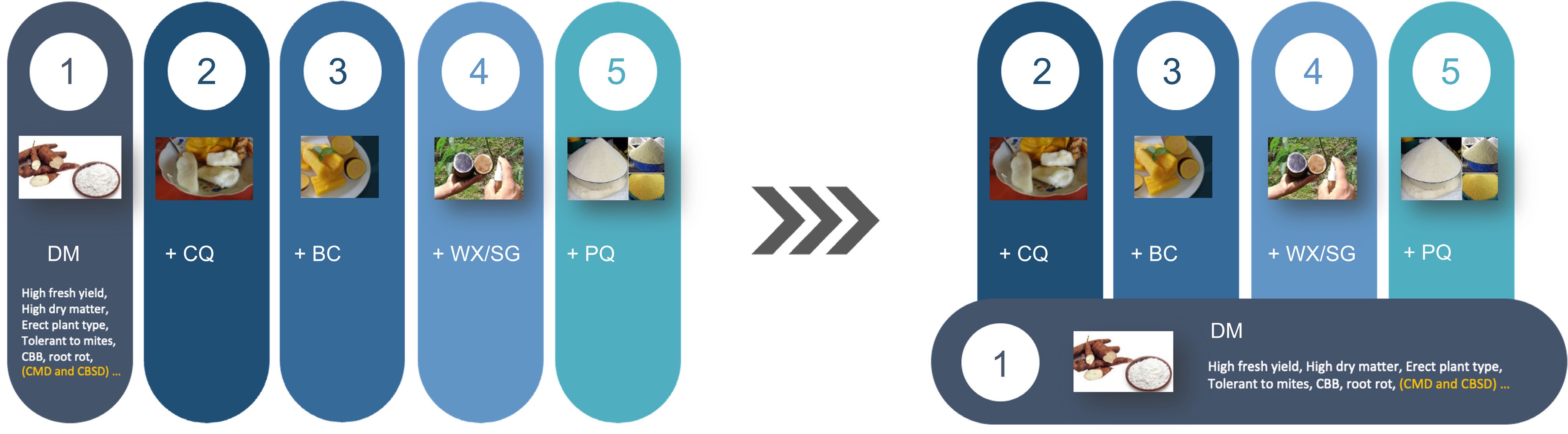
Figure 1.
Transform cassava breeding through inbred-parent-based hybrid breeding. Currently, there are five products in global cassava production: 1) cassava with high dry matter yield; 2) cassava with good cooking quality for human consumption; 3) cassava with high beta-carotene for human consumption; 4) cassava with specialty starch, e.g., waxy starch and small granule starch; 5) cassava with good processing quality for granulated and paste products for human consumption. Using conventional recurrent selection with heterozygous parents, to meet the unique needs of each product, breeding programs need to manage separate breeding populations to select the essential desired traits (left panel). The right panel shows the product development model using inbred-parent-based hybrid breeding. Instead of evaluating separated large populations for different products, breeders will manage one foundation breeding population, coupled with several separate line conversion pipelines to reformulate hybrids to address the unique needs of other products. Moreover, compared with the five full-time equivalent (FTEs) to manage five separate populations, the new product development model only needs 3.5 FTEs, with 1.5 FTEs for the foundation population and 0.5 FTEs for each line conversion pipeline. DM, dry matter; BC, Beta-carotene; CQ, cooking quality; WX, waxy starch; SG, small granule starch; PQ, processing quality; CBB, cassava bacterial blight; CMD, cassava mosaic disease; CBSD, cassava brown streak disease.
By enhancing efficiency, adaptability, and cost-effectiveness, inbred-parent-based hybrid cassava breeding paves the way for sustainable and resilient cassava production, ultimately substantially contributing to ending hunger and reducing poverty during the climate crisis.
-
The cassava breeding programs use conventional recurrent selection for new variety development[20,21]. Each breeding cycle has a single recombination event between heterozygous parents. The selected F1 progeny is then clonally propagated for further evaluation in breeding stages, and potentially, commercial release. Thanks to its simplicity, this approach has been widely adopted. However, breeders working with heterozygous parents encounter two major pain points[4,15,22−24].
First, within individual families (or progeny of individual breeding crosses), the progeny derived from heterozygous parents show considerable segregation, but between families, the variation is relatively small. This observation has been validated in different cassava populations in independent environments[24−30]. The relatively small between-family variation makes parent selection less effective, i.e., identifying superior parents for producing high-performance progeny. On the other hand, the large within-family variation requires extensive screening of large breeding populations, and the performance of selected progeny must be validated across multiple environments[31,32], increasing the yield trialing cost and duration of the breeding process. Recently, genomic selection (GS) has been broadly tested in cassava breeding, which potentially increases breeding efficiency by significantly shortening breeding cycles[9,33,34]. However, releasing varieties to farmers' fields remains time-consuming, requiring multi-year and multi-location yield trials to validate their performance in yield stability, quality, and disease and insect resistance. Even with the advancements of the NextGen Cassava, it took eight years of extensive evaluation before the Game Changer variety (TMS13F1160P0004) was released in Nigeria in 2021, derived from crosses made in 2012.
Second, the inbreeding depression associated with heterozygous parents make it impossible to integrate essential traits via backcrossing[5,22]. For instance, when developing CMD resistant varieties in Southeast Asia, the resistance controlled by a single dominant gene could not be quickly integrated into the local elite varieties due to the absence of backcrossing. This limitation resulted in a lengthy variety development period with at least two cycles of recurrent selection, spanning over 6 years and requiring evaluations across multiple environments[20]. On the contrary, if inbred-parent-enabled backcrossing is available, after 2−3 years of trait introgression with minimal field evaluation, breeders would be able to convert the most popular local varieties into resistant ones. Moreover, farmers, familiar with the existing varieties will be willing to adopt the resistant varieties, which amplifies the benefits of disease resistance. Thus, to achieve enhanced genetic gains in farmers' fields, we need to promote the use of inbred parents in cassava breeding, reducing complexity, shortening breeding cycles, and ensuring the rapid introduction of superior traits into farmer-preferred varieties.
-
Using inbred-parent-based hybrid breeding, we have great opportunities to radically improve the rate of genetic gains while also significantly enhancing the market response of cassava.
First, hybrid cassava varieties will be much more cost-efficient for farmers due to the clonal propagation, especially when compared with seed crops. Unlike seed crops that rely on hybrid seed production systems, cassava does not require such a complex and often costly procedure[35,36]. Moreover, with the ability to multiply desirable varieties directly on the farm, farmers can avoid the recurring cost of purchasing seeds every year. This cost-saving feature makes improved cassava variety more accessible to small-scale producers. However, for breeders, the use of botanical hybrid seeds is a necessary step in cassava hybrid development, offering significant advantages, as discussed below.
Second, one of the primary advantages of inbred-parent-based hybrid breeding is to enable breeders to introgress desired traits quickly, so breeders can promptly respond to the dynamically changed market demands[37−39]. With the inbred-parent-enabled backcrossing, adjustments can be made quickly and efficiently in the popular varieties that farmers and consumers are familiar with. The enhanced adaptability ensures that released cassava varieties meet the evolving needs of consumers and regional markets, which is even crucial in the agricultural landscape during the climate crisis.
Third, inbred-parent-based hybrid breeding enables the establishment of a highly efficient product development model. Instead of evaluating separated large populations for different market segments, we will manage one unified breeding pipeline, coupled with several separate trait conversion pipelines to reformulate hybrids to address different regional markets and/or ever-changing market needs (Fig. 1). This process can be greatly facilitated by genomics tools and self-pollination, followed by establishing a doubled haploid (DH) system[40]. For instance, the large-effect phytoene synthase gene is used in biofortified cassava to increase beta-carotene content, while mutation of the granule-bound starch synthase I gene provides waxy starch in cassava[41,42]. This product development model will enhance resource optimization, redundancy reduction, and acceleration of market-targeted variety development.
Fourth, during the development of inbred parents or doubled haploids, the genetic load in cassava breeding populations will be purged[5,10]. Unlike conventional breeding methods that hide deleterious mutations in heterozygous status, inbred-parent-based hybrid breeding actively removes harmful genetic elements. This process ensures that the developed hybrids possess a cleaner genetic background, and improved overall vigor, productivity, stability, and adaptability of hybrid varieties.
Fifth, once heterotic pools are identified or created, the improvement within each pool will facilitate the systematic use of heterosis in cassava breeding. Heterosis, a phenomenon observed in multiple crops, leads to substantial improvements in yield and yield stability[43−46]. By effectively using heterosis, breeders can use complementary inbred parents to systematically cover up the remaining genetic loads in their genomes, dramatically improving the productivity and stability of cassava hybrid for small-scale producers.
Sixth, inbred-parent-based hybrid breeding offers an advantage through genetically identical clean seeds of inbred. The botanical seeds facilitate germplasm sharing with breeding network partners, reducing the risks associated with spreading quarantine pathogens[47−49]. Moreover, the inbred seeds carrying improved breeding values can be directly used by partners for hybrid development, which is critical for collaborative breeding efforts and the global advancements of cassava breeding.
-
Cassava is the second largest crop for starch production after corn. Also, like corn, cassava is self-compatible[50,51]. This feature makes it possible to self-pollinate and develop inbred parents, which is a fundamental requirement for inbred-parent-based hybrid breeding. Although inbreeding depression has been reported in cassava, the level of depression varies among cassava families[11,12]. Overcoming this challenge and reaching a high percentage of selfing or homozygosity can be achieved at least in some families. On the other hand, inbreeding depression and heterosis are generally considered as the two opposite sides of the same coin[52]. Thereby, heterosis can be expected to recover and boost yield and yield stability in hybrid.
Flower-inducing technology facilitates self-pollination
-
Cassava requires a long growth season, usually 10−12 months, for seed production in crossing nurseries. Obtaining flowers, especially for parents with erect plant architecture, has been challenging for cassava breeding programs[53]. Recent technological advancements have led to the best practice of flower-inducing technology, enabling the selfing of any cassava parent[53−57]. This technology empowers breeders to explore a wider range of genetic diversity for inbred parent development.
Doubled haploid technology, a promising frontier
-
In breeding programs, one cassava generation from seeds to seeds needs at least ten months, even with the flower-inducing technology[54]. Instead of developing inbred parents via 5−6 years of selfing, a promising option is to explore DH technology. DH technology, proven effective in multiple crops, can rapidly produce homozygous or inbred plants[58−60]. In hybrid cassava breeding, DH can dramatically reduce the timeline of inbred parent development from 5−6 years to 1−2 years. Recently DH technology has been used to produce pure lines from maize landrace to unlock their genetic diversity even though they have a heavy genetic load[59,60]. Nowadays, leveraging an AI-based high-throughput method, millions of microspores can be generated and screened for DH development (ScreenSYS, Freiburg, Germany). Exploring the potential of DH technology to develop inbred parents will increase the efficiency of cassava breeding, leading to faster improvements in cassava productivity and resilience.
Gain deeper genome insights through whole genome sequencing
-
Moreover, advancements in genome sequencing have paved the way to a comprehensive understanding of cassava genomics. Cassava HapMap has been developed, and genome-wide deleterious mutations have been located and characterized[5,14]. Due to the reduced sequencing cost and relatively small genome size of 750 Mb[61], whole genome sequencing can be used for the prediction and purging of deleterious mutations[10].
Purge genetic load through GS-based rapid cycling
-
With the broadly adapted genomic selection in hybrid breeding, breeding programs will be able to accurately predict the performance and breeding value of inbred or DH parents. This accurate prediction enables GS-based rapid cycling between F1 and selfing or DH clones[4]. The rapid cycling will enhance the efficiency of purging genetic load and reducing deleterious mutations. Furthermore, using the genomic information of the parental lines, GS-based approaches could also be progressively tested and incorporated into the breeding pipeline for the eventual prediction of hybrid performance with relatively good accuracy. The potential of this tool to predict hybrid performance has been demonstrated in major cereal crops, including maize[62,63], rice[64,65], and sorghum[66,67].
Establish a global cassava breeding network
-
The systematic cassava breeding started with the establishment of the International Center for Tropical Agriculture (CIAT) and International Institute of Tropical Agriculture (IITA) in the 1970s within the Consultative Group on International Agricultural Research (CGIAR), and later National Agricultural Research and Extension Systems (NARES) joined the breeding effort such as India, Thailand, and Brazil[4,68]. As we transition into the era of One CGIAR, a global CGIAR-NARES cassava breeding network is currently in the development phase. This collaborative effort aims to bring together international and national breeding efforts under a unified framework with the same standard operating procedure. This harmonization will not only facilitate the breeding data sharing, and the determination of heterotic groups but will enhance the buildup of a strong heterotic pattern over time.
Therefore, by leveraging self-compatibility, flower-inducing technology, DH technology, genomics advancements, and the global network, cassava breeders are empowered with tools to achieve efficient, precise, and accelerated inbred-parent-based hybrid breeding.
-
To initiate inbred-parent-based hybrid cassava breeding, we proposed four essential areas to concentrate on for the initial phase. By focusing on understanding inbreeding depression, developing inbred or DH parents, purging genetic load, and identifying or creating heterotic pools, we aim to accelerate the development of hybrid cassava breeding to enhance rapid adaptation to market demands and environmental stresses.
First, understanding inbreeding depression and deleterious mutations in cassava. Cassava has a significant number of deleterious mutations[5]. Large-effect deleterious mutations can cause the failure of developing inbred parents or lead to low homozygosity rates in certain regions even after generations of selfing[13]. To pinpoint these large-effect deleterious mutations, we propose selfing dozens of elite breeding parents and investigating resulting populations using whole genome sequencing. Whole genome sequencing will facilitate the discovery of genome-wide markers and enable the prediction of deleterious mutations[15]. Moreover, by associating genome-wide markers with field performance, we can identify the loci of the large-effect deleterious mutations affecting growth vigor and yield, allowing breeders to select against these loci during inbred parent development.
Second, developing inbred or DH parents. Using flower-inducing technology, we can self-pollinate any elite parents and their progeny. In the initial phase, breeding programs should prioritize elite breeding parents of the foundation pipeline (Fig. 1). At each selfing generation, whole genome sequencing will enable selecting against deleterious mutations and selecting for clones with high homozygosity rates[10]. Due to the concerns about the effect of inbreeding depression on flower fertility, flowering ability, along with growth vigor, plant architecture, and yield, will be the selection criteria for selfing clones. Considering that cassava's extended growth cycle required multiple years to develop high-homozygosity inbred parents, we should explore DH technology, especially the Al-based high throughput method that can screen millions of microspores (ScreenSYS, Freiburg, Germany), ultimately accelerating the development of inbred-parent-based hybrid cassava breeding.
Third, purging genetic load using GS-based rapid cycling. Due to the severe effect of inbreeding depression, it might be difficult to produce highly homozygous selfing progeny from some families[69]. To deal with the impact of inbreeding depression, we propose a strategy of rapid cycling between generations. S1 clones within families will be intercrossed to generate F1 clones, which, upon self-pollination, produce new S1 clones. Genomic selection will facilitate the rapid F1−S1 cycling, where the number of deleterious mutations and predicted agronomic performance in flowering, vigor, and yield serve as the primary selection criteria[10]. After heterotic pools are established, the S1 clones within each pool will be intercrossed to generate F1 hybrids, driving the rapid cycling of F1−S1 generations.
Fourth, identifying or creating two distinct heterotic pools. Given the limited knowledge about combining ability and heterosis in cassava, we propose conducting test crosses between S1 selections and 4−6 diverse elite parents[70]. The progeny will be evaluated for general and specific combining ability, which will inform the presence or absence of heterotic pools in cassava. Furthermore, we recommend exploring the combining ability of selfing clones between different cassava breeding programs. If no clear heterotic pattern is observed, breeders will create heterotic pools based on the inheritance of essential traits[71]. For example, one heterotic group could emphasize early branching/flowering for rapid improvement, while the other may focus on late branching/flowering, providing erect plant architecture in the hybrid. We also want to emphasize that the determination of the two heterotic groups should be a collaborative decision within the global cassava breeding network. The effectiveness of the two heterotic groups relies on the collective commitment of cassava breeders working within the selected groups to build a strong heterotic pattern over time. Due to the current state of the mixed pools, it will likely take more than 10 years of collaborative effort from the global team to achieve the high heterotic potential of cassava.
-
The authors confirm contribution to the paper as follows: study conception and design: Zhang X, Holley R, Egesi CN; draft manuscript preparation: Zhang X; manuscript editing: Holley R, Egesi CN, Gemenet DC, Moreta D, Gimode W. All authors reviewed the results and approved the final version of the manuscript.
-
There are no original data associated with this article. Referenced data are available in the literatures.
This research was supported by the 'Next Generation Cassava Breeding Project', OPP1175661, funded by the Bill & Melinda Gates Foundation and the Department for International Development of the United Kingdom Agreement, and 'Genetic Improvement in Cassava' funded by the United States Agency for International Development (USAID). We would like to thank Dr. Hernan Ceballos, Dr. Clair Hershey, Dr. Carlos Iglesias, Dr. Giovanny Covarrubias Pazaran, and Dr. Marlee R. Labroo for their valuable insights on various aspects of the manuscript. We also want to thank the reviewers for their constructive suggestions and corrections, and we acknowledge the active role of the editor during the revision of our manuscript.
-
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Xiaofei Zhang is the Editorial Board member of Tropical Plants who was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of this Editorial Board member and the research groups.
-
Received 27 February 2024; Accepted 8 May 2024; Published online 2 August 2024
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press on behalf of Hainan University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Zhang X, Holley R, Egesi CN, Gemenet DC, Moreta D, et al. 2024. Towards transforming cassava breeding: harnessing inbred-parent-based hybrid breeding strategies. Tropical Plants 3: e025 doi: 10.48130/tp-0024-0024
Towards transforming cassava breeding: harnessing inbred-parent-based hybrid breeding strategies
- Received: 27 February 2024
- Revised: 28 April 2024
- Accepted: 08 May 2024
- Published online: 02 August 2024
Abstract: Genomics-assisted breeding has significantly improved recurrent selection in cassava. However, challenges persist with the use of heterozygous parents, hindering efficient trait introgression to meet the needs of ever-changing markets and environmental conditions. To address this, we propose an innovative approach – inbred-parent-based hybrid cassava breeding, aiming to transform cassava breeding by implementing backcrossing-based trait introgression, effectively purging deleterious mutations, and systematically exploring and utilizing heterosis. This perspective paper discusses the key drawbacks of heterozygous parent-based recurrent selection and outlines how the proposed approach overcomes these challenges. By leveraging the self-compatibility of cassava and advanced technologies like flower-inducing and doubled haploid technologies, along with genomics advancements and a global network, cassava breeding programs can achieve efficient, cost-effective, and accelerated inbred-parent-based hybrid breeding. In conclusion, we emphasize four crucial action areas to focus on for the initial phase to realize this transformation, i.e., understanding inbreeding depression, developing inbred or doubled haploid parents, purging genetic load, and identifying or creating heterotic pools. Through collective efforts and global collaboration, inbred-parent-based hybrid cassava breeding will transform cassava breeding and production, ensuring resilience and adaptability to significantly contribute to ending hunger and reducing poverty during the climate crisis.
-
Key words:
- Cassava /
- Breeding /
- Doubled haploid /
- Genomics /
- Inbreeding depression












