-
Highland barley belongs to the family Poaceae. It is rich in nutrients and has a high content of fiber, vitamin, protein, and a low fat[1]. Among them, dietary fiber (DF), as an active substance, plays a very important role in the regulation of the human body. Dietary fiber includes soluble dietary fiber (SDF) and insoluble dietary fiber (IDF). Examples of SDF are some gums, such as pectin, gum Arabic, guar gum, and glucan, and include some biological polysaccharides and synthetic polysaccharides. IDF includes some components of the structure of cell walls, such as cellulose, hemicellulose, and lignin[2]. Compared with IDF, SDF exhibited many functional advantages[3], such as decreasing blood pressure[4], lowering blood sugar[5], lowering blood lipids[6], and regulating gut flora[7]. In highland barley, IDF accounted for the major proportion of DF and the proportion of SDF was low. Thus, finding an effective method to increase the SDF content of highland barely whole grain flour and improve its functional activity is required.
At present, there are many methods used to increase the SDF content of grains, including physical, chemical and microbial fermentation methods[8]. Among which, microbial fermentation is of interest. Studies had shown that fermentation could significantly increase the content of SDF. Jia et al.[9] used Trichoderma viride to ferment defatted rice bran and found that the yield of SDF increased significantly from 10.5% to 33.4%, and its various functional properties were obviously enhanced. Wang et al.[10] used Penicillium sp. Cis16 to ferment citrus pulp, and the SDF reached a high level after fermentation, from 6.77% to 36.56%. Li et al.[11] fermented millet bran with lactic acid bacteria, and the yield of SDF increased from 4.2% to 7.6% after fermentation, and the odor intensity and flavor characteristics also changed significantly. Chu et al.[12] fermented millet bran with Bacillus natto, and found that the SDF content increased from 2.3% to 13.2%, and the SDF/IDF ratio increased from 3.1% to 19.9%, while the increase in SDF content was based on the production of enzymes during microbial fermentation. Cellulase is a multicomponent enzyme complex containing mainly endo-β-1,4-D-glucosidase, exo-β-1,4-D-glucanase and β-1,4-D-glucosidase[13], which could effectively catalyze the decomposition of cellulose to invert IDF to SDF[14,15]. However, the enzymatic hydrolysis process of cellulase is hindered due to the complex substrate formed by lignin, cellulose and hemicellulose and the high crystallinity of cellulose[16]. Enzymes that could effectively degrade lignin include laccases and peroxidases (lignin peroxidase (LiP), manganese peroxidase (MnP), and versatile peroxidase (VP))[17]. For the complete hydrolysis of insoluble cellulose and lignin to improve the proportion of SDF in total dietary fiber (TDF), synergistic action between the cellulase and ligninase is required. Present research on the fermentation strain is focused on cellulases or ligninases-produing strains[13,18], studies on fermentation strains producing both cellulase and ligninase is relatively rare. Thus, screening and breeding for new lignin and cellulose degrading strains with high effectivity is a suitable method for raising the SDF proportion in TDF through fermentation and improving the added value of agricultural by-products.
Bacillus amyloliquefaciens[19], Bacillus glucosolvans[20], Bacillus velezensis[21], Bacillus methylotrophicus[22] and other Bacillus species[23,24] have all been researched for their function in the breakdown of lignocellulose. However, these strains producing both ligninase and cellulase are generally used for environmental treatment or industrial product production, and are rarely used for fermenting grains to improve its nutritional quality. Therefore, the purpose of this study was to screen and isolate safe and high effective lignin and cellulose degrading strains and investigate the effect of fermentation treatment on SDF content in highland barely whole grain flour. This study will develop a new application area of lignin and cellulose degrading strains and provide a technical basis for the beneficial utilization of cereals and their byproducts.
-
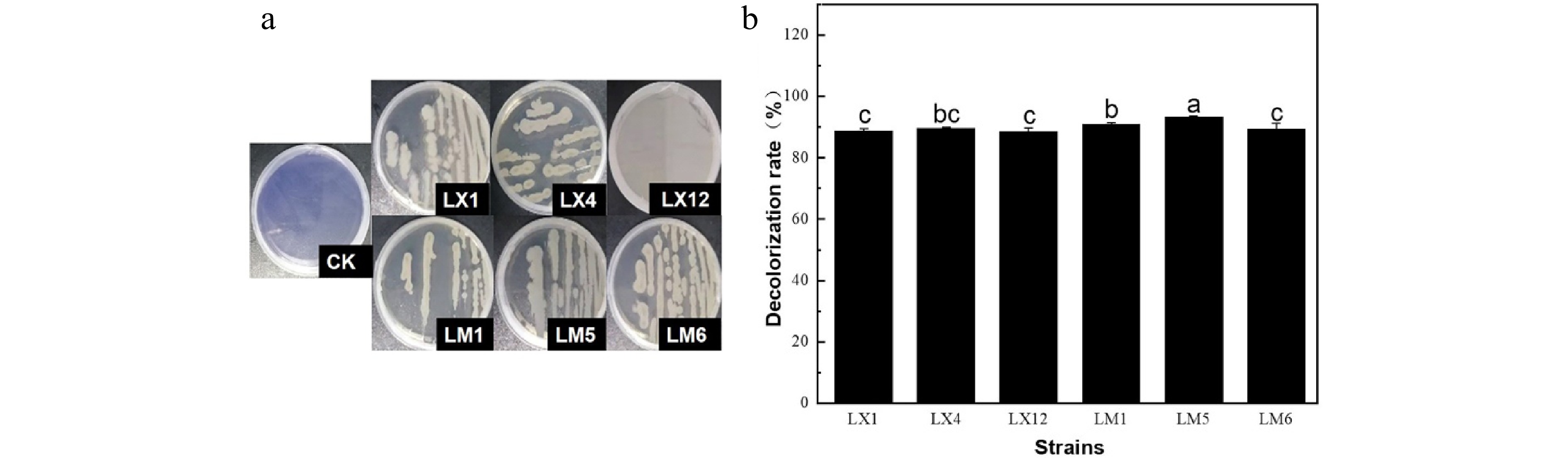
In order to obtain high efficiency lignin and cellulose degrading strains, lignin or cellulose enrichment medium were used and 20 strains were obtained (eight lignin degrading strains and 12 cellulose degrading strains). The diameter ratio of transparent hydrolysis circles to single colony and dye decolorization ratio were used to preliminarily analyze the lignin-cellulose degradation ability of strains[25]. Through aniline blue dye fading, Congo red dye fading circles, filter paper disintegration and cross screening, six strains were obtained and named as LX1, LX4, LX12, LM1, LM5, and LM6, which could degrade both lignin and cellulose. Figure 1 showed that six strains could effectively fade the color of the aniline blue plate. Among them, strains LM1, LM5, and LX4 presented greater decolorization rates, 90.8%, 93.4%, and 89.6%, respectively, indicating their potential to degrade lignin.
The cellulose degradation ability of six strains was evaluated according to the ratio of transparent circle diameter to single colony diameter[26], which was positively proportional to the cellulase-producing capacity of the strain. Results are presented in Table 1. Strains LM1, LM6, and LX4 presented larger ratio of transparent circle diameter to single colony diameter of 5.66, 5.76, and 5.16, respectively, indicating their high cellulase-producing capacity. The results of the filter paper disintegration test further demonstrated that the three strains could efficiently decompose the filter paper, verifying their high cellulase-producing capacities.
Table 1. Congo red transparent circle and filter paper strip disintegration effects of different strains.
Strains Diameter of colony (D/mm) Diameter of transparent circle (d/mm) d/D Degree of disintegration LX1 4.23 ± 0.01b 14.62 ± 0.21d 3.46 ++ LX4 4.61 ± 0.18a 23.79 ± 0.64b 5.16 ++++ LX12 3.55 ± 0.05c 18.07 ± 0.09c 5.10 +++ LM1 4.77 ± 0.01a 27.02 ± 0.02a 5.66 +++++ LM5 4.29 ± 0.04b 9.25 ± 0.52e 2.16 + LM6 4.84 ± 0.18a 27.83 ± 1.39a 5.76 ++++ Lowercase letters represent significant differences in the same column. '+' represents the degree of damage to the filter paper, and the more '+', the stronger the degree of damage. Rescreening of a lignocellulose degrading strain
-
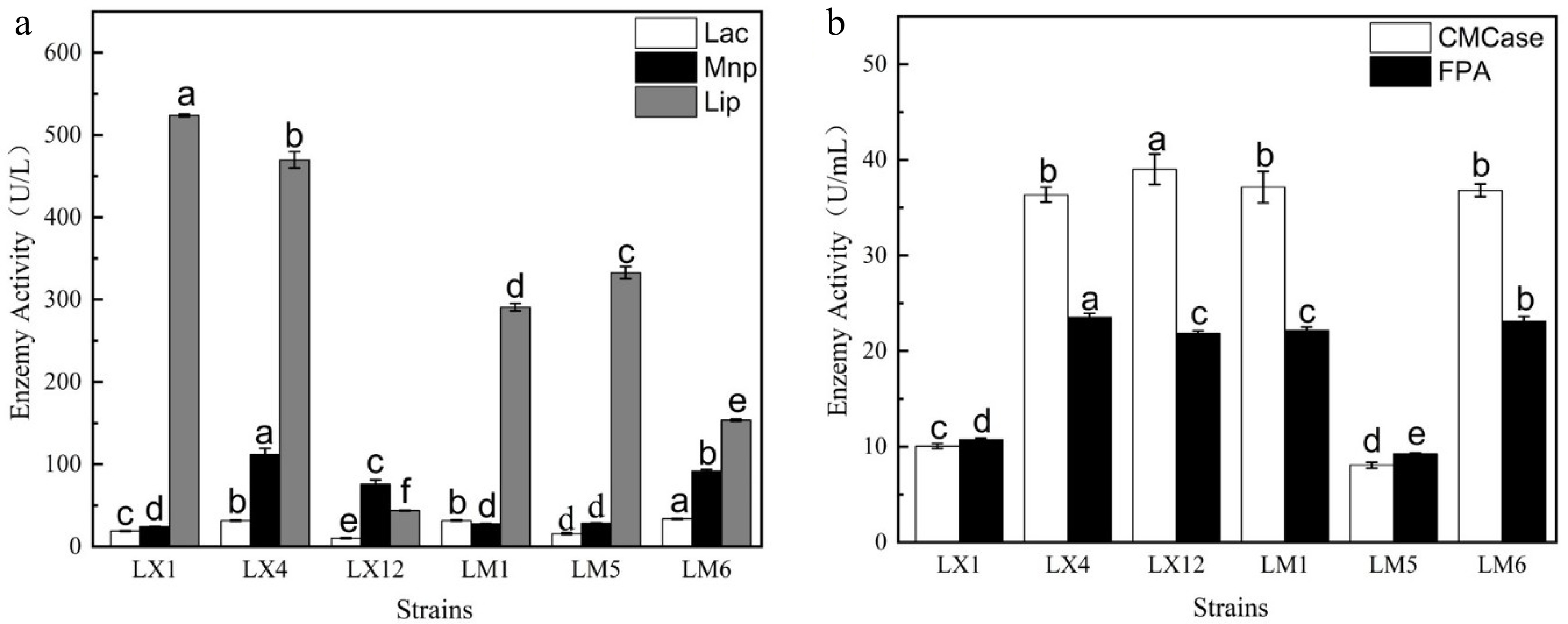
Enzymes play a key role in the degradation of lignocellulose. By monitoring its enzymatic activity, the measurement of ligninase and cellulase activities, including Lac, Lip, Mnp, CMCase, and FPA was used to quantitatively analyze the lignocellulose degradation ability of the screened strains[19]. Figure 2 displayed the enzymatic activities including Lac, Lip, Mnp, CMCase, and FPA. Both strains LX1 and LM5 showed lower ligninase and cellulase activities. Strains LM6 and LX12 presented high cellulase activities and low ligninase activities. Strain LX4 had the highest level of lignocellulase activity, followed by strain LM1. However, considering the comprehensive degradation effect of aniline blue, sodium carboxymethyl cellulose and filter paper in the primary screening, the overall activities of LM1 was the best. So, LM1 was selected as the final experimental strain. The enzyme activity of Lac, Mnp, Lip, CMCase, and FPA produced by strain LM1 reached 31.68 U/L, 27.78 U/L, 290.8 U/L, 37.16 U/mL, and 22.16 U/mL respectively. All the strains examined by Zhang et al.[27] produced lower enzymatic activities of Lac, Mnp, and Lip than that of strain LM1. When fermented for a longer period of time, the CMCase and FPA activities for the strains examined by Dobrzyński et al.[28] reached 0.617 U/mL and 0.903 U/mL, respectively, which were far less than that of the strain LM1 examined in the present study. The outcomes demonstrated that strain LM1 could produce high level lignocellulase to break down lignocellulose.
Strain identification
Colony and cell morphologies
-
Strain LM1 was spread over an LB agar plate and incubated at 30 °C for 12 or 24 h. The strain grew well on LB plates and produced white colonies. Results showed that, when cultured for 12 h (Fig. 3a), the colony edge was irregular, the surface was smooth and protruding, and the texture was sticky. When cultured for 24 h (Fig. 3b), the colony edge was irregular, the surface was wrinkled and flat, and the texture was dry. Gram staining indicated that the selected strain LM1 was a Gram-positive, rod-shaped bacterium with a length of 0.94−1.13 × 4.42−10.47 µm (width × length) (Fig. 3c). Spore staining showed that strain LM1 could form dormant spores (Fig. 3d).
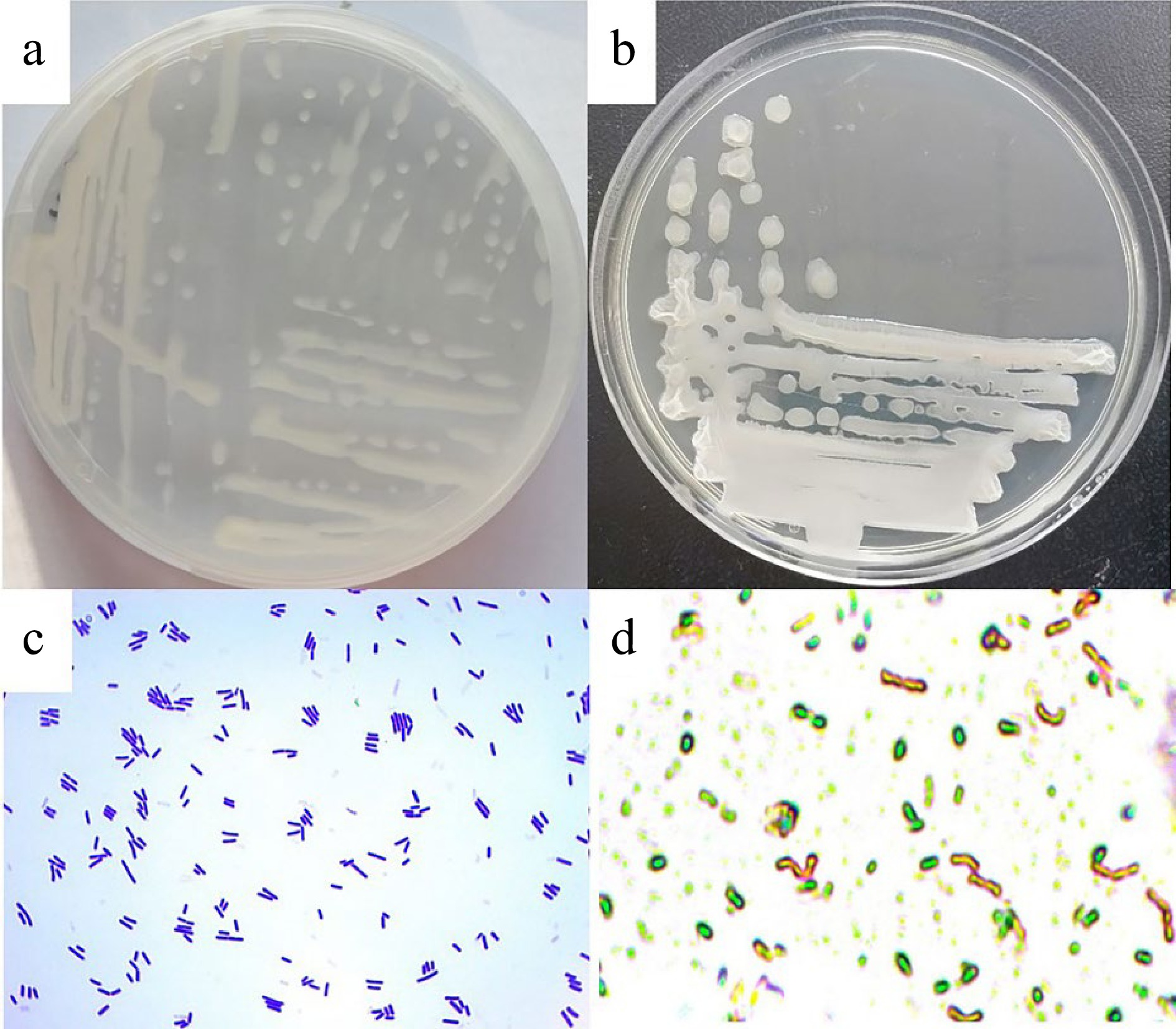
Figure 3.
Colony, cell morphologies and spore of the isolated ligninase and cellulase-producing strain LM1. (a) Colony morphology 12 h, (b) colony morphology 24 h, (c) cell morphology, (d) spore.
Physiological and biochemical identification
-
Physiological and biochemical tests showed that strain LM1 had the ability to produce catalase and oxidase, use starch and gelatin but the methyl red test was negative (Supplemental Table S1). Based on the morphological, biochemical and physiological characteristics, strain LM1 belonged to the genus Bacillus.
Molecular biological identification
-
Further identification was performed using 16S rRNA and gyrA gene sequencing. The 16S rRNA gene sequences (1348 nt) were subsequently compared with sequences available in the GenBank database. A phylogenetic tree based on16S rRNA gene sequences was constructed and shown in Fig. 4a. Strain LM1 was closely related to B. velezensis LHNL1 (99.93%), B. velezensis JE3 (99.93%), B. amyloliquefaciens 262XY1 (99.93%), B. amyloliquefaciens LXZ (99.86%), B. amyloliquefaciens LD5 (99.86%), Bacillus sp. G1 (99.86), B. amyloliquefaciens HS8 (99.86), B. velezensis EH9 (99.79), Bacillus sp. Z44 (99.79%), Bacillus sp. DYJL30 (99.86%), B. vallismortis 263XY1 (99.86%), and B. amyloliquefaciens Y6 (99.86%). The results confirmed that strain LM1 belonged to the genus Bacillus. A further gyrA (DNA gyrase subunit A) gene sequence analysis was conducted to clarify the degree of strain LM1's relatedness with its nearest phylogenetic neighbors. The phylogenetic analysis of gyrA gene sequence (949 nt) positioned the strain LM1 within the blade of B. velezensis (Fig. 4b). Based on the morphological, biochemical, and physiological characterization of strain LM1 and the phylogenetic analyses based on16S rRNA and gyrA gene sequences, strain LM1 was identified as B. velezensis.
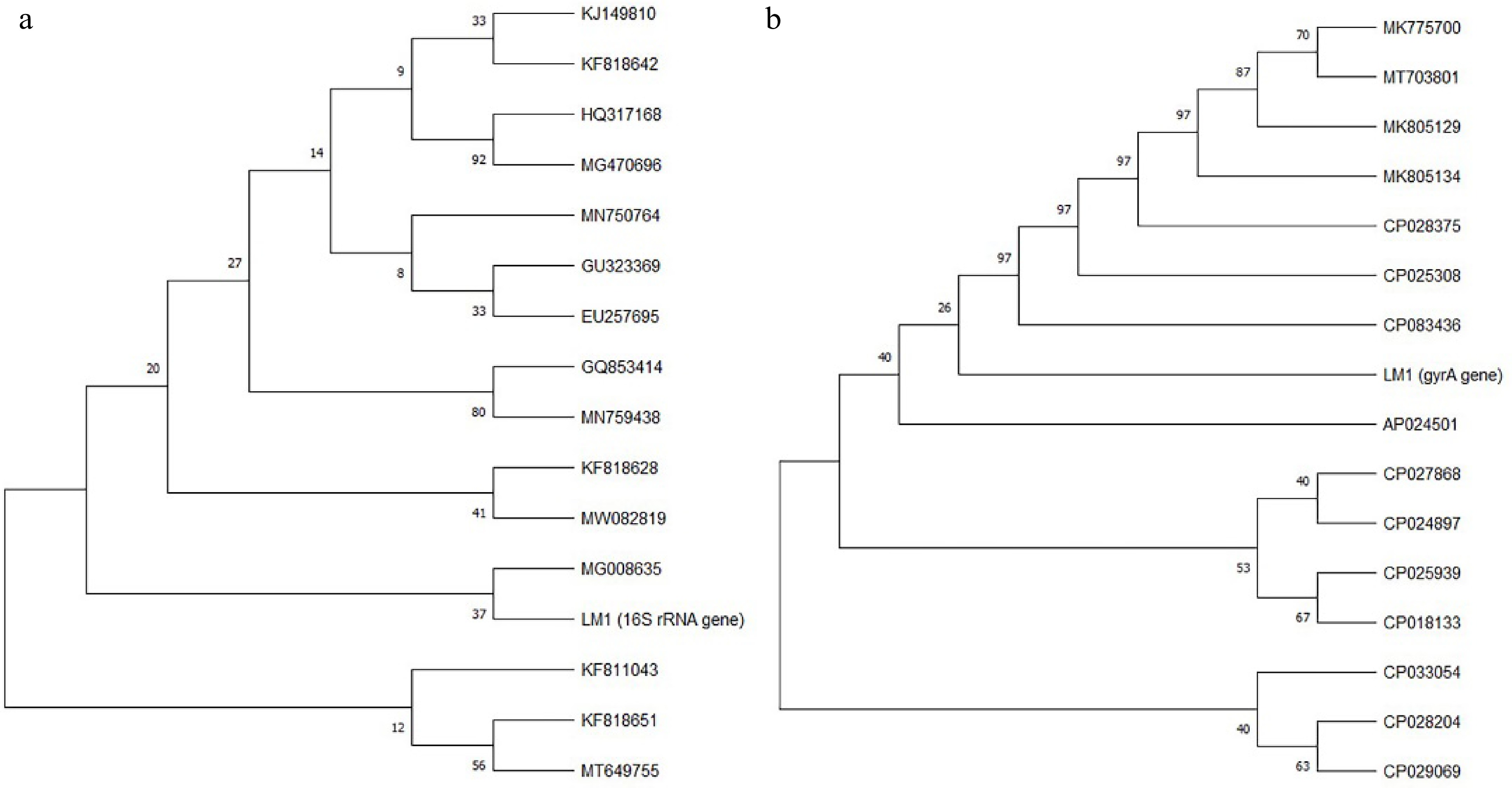
Figure 4.
Phylogenetic trees based on (a) 16S rRNA gene sequence and (b) gyrA gene sequence of strain LM1.
Strain safety test
-
The safety of B. velezensis LM1 was evaluated by the antibiotic sensitivity and hemolysis tests. The antibiotic sensitivity was evaluated using the disc diffusion method and the zones of inhibition around antibiotic-paper discs from the back of the plates were measured. The sensitivity or resistance status of the bacteria was determined by comparing the values with standard breakpoint tables. It was observed from Supplemental Table S2 that B. velezensis LM1 was sensitive to all tested antibiotics, indicating its safety.
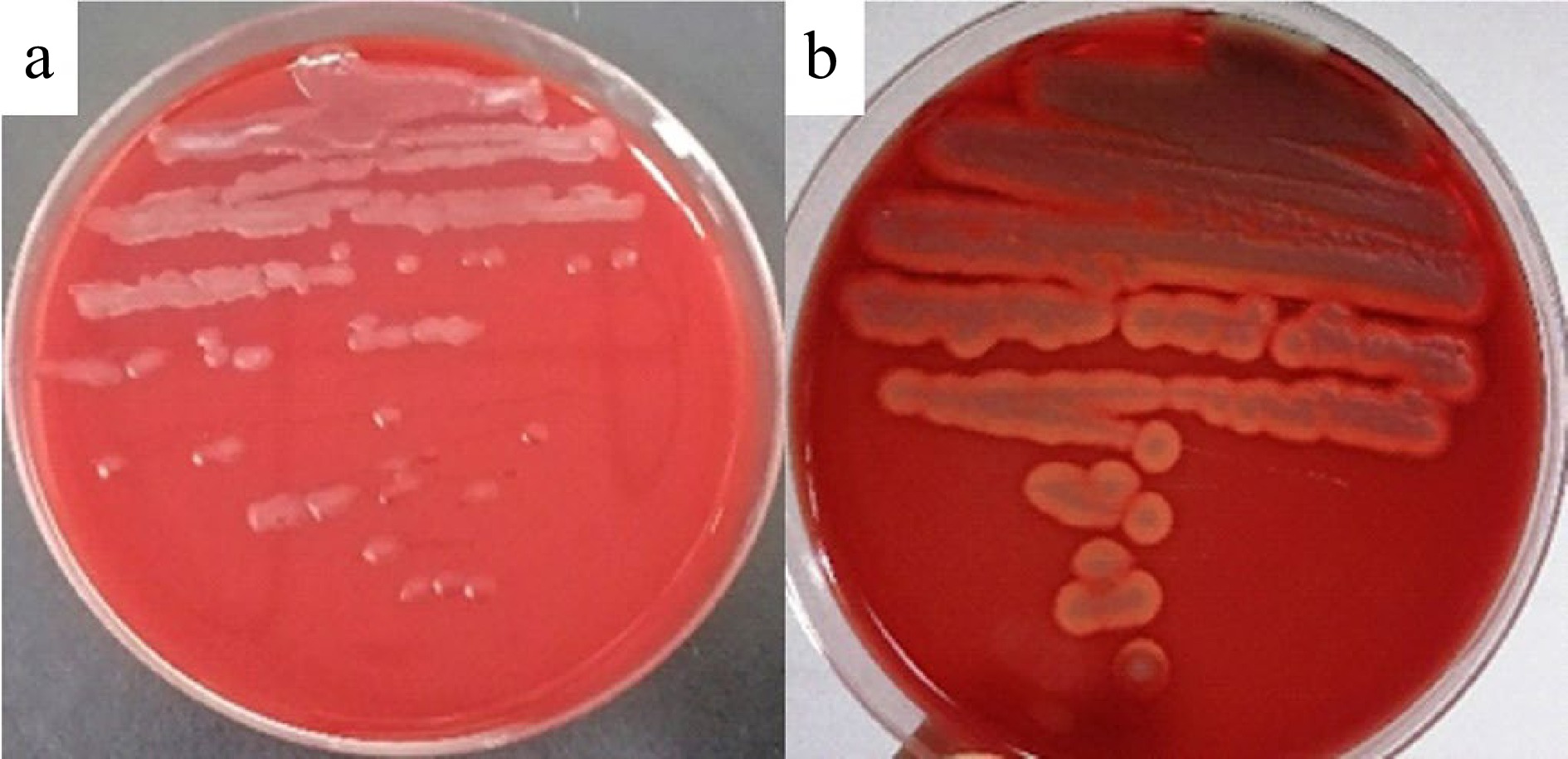
The blood agar hemolysis test showed that the positive control bacteria B. cereus ATCC14579 produced an obvious β-hemolytic ring, while B. velezensis LM1 was unable to produce hemolysin activity on plates (Fig. 5), further confirming the safety of the strain.
Extracellular enzyme profiling analysis
-
B. velezensis LM1 was incubated in LB medium at 37 °C and 200 rpm. Cell growth was monitored at 600 nm by taking sample regularly. It took about 20 h for the culture to grow to the highest optical density (Fig. 6a). The enzymatic activity curves of B. velezensis LM1 lagged the cell growth curve, and reached a plateau at different incubation time for different enzymes (Fig. 6b−6d). It was found that CMCase, FPA, Lac, LiP and MnP activities changed obviously over time, and exhibited a trend of first increasing and then decreasing. This was consistent with the results of Mei et al.[19] The highest Lac activity was up to 22.75 U/L at 72 h, the highest Lip activity was 698.49 U/L at 24 h, and the highest Mnp activity was 85.07 U/L at 60 h. The highest CMCase and FPA activities were 34.91 U/mL and 23.16 U/mL, respectively, at 72 h. It was observed that B. velezensis LM1 presented high lignin-degrading enzyme activity at an earlier time, then followed by the cellulose-degrading enzyme activity, indicating that lignin was first decomposed, thus contributing to the exposure of cellulose-degrading enzymes to cellulose and resulting in the degradation of cellulose. This will facilitate the conversion of IDF into SDF, thereby increasing the proportion of SDF. The changes of amylase and protease activities over time are shown in Fig. 6d. It was found that amylase activity reached a maximum of 136.58 U/mL at 36 h, and the protease activity reached a maximum of 231.95 U/mL at 72 h.The high content of starch and a certain amount of protein in highland barley whole grain flour would interfere with the purity of DF during its preparation process. Therefore, the amylase and protease produced by the B. velezensis LM1 could hydrolyze the starch and protein in highland barley whole grain flour, and improve the purity of DF.
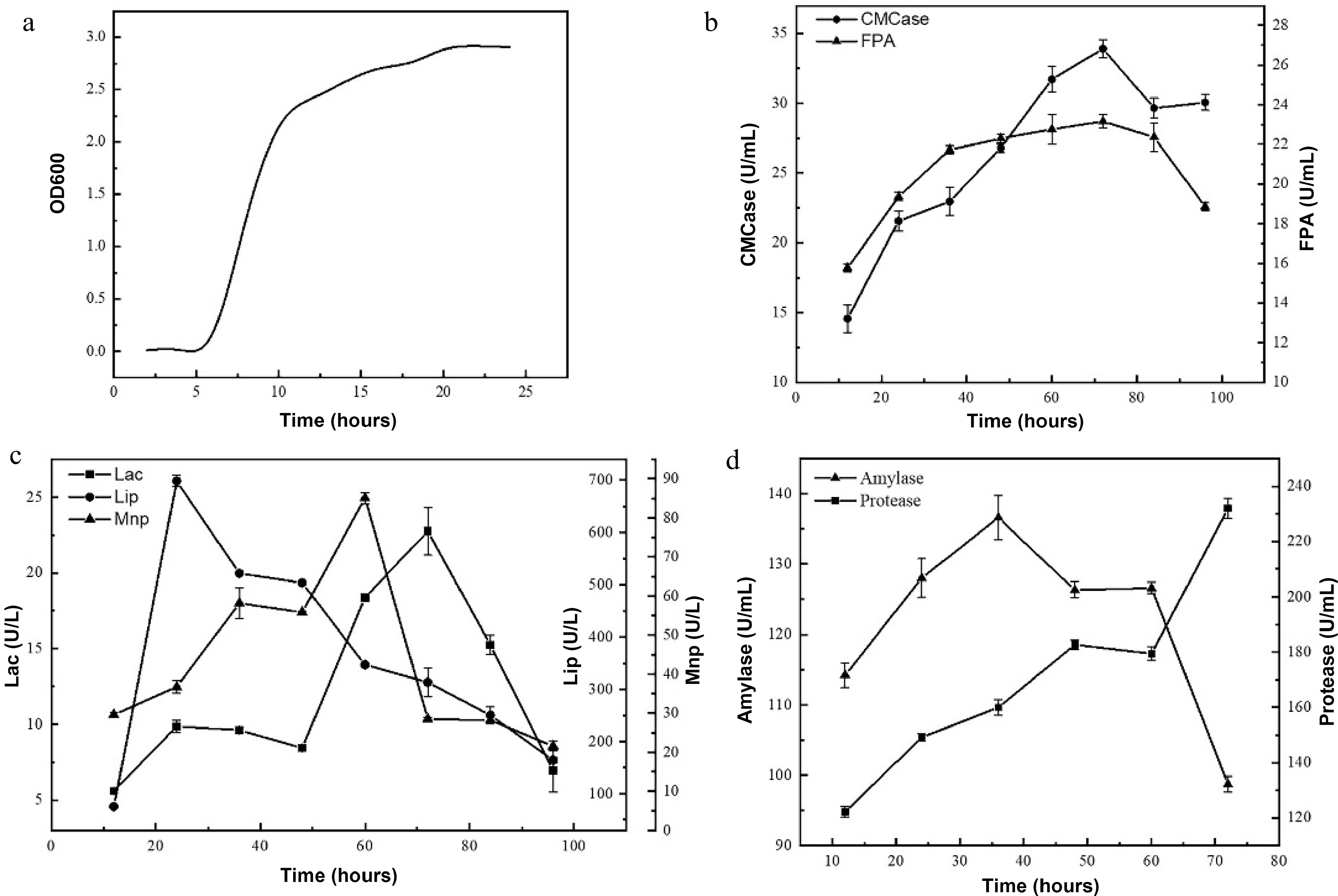
Figure 6.
(a) Growth curve and the enzyme activities of (b) CMCase and FPA, (c) Lac, Lip and Mnp, (d) amylase and protease of B. velezensis LM1.
Effect of B. velezensis LM1 fermentation on DF content and purity of highland barely
-
B. velezensis LM1 was used for the fermentation of highland barely whole grain flour and the yield and purity of DF was determined. The results were shown in Table 2. In terms of yield, the yield of FSDF reached 12.57%, which was 2.07 times higher than that of RSDF (6.07%). At the same time, the yield of FIDF was 18.68%, which was significantly lower than that of RIDF (22.45%), indicating the conversion of IDF into SDF. The ratio of SDF/IDF increased from 27.04% to 67.29%, indicating that the lignocellulosic enzymes produced during the fermentation process of B. velezensis played a key role in increasing the yield of SDF. In terms of purity, the purity of FSDF was 87.13%, which was equivalent to the purity of RSDF (87%), indicating that the amylase and protease produced in the fermentation process of B. velezensis had effectively hydrolyzed the starch and protein.
Table 2. The yield and purity of dietary fiber.
Sample Yield (%) Purity (%) RSDF 6.07 ± 0.01B 87.00 ± 0.30A FSDF 12.57 ± 0.22A 87.13 ± 0.99A RIDF 22.45 ± 0.02a 91.70 ± 0.82a FIDF 18.68 ± 0.21b 77.27 ± 0.50b A, B reflect the significance of SDF, and a, b reflect the significance of IDF.
P < 0.05. -
In this study, a high efficiency lignin and cellulose-degrading strain was screened and identified as B. velezensis LM1 after morphological, physiological, biochemical, and molecular biology identification. Fermentation of highland barely with the strain increased the SDF content, indicating microbial fermentation is a beneficial way to improve the content of SDF. Moreover, the screening and breeding of lignin and cellulose degrading strain enrich the types of fermentation strains and promote the development and utilization of cereals and their by-products.
-
The soil sample was taken from 5−10 cm under the plane tree of Nanjing Agricultural University (Nanjing, China). Alkaline lignin was purchased from Solarbio Biotechnology Co., Ltd (Beijing, China). Aniline blue and DNS reagents were purchased from Shanghai Yuanye Biotechnology Co., Ltd (Shanghai, China). Congo red and cellulose were purchased from Shanghai Macklin Biochemical Technology Co., Ltd (Shanghai, China). Highland barley (Tibetan Qing 2000) was obtained from Tibet. Filter paper (Whatman 1) was purchased from the Whatman Company. The drug sensitive discs and blood agar plates were purchased from Beekman Biotechnology Co., Ltd. Other chemicals and reagents were analytical grade and obtained from Nanjing chemical reagents Co., Ltd (Nanjing, China). B. velezensis LM1 has been preserved in the China Culture Collection Center with the preservation number CGMCC NO.24989.
Screening of lignin and cellulose degrading strain
Purification of lignin or cellulose degrading strain
-
Ten grams of soil sample were weighed, mixed with 90 mL of sterile water in a conical flask and shaken for 30 min at 30 °C and 200 rpm. The supernatant was added to the lignin and cellulose containing enrichment medium (alkaline lignin 2 g, ammonium sulfate 2 g, magnesium sulfate 0.5 g, potassium dihydrogen phosphate 1 g, sodium chloride 5 g, peptone 1 g, water 1 L; cellulose 2 g, peptone 1 g, sodium chloride 5 g, calcium chloride 0.1 g, potassium dihydrogen phosphate 1 g, ammonium sulfate 2 g, sulfuric acid magnesium 0.5 g, filter paper strip 5 pieces of 1 × 5 cm, water 1 L) at a ratio of 5%, respectively. The lignin-containing enrichment culture was conducted at 30 °C and 200 rpm for 3 d, and the cellulose enrichment culture was cultured at 30 °C and 200 rpm until the filter paper strips were completely dissolved.
The enriched culture solution was serially diluted to 10−6 with sterile water , and 100 µL of the diluted solution was spread over LB agar plates (yeast extract 5 g, tryptone 10 g, sodium chloride 10 g, agar 15 g, water 1 L) supplemented with nystatin and incubated at 30 °C for 24 h. The obtained single colonies of different shapes were streaked onto LB agar plates for further purification. The purified colonies were separately inoculated into LB liquid medium (yeast extract 5 g, tryptone 10 g, sodium chloride 10 g, water 1 L) and incubated at 30 °C and 200 rpm for 12 h. The obtained bacterial liquid was stored in glycerol at −80 °C.
Preliminary screening of lignin and cellulose degrading strain
-
Aniline blue agar medium (yeast extract 5 g, tryptone 10 g, sodium chloride 10 g, agar 15 g, aniline blue 0.1 g, water 1 L) and liquid medium (yeast extract 5 g, tryptone 10 g, sodium chloride 10 g, aniline blue 0.1 g, water 1 L) were used for the screening of lignin-degrading strain. Pure lignin-degrading strains with different shapes were streaked onto or inoculated into the above two media and incubated at 30 °C and 200 rpm for 3 d. The fading results were recorded and the fading rates were calculated according to the absorbance value change at 590 nm. At the same time, pure cellulose-degrading strains were spotted in CMC-Na medium (CMC-Na 2 g, ammonium sulfate 2 g, dipotassium hydrogen phosphate 1 g, magnesium sulfate 0.5 g, sodium chloride 0.5 g, water 1 L) and incubated at 30 °C for 3 d, then stained with Congo red solution for 20 min and decolorized with 1 M sodium chloride solution for 10 min. The diameters of transparent circles and colonies were measured[29]. Pure cellulose-degrading strains were inoculated into filter paper strip disintegration medium (filter paper strips 30 pieces of 2 × 6 cm, magnesium sulfate 0.1 g, ammonium sulfate 4 g, dipotassium hydrogen phosphate 2 g, sodium chloride 5 g, peptone 10 g, yeast extract powder 10 g, water 1 L) and cultured at 30 °C and 200 rpm for 3 d, to record the degree of disintegration of filter paper strips[30].
Cross screening of lignin and cellulose-degrading strain
-
The effective lignin-degrading bacteria obtained in the preliminary screening were cultivated in the CMC-Na plates and filter paper disintegration medium, and simultaneously the effective cellulose-degrading bacteria obtained in the preliminary screening were cultivated in the aniline blue medium. Culture conditions and treatment methods were the same as above. Then, strains that could degrade both lignin and cellulose were screened out.
Repetitive screening of lignin and cellulose degrading strains
-
The strains obtained from the final cross-screening were separately inoculated into LB liquid medium and cultured at 30 °C and 200 rpm for 24 h to obtain seed liquid. The seed liquid was inoculated into ligninase-producing medium (lignin 10 g, magnesium sulfate 0.5 g, potassium dihydrogen phosphate 2 g, ammonium sulfate 4 g, peptone 10 g, yeast extract 5 g, sodium chloride 0.5 g, water 1 L) and cellulose-producing medium (CMC-Na 10 g, magnesium sulfate 0.5 g, potassium dihydrogen phosphate 2 g, ammonium sulfate 4 g, peptone 10 g, yeast extract 5 g, water 1 L), respectively, and shaken at 30 °C and 200 rpm for 3 d. The bacterial culture was centrifuged at 8,000 rpm and 4 °C for 10 min to obtain the crude enzyme solution for enzyme activity assay. Laccase (Lac)[31], lignin peroxidase (Lip)[27], manganese peroxidase (Mnp)[32], carboxymethyl cellulase (CMCase)[27] and filter paper lyase (FPase) activities[33] were analyzed for all selected strains.
Lac activity: 1.5 mL of 100 mM sodium malonate buffer (pH = 4.5) was mixed with 1.5 mL of 0.6 mM ABTS and 150 µL of the crude enzyme solution, and the mixture were incubated at 30 °C. The absorbance changes at 420 nm of the reaction solution within 1 min were recorded. One unit of enzyme activity was expressed as the amount of enzyme that catalyzed 1 μmol of substrate to product per minute.
Mnp activity: 1.5 mL of 100 mM sodium malonate buffer (pH = 4.5), 300 µL of 10 mM MnSO4 solution, 150 µL of crude enzyme solution and 1.02 mL of deionized water were mixed. 30 µL of 10 mM H2O2 was added to initiate the reaction at 30 °C, and the change in absorbance at 270 nm within 1 min was measured. One unit of enzyme activity (U) is defined as the amount of enzyme that oxidizes 1 μmol of MnSO4 within 1 min.
Lip activity: 1.5 mL of 200 mM sodium tartrate buffer (pH = 3.0), 300 µL of 40 mM veratryl alcohol, 150 µL of supernatant and 1.02 mL of deionized water were mixed. The change in absorbance at 310 nm within 1 min was measured after adding 30 µL of 20 mM H2O2 at 30 °C. The amount of enzyme required to oxidize 1 μmol of veratryl alcohol in 1 min is defined as one unit of enzyme activity (U).
CMCase activity: 50 µL of crude enzyme solution and 100 µL of 1% CMC-Na in citric acid buffer solution (0.1 M, pH 5.0) were mixed totally and incubated at 50 °C for 30 min. Then the reaction was terminated by adding 200 µL of DNS solution, boiled for 5 min in a water bath and chilled with ice water. 1 mL of distilled water was added. 200 µL of mixture were taken and measured using a microplate reader (InfiniteF200, Tecan Co., Switzerland) at 540 nm. One unit of enzyme activity (U) is defined as the amount of enzyme that hydrolyze sodium carboxymethyl cellulose to yield 1 µg of reducing sugar within 1 min.
FPA activity: A strip of 1 × 3 cm filter paper was mixed with 200 µL of citric acid buffer solution and 100 µL of crude enzyme solution, which was incubated at 50 °C for 30 min. Then the reaction was terminated by adding 400 µL of DNS solution, and boiling in a water bath for 5 min, and cooled with ice water. 2 mL of distilled water were added to dilute the reaction mixture. 200 µL of reaction mixture was taken and its absorbance at 540 nm was measured. The amount of enzyme that hydrolyze filter paper to produce 1 µg of reducing sugar within 1 min, is defined as one unit of enzyme activity (U).
Identification of a lignin and cellulose degrading strain
Morphological, physiological, and biochemical tests
-
The selected strain was spread over LB agar plates, and cultured at 30 °C for 12 or 24 h to observe the colony's morphological characteristics, such as size, color, shape, and texture. Physiological and biochemical tests were performed and included Gram staining, gelatin liquefaction, starch hydrolysis, methyl red, catalase, oxidase, and so on[34]. A single colony was inoculated into LB liquid medium and shaken at 30 °C and 200 rpm for 4 d to obtain bacterial solution. The bacterial solution was treated in a water bath at 80 °C for 15 min[35], and observed whether spores were formed under the microscope through staining with malachite green.
Molecular identification
16S rRNA gene sequencing
-
The selected strain was further identified by 16S rRNA gene sequencing. DNA was isolated using a Bacterial DNA Kit (Omega Bio-Tek, America), and PCR amplification was performed using universal primers 1492R (5′-TAGGGTTACCTTGTCGACTT-3′) and 27F (5′-AGAGTTTGATCATGGCTCAG-3′). PCR reaction conditions: pre-denaturation at 94 °C for 5 min, deformation at 94 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 60 s, 35 cycles, and finally, extension at 72 °C for 10 min. The amplified products were analyzed and sent to GENEWIZ Co. Ltd (Nanjing, Jiangsu, China) for sequencing. The resulting partial sequence of selected strain was compared with reference sequences from the GenBank database (Supplemental Table S3) by BLAST analysis, and the neighbor-joining method was used for phylogenetic tree construction using software MEGA-X[36].
gyrA gene sequencing
-
We also attempted to amplify and sequence the gyrA gene (DNA gyrase subunit A). DNA extraction and PCR reaction conditions were the same as 4.3.2.1. Amplification primers were gyrA-5p (5′-CAGTCAGGAAATGCGTACGTCCTT-3′) and gyrA-3p (5′-CAAGGTAATGCCTCCAGGCATTGC T-3′). The amplified products were analyzed and sent to GENEWIZ Co. Ltd (China) for sequencing. The resulting partial sequence was also compared with those reference sequences from the GenBank database (Supplemental Table S4) by BLAST analysis, and the neighbor-joining method was used for phylogenetic tree construction using software MEGA-X.
Nucleotide sequence accession number
-
The 16S rRNA and gyrA gene sequences were deposited in the GenBank databases with the accession numbers OP828653 and OP881940, respectively.
Safety test of the lignin and cellulose degrading strain
-
One hundred microliters of the bacterial solution were spread on an LB agar plate. Different types of drug-sensitive discs (tetracycline, penicillin, erythromycin, chloramphenicol, gentamicin, vancomycin, norfloxacin and cefoxicil) were adhered individually to the surface of an agar plate before being incubated at 37 °C for 2 d. The diameters of inhibition zones were recorded[37].
The selected strain was dropped on blood agar plates and cultured at 37 °C for 2 d to observe whether there was a hemolytic ring[37]. Bacillus cereus ATCC14579 was used as the positive control.
Enzyme activities assay
-
Bacterial growth is closely related to the enzyme activity, so the growth curve of the selected strain was plotted. The selected strain was inoculated into LB liquid medium and grown to log phase to obtain seed liquid. The seed liquid was inoculated with a proportion of 3% into medium for ligninase, cellulase, amylase and protease production. During the incubation at 30 °C and 200 rpm, the Lac, Lip, Mnp, CMCase, FPA, amylase and protease activities in the crude enzyme solution were determined. The crude enzyme solution was harvested by collecting and centrifuging 2 mL of the bacterial culture every 12 h. The changing trend in enzyme activity was used to provide reference for fermentation time. The ligninase and cellulase assay procedures were the same as above. The method for the determination of amylase activity refers to the method of Benabda et al.[38] with a slight change. One hundred microlitres of crude enzyme solution was mixed with 200 µL of 1% soluble starch solution (prepared from sodium citrate buffer) and reacted at 55 °C for 30 min. After that, DNS method was used to detect the amount of reducing sugar produced. The protease activity was determined according to the method of Ma et al.[39].
Fermentation of highland barely and analysis of DF
Preparation of fermented highland barley
-
The highland barley grains were ground into powder and passed through a 60-mesh sieve. Highland barley flour fermentation medium (highland barley flour 10 g, K2HPO4 0.2 g, MgSO4 0.03 g, CaCl2 0.03 g, CMC-Na 1 g, MnSO4 0.016 g) was autoclaved at 115 °C for 30 min. The screened strain was inoculated into LB liquid medium, and cultivated at 30 °C and 200 rpm to the log phase to obtain the seed liquid. Three percent seed liquid was mixed with Highland barley flour fermentation medium and continuously cultured at 30 °C and 200 rpm for 3 d.
Analysis of DF content in highland barley flour
-
The AOAC procedures with some modifications were used to determine SDF contents of raw highland barley. Ten grams (recorded as M1) of highland barley flour was mixed with 100 mL of deionized water. In order to remove starch, 250 µL of thermostable α-amylase (20,000 U/mL) was added and incubated in a water bath at 95 °C for 35 min. Then 4 mg of alkaline protease (200U/mg) was mixed with the above mixture and incubated at 60 °C for 30 min to remove protein. Twenty microliters of saccharifying enzyme (10,000 U/mL) was added to the mixture and incubated at pH 4.5 ± 0.2, 60 °C for 30 min. After cooling to room temperature, the mixture was centrifuged at 4 °C and 8,000 rpm for 10 min to obtain the pellet and the supernatant. The pellet was washed with deionized water and freeze-dried to get IDF (recorded as M2), which was named as RIDF. Four times volume of 95% ethanol was added to the supernatant and precipitated overnight at room temperature. The precipitate was rinsed three times with 95% ethanol and dried (recorded as M3). The target product SDF was obtained and named as RSDF. The yield of RIDF and RSDF were calculated as follows:
$\rm RIDF\; (g/100\; g) = (M_{2}/M_{1}) \times 100 $ $\rm RSDF\; (g/100\; g) = (M_{3}/M_{1}) \times 100 $ The weight of highland barley flour used to prepare the fermentation medium was recorded as G1. For fermented highland barley flour, the fermented mixture was centrifuged at 4 °C and 8000 rpm for 10 min to obtain the pellet and the supernatant, the following treatment was the same as above. The target product IDF (recorded as G2) and SDF (recorded as G3) were obtained and named as FIDF and FSDF, respectively. The yield of FIDF and FSDF were calculated as follows:
$\rm FIDF \;(g/100\; g) = (G_{2}/G_{1}) \times 100 $ $\rm FSDF\; (g/100\; g) = (G_{3}/G_{1}) \times 100 $ Analysis of DF purity
-
RIDF, RSDF, FIDF and FSDF obtained previously were used to measure their purity according to the methods above.
Statistical analysis
-
All experiments were replicated three times and the results were expressed as mean values ± standard deviation. SAS Version 8.0 software was used to perform one-way analysis of variance (ANOVA). Different lowercase letters indicated statistically significant difference at the level of P < 0.05.
The authors declare no competing financial interest, and this work was supported by the 'Fundamental Research Funds for the Central Universities' [No. Y0202100226].
-
The authors declare that they have no conflict of interest.
- Supplemental Table S1 Physiological and biochemical properties of LM1.
- Supplemental Table S2 Drug susceptibility test of B. velezensis LM1.
- Supplemental Table S3 16S rRNA reference strains.
- Supplemental Table S4 gyrA gene reference strains.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press on behalf of Nanjing Agricultural University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Wang A, Li L, Zhang S, Wang J, Lu Z, et al. 2023. Screening and identification of a lignocellulose-degrading strain and its application in highland barley fermentation. Food Materials Research 3:6 doi: 10.48130/FMR-2023-0006
Screening and identification of a lignocellulose-degrading strain and its application in highland barley fermentation
- Received: 03 February 2023
- Accepted: 15 March 2023
- Published online: 14 April 2023
Abstract: This study aimed to screen out bacterial strains with higher lignocellulolytic degradation enzyme activity and explore the effect of this strain on the highland barely soluble dietary fiber content through fermentation. Soil samples were collected from the ground of Nanjing Agriculture University (Nanjing, China) and used for enrichment culture. The methods used to screen the bacteria with high lignocellulose-degrading ability included decolorization test of aniline blue, hydrolyzation circle measurement of sodium carboxymethyl cellulose, disintegration test of filter paper strip and detection of enzyme activity. Six bacteria were isolated with lignocellulose degrading ability, of which LM1-strain possessed high lignocellulose degrading ability. The LM1 strain was identified as Bacillus velezensis by morphological, physiological, biochemical characteristics and molecular sequence analysis. Antibiotic sensitivity tests and hemolysis tests showed that strain LM1 was sensitive to all antibiotics and caused no hemolysis, indicating the safety of strain LM1. Strain LM1 was used to ferment highland barley whole grain flour and the content of soluble dietary fiber reached high levels (12.57 g/100 g), which was 2.07 times that of the raw barley soluble dietary fiber (6.07 g/100 g). The purity of soluble dietary fiber also reached the level of enzymatic extraction. The results will enrich the types of fermentation strains and promote the development and utilization of cereals and their by-products.
-
Key words:
- Lignocellulose degradation /
- Bacillus velezensis LM1 /
- Screening /
- Identification /
- SDF /
- Highland barley