-
Tea plant is a perennial woody economic crop plant. Tea is preferred by people around the world due to its unique flavour[1]. Tea has an abundance of secondary metabolites which endows tea infusion taste, aroma, and color, as well as providing multiple health benefits[2]. Thus, tea science research has become increasingly popular. In the past decade, much progress has been made in the study of tea plant functional genomics, tea plant origin and genetic diversity, and the formation mechanism of tea characteristic secondary metabolites[3]. However, due to the limitations of genetic transformation, we cannot overexpress or knockdown genes in tea plant. This problem has hindered tea plant biology studies. Understanding the cell type of gene expression is critical for revealing the gene functions in biological processes.
To detect the cell type of gene expression, one of the following methods are usually used: 1) generating transgenic plants with gene promoter-reporter fusions[4]; 2) combining the isolation of single cells or single cell contents and qPCR detection of genes[5]; 3) using the hybridization of RNA/cDNA in situ technique[6]. Each of these three approaches has its advantages and disadvantages. The first method is able to detect the cell type of gene expression in living plants and can allow observation over a long period of time. However, this method has a limited scope of application which is not suitable for some plants that cannot be genetically modified[4]. The second method, although is suitable for most plants, it is a method of indirectly obtaining information about the cell type of gene expression[5]. The third method, based on in situ hybridization techniques, can intuitively show the cell type of gene expression, is suitable for most plants, and does not depend on plant transgenic systems. But this approach has limitations that require specific probes that bind efficiently[6]. After the advent of in situ hybridization techniques, researchers combined in situ hybridization with PCR techniques to develop in situ PCR[7]. In situ PCR not only has the technical advantages of in-situ hybridization, but also importantly this technology does not require the synthesis of special detection probes. In situ PCR only requires a pair of primers that can specifically amplify a gene fragment to detect gene expression patterns[7].
Although in situ PCR technology has been around for a time, it is not yet mature in some plants, including tea plants, indicating that the method still needs to be optimized for application in tea plant tissues. In our recent studies, we successfully applied this method in tea root tissues. Based on single cell transcriptome of tea roots, we identified epidermis cells, cortex cells, endodermis cells, pericycle cells, xylem, and phloem cells[8]. In situ RT-PCR results and single cell transcriptome data have consistency. Here, we comprehensively describe the in situ RT-PCR method of tea plant root. Just as all techniques have limitations, in situ PCR also has disadvantages such as false positives. This disadvantage can however be avoided. Suggestions to solve the problems that may arise in the experiments are also provided.
-
The detailed descriptions of the reagents, equipment, and setup required for the experiment are shown in Supplementary File S1.
Procedure
-
Step 1 and 2: Sample preparation and fixation
The experimental material was primary root of a 2-month-old 'Shuchazao'.
(1) The young root tissue was cut into pieces and soaked in fresh FAA solution.
(2) The sample was evacuated for 15−20 min, then put at 4 °C for 12 h.
Step 3 and 4: Embedding and sectioning
(3) The samples were eluted three times for 10 min each in a mixture solution (63% ethanol and 5% acetic acid), and two times for 5 min each in 1× PBS.
(4) Embedded root tissue in 5% low-melting point agarose (agarose dissolved in 1× PBS).
(5) The embedded blocks were sectioned using Leica RM2255 microtome. Root section is 30 to 50 μm thick. Each section was put in a separate 0.2 ml centrifuge tube for independent test. After that, the experimental procedures of steps 5−9 were carried out in 0.2 ml centrifuge tubes.
Step 5 and 6: Removing genomic DNA
(6) The root tissue sections were separated from agarose by rinsing twice with RNase-Free water.
(7) Added 3 μg/ml proteinase K, and left at 25 °C for 30 min. Heat at 85 °C for 2 min to inactivate proteinase K,
(8) Wash once with 1 × PBS and RNase-Free water for 5−10 min, separately. Added 1 U/μl DNase I at 37 °C for 20 min or overnight.
Step 7: Reverse transcription
(9) Removed liquid and added 15 mM EDTA (pH 8.0) at 75 °C for 10 min.
(10) Washed 1−2 times with RNase-Free water to remove excess liquid.
(11) Reverse transcription reaction was performed using the PrimeScript II 1st strand cDNA Synthesis Kit.
Step 8: In situ PCR
(12) Carried out liquid and washed once with RNase-Free water.
(13) Mixed tissue sections and reaction reagents: 2 μl 10× Tag DNA polymerase buffer, 0.4 μl Tag DNA polymerase (5 U/μl), 1.6 μl dNTP, 0.32 μl DIG-11-dUTP (25 nmol), 1 μl Forward Primer, 1 μl Reverse Primer, 0.6 μl MgCl2 (1.5 mM) and 13.08 μl ddH2O.
(14) PCR program: 95 °C for 30 s, 95 °C for 30 s, 55 °C for 50 s, 68 °C for 45 s, 30−35 cycle, 68 °C for 5 min 10 °C.
Step 9: Immunoassay
(15) Wash twice with 1× PBS for 5 min each.
(16) Block with confining liquid (5% skim milk) for 30 min.
(17) Anti-Digoxigenin-AP was diluted in confining liquid at 1:500, 50 μl was added and stand for 1 h.
(18) Wash twice for 15 min each time using 10× Washing Buffer.
(19) Staining was performed using BM purple AP substrate, precipitating for 30 min or 1 h.
Step 10: Microscopy
(20) Washed twice with RNase-Free water. Then, placed sections on slides and observed under a microscope and photographed.
-
The main steps of in situ RT-PCR include sample (tissue) preparation, fixation, embedding, sectioning, proteinase K treatment for digestion, gDNA removing, reverse transcription, PCR amplification, immunoassay, and microscopy imaging (Fig. 1).
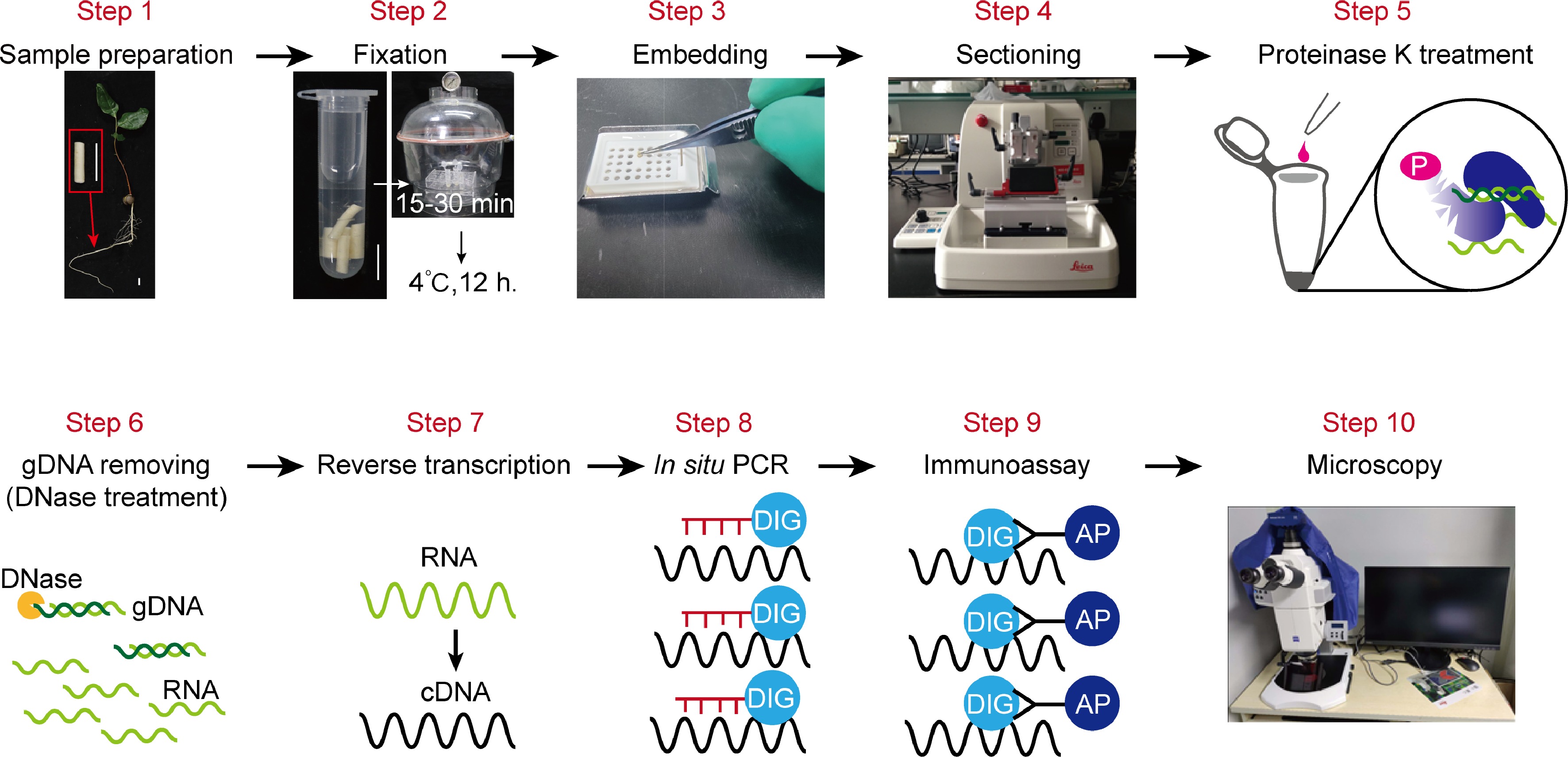
Figure 1.
An overview of the in situ RT-PCR procedure. This illustration summarizes all the steps to carry out experiment. This includes: Step 1, Sample preparation; Step 2, fixation; Step 3, embedding; Step 4, sectioning; Step 5, proteinase K treatment for digestion; Step 6, Genomic DNA removing through DNase treatment before reverse transcriptase-PCR (RT-PCR); Step 7, reverse transcription; Step 8, PCR-amplification of target gene fragments; Step 9, immunoassay; Step 10, imaging of sections by using microscopy.
Tissue preparation
-
Tender or moderately tender tea plant roots are recommended as experimental tissue. Tea plant roots with high lignification have thick cell walls, making reagent penetration into cells difficult. Sample fixation is the first step of this experiment. The choice of fixation solution should be based on the type of sample. Formaldehyde-acetic acid-ethanol (FAA) is a common fixative. Alcohol and formaldehyde can induce tissue contraction, whereas glacial acetic acid has the propensity to cause tissue expansion[9]. For young tissue, as low a concentration of alcohol was chosen to avoid material shrinkage. In the experimental system, FAA solution that contained 63% alcohol was chosen for fixing tea plant root.
Common tissue embedding methods include paraffin embedding[10], frozen embedding[11], and agarose embedding[12]. Compared with frozen or agarose-fixed sections, paraffin-fixed sections could maintain the best morphology of the tissue undergoing the PCR process. However, the RNA in paraffin-fixed sections is generally not as stable as with agarose-fixed sections. Furthermore, paraffin embedding and frozen embedding require pre- and post-treatment steps and specialized equipment, while agarose embedding dose not. Therefore, agarose embedding is an efficienct and easy way to fix sections in this experiment. Embedding should prevent root tissue exposure, which will increase the risk of root RNA degradation. Thus, cutting the appropriate root length according to the specification of the embedded box. Normally, agarose-fixed sections of tea plant root can be preserved for one week at 4 °C.
Proteinase K digestion
-
Digested tissue cells with protease K can increase cell permeability[13], allowing all components of the reaction system to enter the cells fully and effectively expose the target sequences for amplification. The appropriate protease concentration is crucial for the success of the experiment and the reliability of the results. If the concentration is too low, digestion is incomplete and cell permeability cannot be effectively increased. Conversely, if the concentration is too high, excessive digestion will lead to the loss of cell permeability, and high background or false positive. Three to six mg/ml protease K for protease digestion is recommended in tea plant root. It is important that proteases must be inactivated by overheating after digestion.
Primer design
-
Although in situ PCR does not require special labeled probes, it is still necessary to design primers for specific amplification. The design principles of in situ PCR primers generally follow those of quantitative PCR primers[14]. The length of primer is 18–22 bp; amplified fragment is 180–230 bp and not more than 300 bp; G/C content of 40%–60%. During the PCR amplification extension, in order to guarantee that the amplified fragment can represent the transcription level of the gene, the short fragment of cDNA can be fully amplified and the long fragment of gDNA cannot be amplified. Therefore, the forward and reversed primers are respectively on two exons. Single primer is ideally capable of being designed across exons (Fig. 2).
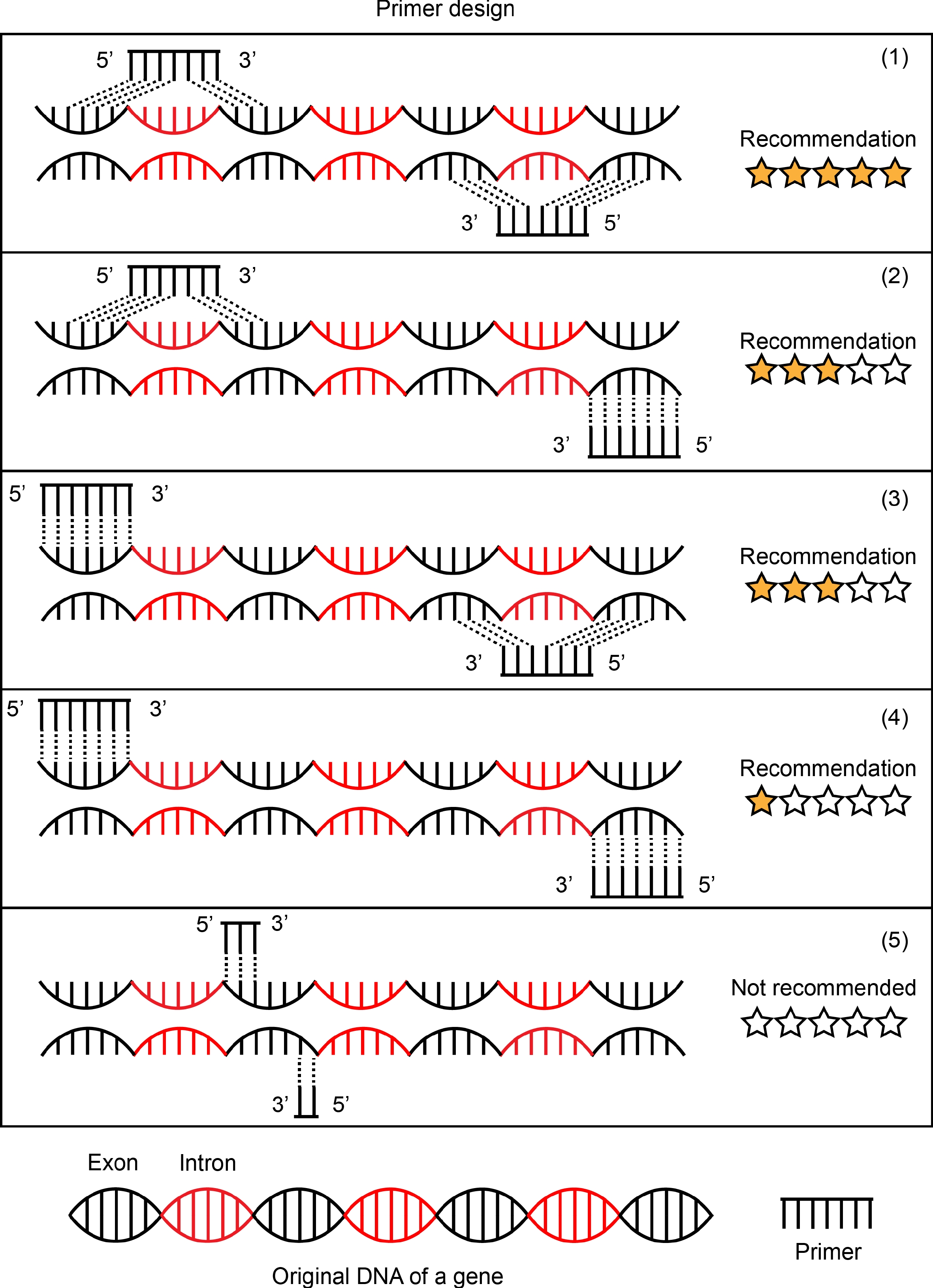
Figure 2.
Schematic representation of primer design. Five forms of primer design are listed in this picture. (1) Forward primer and reverse primer targeted mRNA sequences by designing the primers to span introns in the genomic DNA. This is an ideal primer design and is recommended. (2) and (3) are common forms of primer design and are recommended in most cases. Forward primer and reverse primer, one of two, to be designed to targeted mRNA sequences and span introns in the genomic DNA. The primer design of (4) is chosen if there is no better design scheme of a target gene. Forward primer and reverse primer were designed to target two exon sequences separated by a distance respectively. (5) Forward primer and reverse primer were designed in the same exon sequences. This is not to be recommended.
Reverse transcription
-
To ensure the transcription level of the gene reflected by PCR amplification, gDNA is removed using DNase prior to cDNA synthesis. Following this, the mRNA is converted to cDNA via reverse transcription. There are two ways to choose in reverse transcription reaction based on random primer or specific primer. Random primer is a set of very short random sequence oligonucleotides that are 'hexamers' only six bases long. Random primer converted almost all RNA molecules (mRNAs, rRNA, and ncRNAs) into cDNA. Sometimes, we could choose Oligo (dt) primer instead of random primer in order to convert only mRNA into cDNA. Specific primer converted only the gene fragment of interest from the mRNA into cDNA. Then, in the following PCR using either a pair of nested gene-specific primers to amplify a fragment of this transcript. The specific primer-based method is useful for detecting some genes with low expression levels. In most cases, the Oligo (dt) primer- or specific primer-based methods in reverse transcription reactions were chosen.
PCR
-
In situ PCR is different from conventional PCR, DIG-11-dUTP labeled probe needs to be added into the reaction system[15,16]. During the PCR reaction process, the target fragment is labeled with digoxin, which is convenient for subsequent in situ detection[15,16]. In PCR reaction, the concentration of magnesium ion has an important effect on the efficiency and specificity of the reaction[17]. Low concentrations of magnesium ions can cause the polymerase to be unable to stabilize on the DNA template, thus affecting the amplification effect. However, high concentration of magnesium ions will increase the production of non-specific products and reduce the specificity of PCR products. In addition, considering that the amplified efficiency of PCR in tea plant tissue is not as high as it is in solution and some transcripts exhibit low levels of expression, the cycle number can be appropriately added. However, it should be noted that a higher cycle number may cause the amplification of non-specific fragments. Meanwhile, excessive number of cycles results in prolonged heating of root tissue to heat, leading to greater damage and detachment or separation of tissue in subsequent experimental processes. Therefore, to obtain the best PCR reaction conditions, the magnesium ion concentration, cycle number, annealing temperature, etc. needs to be optimized. Here, we provided an optimized reaction conditions for in situ RT-PCR of tea plant root in the 'Procedure' section.
Strict repetition and control are necessary for evaluating experimental results of in situ RT-PCR. To confirm the expression pattern of a gene, in situ RT-PCR was performed based on not less than four technical replicates and three independent experiments. A negative control setting is that using water or single gene primer replaced a pair of gene primer in PCR. A positive control setting is based on the amplification of 18S transcript. 18S transcript is widely expressed in all cells of most plant species. The negative control is used for testing the signal of digoxin is true or not when root section has a gene expressed signal. The positive control is used for testing that the experimental conditions are reasonable or not when root sections don't result in any signal. Sometimes, a correct negative control is a better indicator of the reality of in situ RT-PCR than a positive control when root sections display a gene expressed signal. Thus, a negative control is indispensable in every time independent experiment.
Immunoassay
-
Use sealing fluid for sealing to avoid high background and non-specific banding. Alkaline phosphatase antibodies are bound to target proteins, stained, and imaged under a microscope. Control the dyeing time within 1−1.5 h, too long dyeing will lead to high background. After a series of operations, the final tissue section of the imaging is fragile and extremely prone to rupture. Therefore, 50%−70% glycerin can be used for sealing when imaging. Because the liquid surface tension of glycerin is smaller than that of water, it can effectively protect the section from rupture due to large tension when pressing the tablet.
Possible problems and solutions
Poor morphology
-
High quality morphology of tissue sections makes the result and image of in situ RT-PCR clear. Several possible causes and solutions for poor morphology are as follows. (1) The adhesion between the root tissue and agarose is not close enough, causing that root tissue in embedding block occurs displacement when cutting sections. During tissue embedding, agarose, and root tissue will be closely jointed if the temperature is controlled at about 40 °C. To fix this problem, we can utilize the temperature control platform to ensure the stability of temperature when embedding. (2) Excessive proteinase digestion and thermocycling causes cell structure change and terrible tissue morphology. Adjusting protease K concentration appropriately or shortening the digestion time. The number of PCR cycles should not exceed 35, 28−32 is preferable. (3) The action of removing or adding liquid is not gentle enough. After proteinase digestion and PCR thermocycling, the root sections become thin and brittle. Thus, we should add or remove liquid carefully and slowly and along the wall of the tube.
High background, false positive, and nonspecific amplicons
-
High background, false positive and nonspecific amplicons may arise in following situations: excessive proteinase digestion leads to loss of cell permeability; gDNA is not removed cleanly and the sealing is not complete; unreasonable primer design; long-time dyeing; too much residual reaction liquid exists in the tube. In order to address this problem, we propose several recommendations. Firstly, conducting the necessary pre-experiment to find out the appropriate concentration of protease K, and adjusting it according to the degree of lignification of tissue cells. For tissue with low lignification degree, reducing the concentration of protease K. Besides, we may explore the substitution of protease K with other proteases (such as trypsin) or consider employing a combination thereof for better inactivation and to prevent excessive proteinase digestion. Secondly, although experimental steps to remove gDNA are included in several reverse transcription kits, it is recommended that DNase treatment alone is necessary before reverse transcription. Sealing with 5% skim milk on the basis of 0.5% BSA, or appropriate extension of the sealing time. Thirdly, checking whether the primer design is specific, and design multiple pairs of primers for experimental comparison. Furthermore, controlling dyeing time limit to one hour. But if the experimental steps before dyeing are standard, long-time dyeing should not result in high background value. Lastly, when removing liquid from tubes before proceeding with subsequent experimental steps, complete removal of liquid should be ensured as this detail can easily be overlooked during experiments.
Weak signal
-
The expression levels of several genes in root are low or there is RNA degradation in root, which likely results in a weak signal from in situ RT-PCR results. To address this problem, we can increase PCR cycle number or lower the annealing temperature. Additionally, it is essential to perform certain steps on ice as outlined in the 'Procedure' section. Furthermore, increasing the concentration of protease K and improving cell permeability facilitate easier entry of regents into root cells, thereby maximizing their effectiveness.
An example of in situ RT-PCR in tea plant roots
-
Next, applying the above mentioned method of in situ RT-PCR, we detected expressed patterns of GLABRA 3 (GL3) and CATIONIC AMINO ACID TRANSPORTER 2 (CAT2) encoding genes in tea plant root cell. We took two different ways to design the detected primers of CsGL3 (CSS0032319) and CsCAT2 (CSS0004634) (Fig. 3 & Supplementary Fig. S1). CsGL3 are homologs of epidermal cell marker gene AtGL3[18]. In Arabidopsis thaliana, epidermal cells include trichoblasts and atrichoblasts[18,19]. AtGL3 is expressed in trichoblasts of epidermal cells[18,19]. CsGL3 transcript is detected by forward primer 5'-CGATGACTGTGGTATGCTTCC-3' and reversed primer 3'-CTGTTGATGAAGTCTGGTCCTG-5' (Fig. 3a). The results of in situ RT-PCR showed that the expression of CsGL3 is discontinuous in epidermal cells (Fig. 3b & Supplementary Fig. S1), where its expression pattern is similar to AtGL3[18]. Our previous studies reported that CsCAT2 localized in tonoplast and may mediate theanine storage in tea plant roots[20]. Recently, we annotated cell types of tea plant root based on single cell RNA sequencing and identified CsTSI was specifically-expressed in pericycle cells of tea plant root. At the same time, we found CsCAT2 and CsTSI were highly expressed in the same cell cluster according to the results of single cell RNA sequencing. This means that CsCAT2 was also likely expressed in pericycle cells[8]. In addition, CsCAT2 also expressed in cluster 4 that was annotated as cortex or endodermis cell. We used forward primer 5'-TCCTGATACTCCCATTTCTTCTGC-3' and reversed primer 3'-GCTCTTGATAGGAACTTGGGTTCG-5' to detect CsCAT2 transcript in tea plant root (Fig. 3a). The present experimental results suggested that CsCAT2 was expressed in pericycle, cortex, and endodermis cells (Fig. 3b & Supplementary Fig. S1), which is consistent with the results of single cell RNA sequencing[8]. AMINO ACID PERMEASE 4 (CsAAP4, CSS00133085) encoding a theanine transporter that barely expressed in the root of tea plant[8,21]. As expected, the expression of CsAAP4 in root cells was not detected, which is consistent with the negative control (Fig. 3 & Supplementary Fig. S1). The presence of 18S rRNA transcript, a positive control, was however observed in all cells (Fig. 3). These results suggest that the validation and controls are reliable.
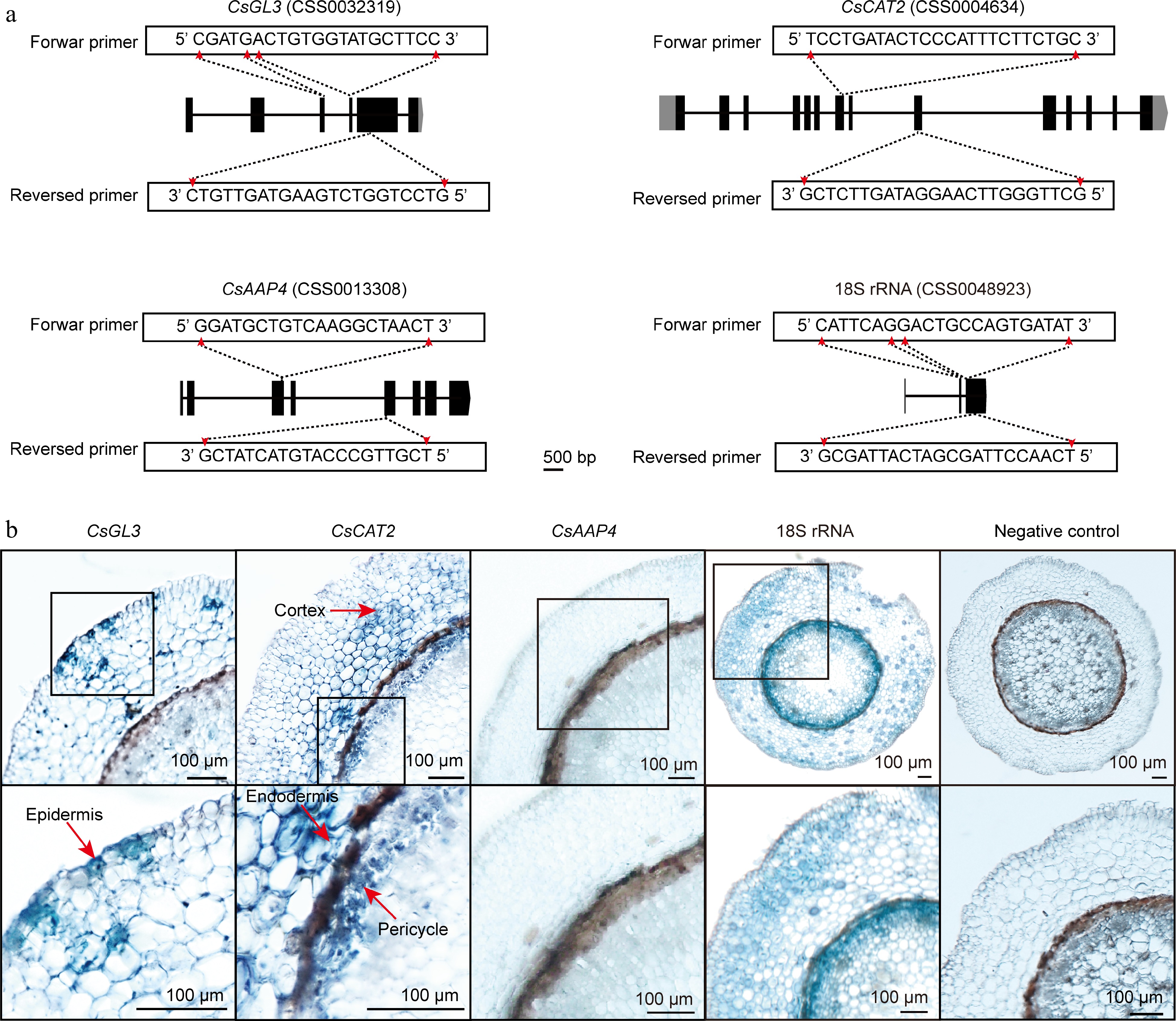
Figure 3.
In situ RT-PCR detection of genes mRNA in tea plant root sections. (a) Primer design of CsGL3, CsCAT2, CsAAP4, and 18S rRNA. (b) The blue areas of sliced tissue represent regions where CsGL3, CsCAT2, CsAAP4, and 18S rRNA are expressed. The black box represents the magnified areas of root sections. Red arrows point to the cell type in which the gene is expressed. Using water replaced gene specific primers in PCR as negative control. Scale bar = 100 μm. Each sample is an independent root section and to be detected in separate 0.2 ml centrifuge tubes.
-
In recent years, people have tried to identify the function of a gene in vivo using hair roots, antisense oligonucleotide silencing, and virus-induced gene silencing (VIGS) technologies. However, many studies lack experimental evidence of the cell type of a gene expressed in vivo. In this study, the method of in situ RT-PCR in tea plant root to detect gene expression is described in detail. Possible problems in experiments are listed and suggested solutions provided to solve these problems. The present results showed that this method can easily and efficiently characterize cell heterogeneity of gene expression in tea plant root. In future research, this method will be further improved so that it can be applied to more tea plant tissues, such as leaves, stems, and roots with higher lignification.
This work was supported by the National Natural Science Foundation of China (32072624), Anhui Provincial Major Science and Technology Project (202103b06020024) and Anhui Educational Committee Excellent Youth Talent Support project (gxyqZD2022018).
-
The authors confirm contribution to the paper as follows: study conception and design, manuscript fianlizing: Zhang Z, Wang X, Zhang S; materials collection, experiment conduction: Lin S, Hu X; draft manuscript preparation: Lin S. All authors reviewed the results and approved the final version of the manuscript.
-
All relevant data in this study are provided in the article and its supplementary file.
-
The authors declare that they have no conflict of interest.
- Supplementary File S1 Supplementary information of reagents, equipment and reagent setup.
- Supplementary Fig. S1 In situ RT-PCR replicates. (a) The global perspective of root sections in figure 3. (b) Replicated samples of in situ RT-PCR of tea plant root. The black box depicts the enlarged regions of root sections, while red arrows indicate the cell types expressing the gene. Scale bar = 100 μm.
- Copyright: © 2025 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Lin S, Hu X, Zhang Z, Wan X, Zhang S. 2025. A simple and efficient in situ RT-PCR method for detecting gene expression in specific cells of tea plant (Camellia sinensis) root. Beverage Plant Research 5: e001 doi: 10.48130/bpr-0024-0033
A simple and efficient in situ RT-PCR method for detecting gene expression in specific cells of tea plant (Camellia sinensis) root
- Received: 31 July 2024
- Revised: 02 September 2024
- Accepted: 11 September 2024
- Published online: 09 January 2025
Abstract: Currently, the field of tea plant biology is rapidly advancing, with numerous significant scientific inquiries being raised and investigated. Meanwhile, a substantial number of functional genes have been reported. However, due to the lack of certain in vivo validation techniques, much of the expression information for these functional genes is at the tissue level in tea plants and remains unclear at the cell-type level. In this study, an in situ PCR method for detecting gene expression heterogeneity in tea plant root cells is presented. A detailed description of the procedure and precautions involved in this method is provided and suggestions offered for addressing potential experimental challenges. Finally, the expression patterns of CsGL3, CsCAT2, and CsAAP4 in tea plant root cells were taken as examples. The present results showed that CsGL3 was predominantly expressed in root epidermal cells, while CsCAT2 shows strong expression in pericycle and cortex. The expression of CsAAP4 was not detected in root cells. These findings are consistent with previous reports, indicating that this method is feasible for the detection of gene expression patterns in tea plant root cells.
-
Key words:
- In situ RT-PCR /
- Gene expression /
- Camellia sinensis /
- CsGL3 /
- CsCAT2 /
- CsAAP4














