-
Inorganic phosphate (Pi) is involved in the metabolism of many substrates and activities in plants[1]. Pi is directly involved in all aspects of photosynthesis, and the metabolism and transport of carbohydrates in plants[1,2]. Applications of phosphate fertilizer during production play an important role in maintaining plant growth and development, as well as the high yields of fruit trees[3].
The P in soil that can be taken up by plants is mainly inorganic phosphate (
${{\text{H}_2}{\text{PO}_4^-}} $ ${\text{HPO}_4^{2-}} $ To maintain growth and development under Pi deficiency, plants have developed complex physiological and morphological adaptations and molecular regulatory mechanisms to ensure that they can obtain more Pi from the soil and remobilize and redistribute the Pi in vivo to maintain Pi balance, i.e., the phosphorus starvation response (PSR)[10]. Roots are highly responsive to PSRs, and the main strategy is an adaptive change in root structure that promotes root access to soil Pi[11]. Lateral roots enhance adaptation to a Pi deficiency. The increase in the number of lateral roots, in addition to increasing the volume of the root system in contact with the soil, creates more root tip area to sense and absorb more exogenous Pi[12,13]. Plants also respond to the phosphate starvation by increasing the uptake of Pi from the soil through the root system and translocating it to aboveground parts[14]. The PHOSPHATE TRANSPORTER (PHT) protein family plays an important role in the acquisition of Pi from the soil and the transport of Pi within cells. PHT proteins have been divided into four subfamilies, including PHT1/2/3/4[15,16]. PHT1 is a high-affinity phosphorus transporter protein localized on the cytoplasmic membrane of the root cortex where it plays a major role in Pi uptake and intracellular Pi homeostasis[17]. The SPX domain is called the SYG1/Pho81/XPR1 protein domain, which is present in the N-terminal peptides of yeast SYG1 and PHO81, as well as human XPR1 proteins[18−21]. Genes containing the SPX functional domain play an important role in plant Pi signaling and Pi homeostasis. PHOSPHATE1 (PHO1) proteins contain the SPX and EXS (ERD1, XPR1, and SYG1) structural domains, which are responsible for the upward transport of Pi absorbed by roots to above ground tissues. Another class of SPX proteins contains only the SPX domain, which is essential for maintaining Pi homeostasis in vivo when the PSR occurs[22].
Transcriptional regulation is a very important regulatory node in the plant PSR regulatory network and is an early event during gene expression. A variety of transcription factors involved in the fine regulation of PSRs in plants[23]. The important TF families involved in PSRs are the v-myb avian myeloblastosis viral oncogene homolog (MYB) family, the basic helix-loop-helix (bHLH) family, and the WRKY family[10]. PHOSPHATE STARVATION RESPONSE 1 (PHR1), which belongs to the MYB-CC subfamily of the MYB family, is the first Pi starvation-induced (PSI) transcription factor identified in plants and a central transcriptional regulator of the Pi signaling pathway[24]. Additionally, MYB1, MYB2, and MYB62 have been identified, and their expression are induced by low Pi[25−27]. In the WRKY transcription factor family, WRKY75 is induced by low Pi and regulates PSI gene expression and root development[28]. Among the bHLH transcription factor family, bHLH32 is induced by Pi deficiency but is involved as a negative regulator of root hair development under the PSR[29]. Other types of transcription factors, such as zinc finger transcription factor 6 (ZAT6), auxin response factor 16 (ARF16), and SQUAMOSA promoter binding protein-like 3 (SPL3), are also involved in the PSR and regulate Pi homeostasis in vivo[30, 31].
Recent studies have identified a membrane-localized bHLH transcription factor, called Glycine max symbiotic ammonium transporter 1 (GmSAT1). The G. max sat1 mutant has a loss of sensitivity to Pi supply compared to the wild-type. SAT1 regulates PSI genes, such as PHT1;1, RNS2, HAD1, MGD2, and SPX2 at the transcriptional level, thereby regulating root Pi homeostasis[32]. Our laboratory has researched MdSAT1, the homologous transcription factor in apples. The findings revealed a role for MdSAT1 in abiotic stress, in particular to saline and drought stress. MdSAT1 functions in regulating plant growth and development, particularly root architecture and the accumulation of reactive oxygen species[33]. They also explored the function of MdSAT1 in regulating nutrient absorption, particularly the absorption and accumulation of ammonium-nitrogen[33]. These data have laid the cornerstone for further research on the function of MdSAT1.
Pi deficiency in the soil environment seriously affects the development and yield of apples. In this study, MdSAT1 encoded a transcription factor that was responsive to Pi. MdSAT1 regulated root architecture and increased gene expression and enzyme activity related to Pi uptake, and ultimately promoted Pi utilization by the plants. Additionally, MdSAT1 also affects the flowering and senescence states in the later stages of plant development. Overall, the present study provides a framework for further research on the molecular mechanisms of MdSAT1 during the PSR.
-
qRT-PCR was used to examine the expression of MdSAT1 in response to the Pi concentration. Under different concentrations of phosphate (0, 1.25, 25, 625, and 1250 μM K2HPO4), MdSAT1 expression was inhibited with increasing Pi concentrations in the roots and shoots and was more pronounced in the roots (Fig. 1a, b). Then, 1.25 μM K2HPO4 and 1,250 μM K2HPO4 were selected as the low Pi (LP) and high Pi (HP) treatments, respectively (Fig. 1c), to detect MdSAT1 expression: Under LP treatment, the expression of MdSAT1 first increased and then decreased with the prolongation of treatment time; while under HP treatment, it decreased with the prolongation of treatment time. In addition to showing that MdSAT1 is a gene induced by LP, the proMdSAT1::GUS transgenic Arabidopsis plant was treated with different Pi concentrations, and the results suggest that GUS activity increases and then decreases with increasing Pi content (Supplemental Fig. S1). These results confirm that MdSAT1 is a Pi-responsive gene induced by Pi deficiency.
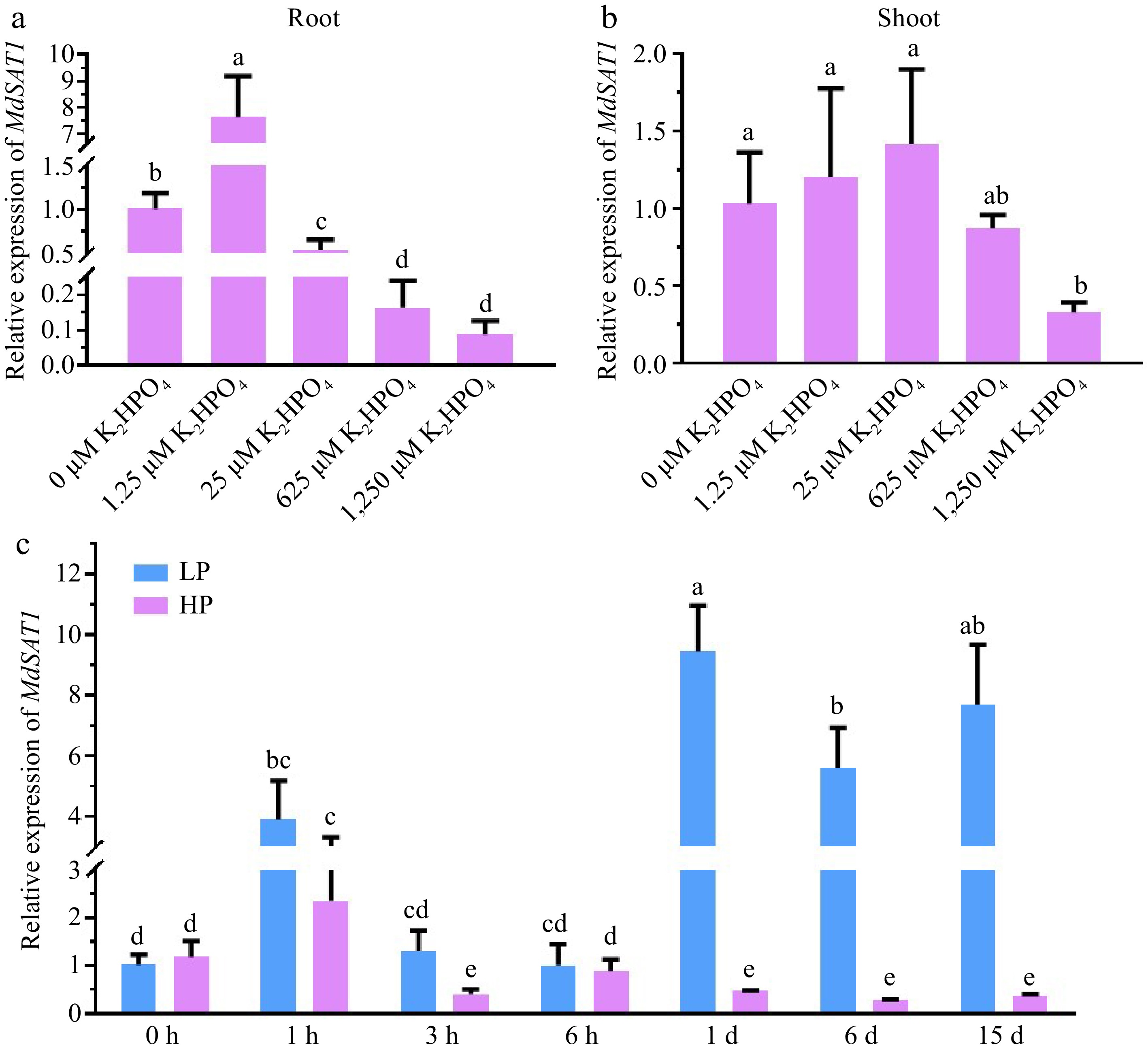
Figure 1.
MdSAT1 gene expression analysis in response to phosphate. (a) Root and (b) shoot MdSAT1 expression levels under different phosphate concentrations (the different Pi concentrations the apple seedlings were treated with were 0, 1.25, 25, 625, and 1,250 μM K2HPO4 for 15 d, supplemented with 2,500 μM K+ using the corresponding concentration of K2SO4). The seedlings treated with 0 μM K2HPO4 (1.25 mM K2SO4) were used as a negative control. (c) MdSAT1 expression levels under the 1.25 μM K2HPO4 (LP) and 1.25 mM K2HPO4 (HP) conditions. Plants grown at 23 °C/21 °C for day/night with 16L/8D, with an irradiance of 150 μmol·m−2·s−1 and relative humidity of 60%−70%. The seedlings treated with LP (0 h) were used as the negative control. Md18S was used as the reference gene. Error bars represent standard deviations (n ≥ 3). Different letters above the bars indicate significantly different values (p < 0.05).
MdSAT1 regulates the root architecture
-
We treated MdSAT1-OE and Col seedlings in 1/2 MS medium with LP or HP for 7 d to investigate the effect of the MdSAT1 gene on root growth. Under LP treatment, no significant difference was observed in the lateral root length between transgenic plants and Col seedlings, while under HP conditions, there was a significant difference (Fig. 2e). The primary root length, the total number of root tips, and the number of lateral roots increased significantly in the MdSAT1-OE lines compared with the Col (Fig. 2c, d & f), and this difference in root configuration was more pronounced under the LP treatment (Fig. 2a, b). These results suggest that overexpression of MdSAT1 enhances the adaptability of root development to Pi deficiency.
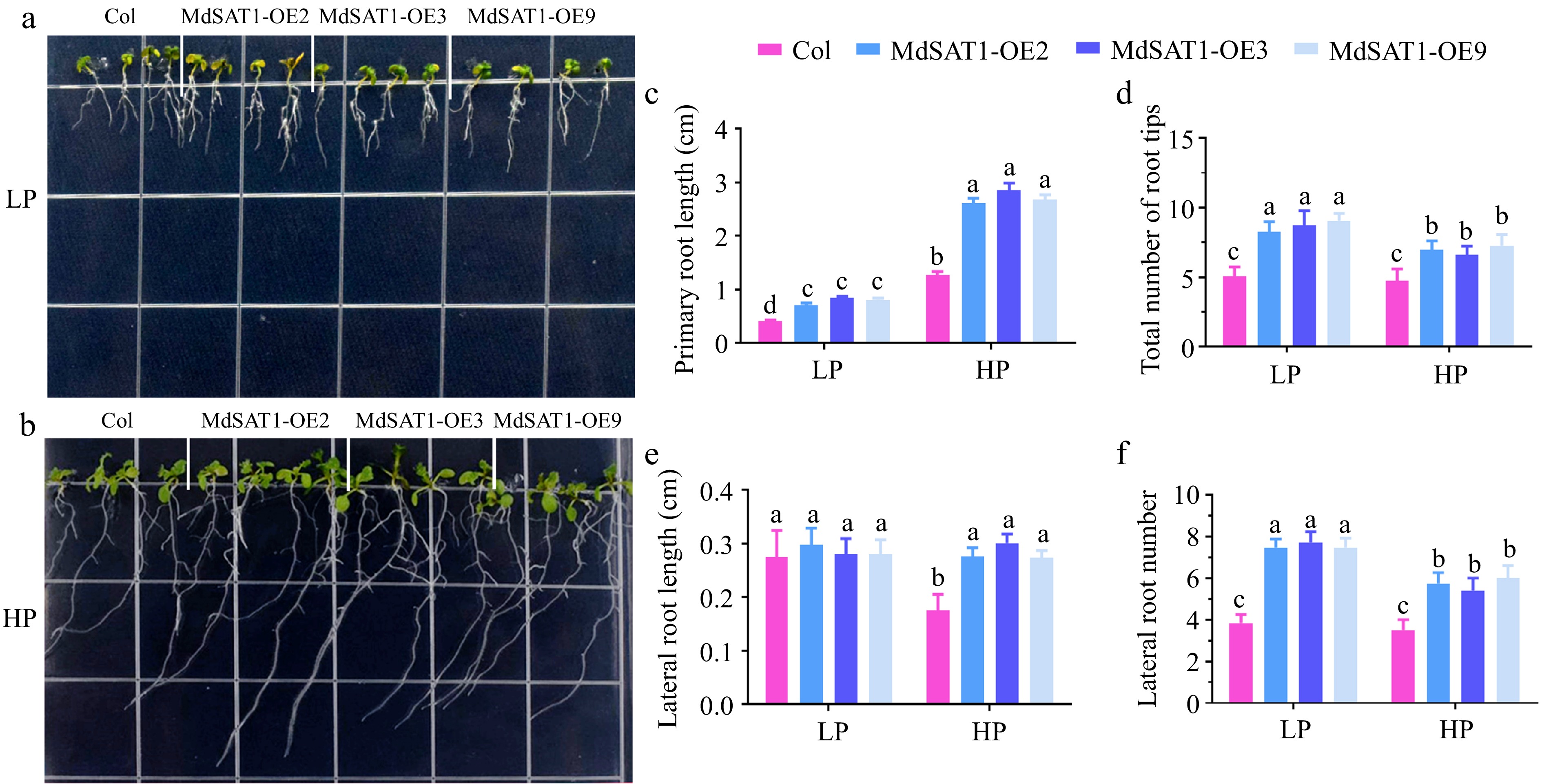
Figure 2.
MdSAT1 regulates root architecture. MdSAT1-OE and Col plants were grown for 7 d under LP (1.25 μM K2HPO4, 1.5% sucrose and 0.8% agar powder, pH 5.9) or HP (1.25 mM K2HPO4, 1.5% sucrose and 0.8% agar powder, pH 5.9) conditions. Plants grown at 23 °C/21 °C for day/night with 16L/8D, with an irradiance of 150 μmol·m−2 ·s−1 and relative humidity of 60%−70%. (a), (b) Morphological changes, (c) primary root length, (d) the total number of root tips, (e) lateral root length, and (f) lateral root number are presented. Error bars represent standard deviations (n ≥ 3). Different letters above the bars indicate significantly different values (p < 0.05).
Overexpressing MdSAT1 promotes Pi assimilation and utilization in response to Pi deficiency
-
Since overexpressing MdSAT1 promoted the lateral root development under LP conditions (Fig. 2), we next asked whether MdSAT1 was involved in Pi assimilation and utilization. MdSAT1-OE and Col were treated in Hoagland's nutrient containing LP or HP for 3 weeks. The results showed that seedling growth was promoted in the MdSAT1-OE lines compared to Col, as evidenced by the significant increase in fresh weight and Pi content (Fig. 3a−c). This effect of MdSAT1 was more pronounced under the LP than the HP treatment (Fig. 3). Further experiments revealed that overexpressing MdSAT1 promoted Acid phosphatase (APase) activity in the roots (Fig. 3d), thereby facilitating Pi assimilation in vivo. These results indicate that MdSAT1 promotes Pi assimilation and utilization, thereby maintaining stronger growth under Pi deficiency.
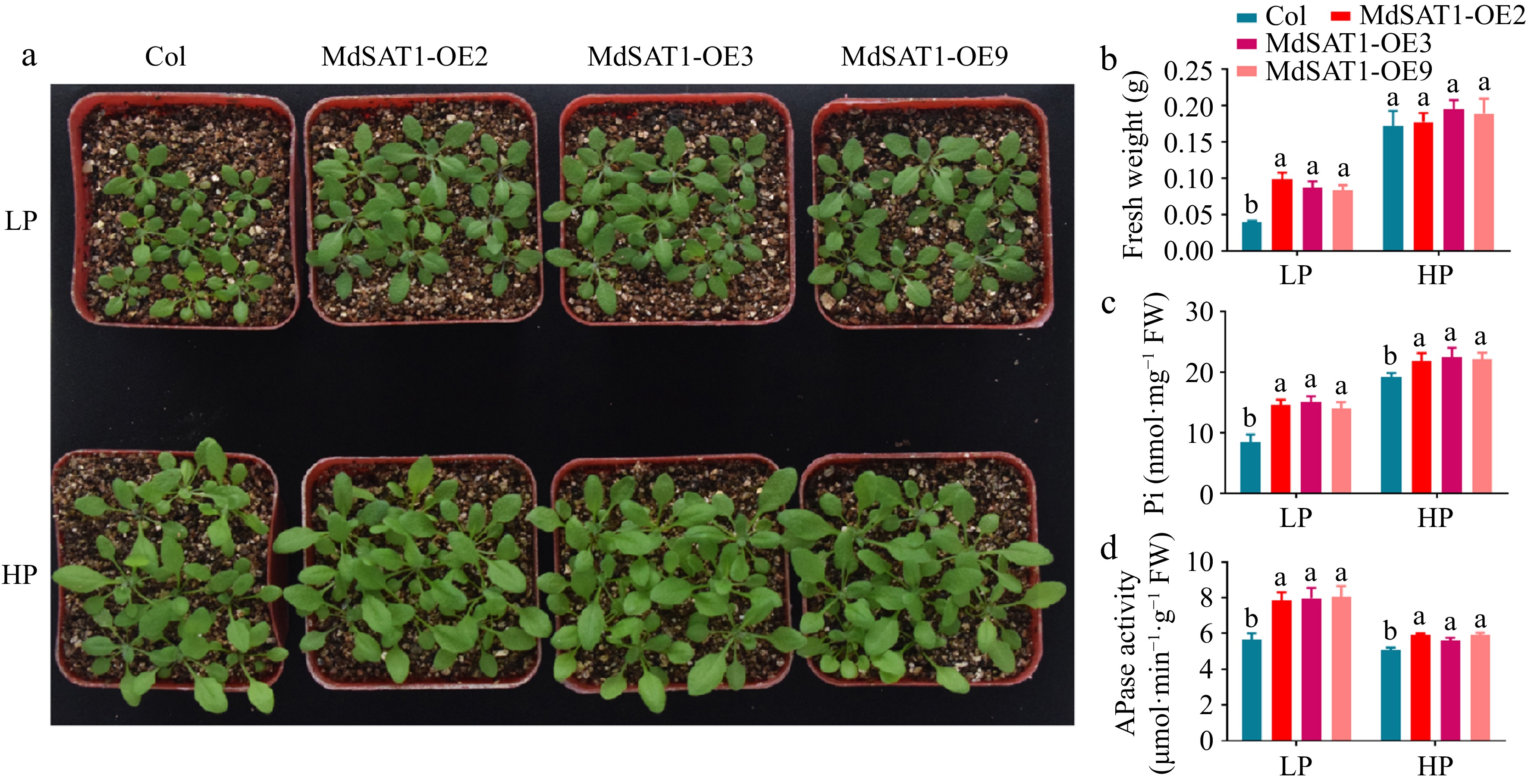
Figure 3.
MdSAT1 promotes phosphate uptake and regulates plant growth. MdSAT1-OE and Col plants were grown for 3 weeks under LP (1.25 μM K2HPO4) or HP (1.25 mM K2HPO4) conditions. Plants grown at 23 °C/21 °C for day/night with 16L/8D, with an irradiance of 150 μmol·m−2·s−1 and relative humidity of 60%−70%. (a) Morphological changes, (b) fresh weight, (c) phosphate content, and (d) APase activity are presented. Error bars represent standard deviations (n ≥ 3). Different letters above the bars indicate significantly different values (p < 0.05).
Overexpression of MdSAT1 affects the expression of PSI genes
-
To further evaluate the role of MdSAT1 in Pi uptake and transport in plants, the effect of MdSAT1 on the expression of PSI genes was analyzed. The expression of genes for Pi translocation to the ground (AtPHT1;2, AtPHT1;8), marker genes induced by Pi-starvation (AtPHT1;4, AtPHR1 and AtPT1), and genes regulating Pi dynamic balance (AtSPX2) were significantly induced in MdSAT1-OE plants (Fig. 4). MdSAT1 was a stronger inducer of these PSI genes under the LP treatment than the HP treatment (Fig. 4). For example, the difference in the upregulated multiple of AtPHT1;2, AtPHT1;8 and AtSPX2 expression between MdSAT1-OE and Col was smaller under the HP than the LP treatment (Fig. 4a, b & e). These data indicate that MdSAT1 regulates a series of PSI genes that are more pronounced under Pi deficiency.
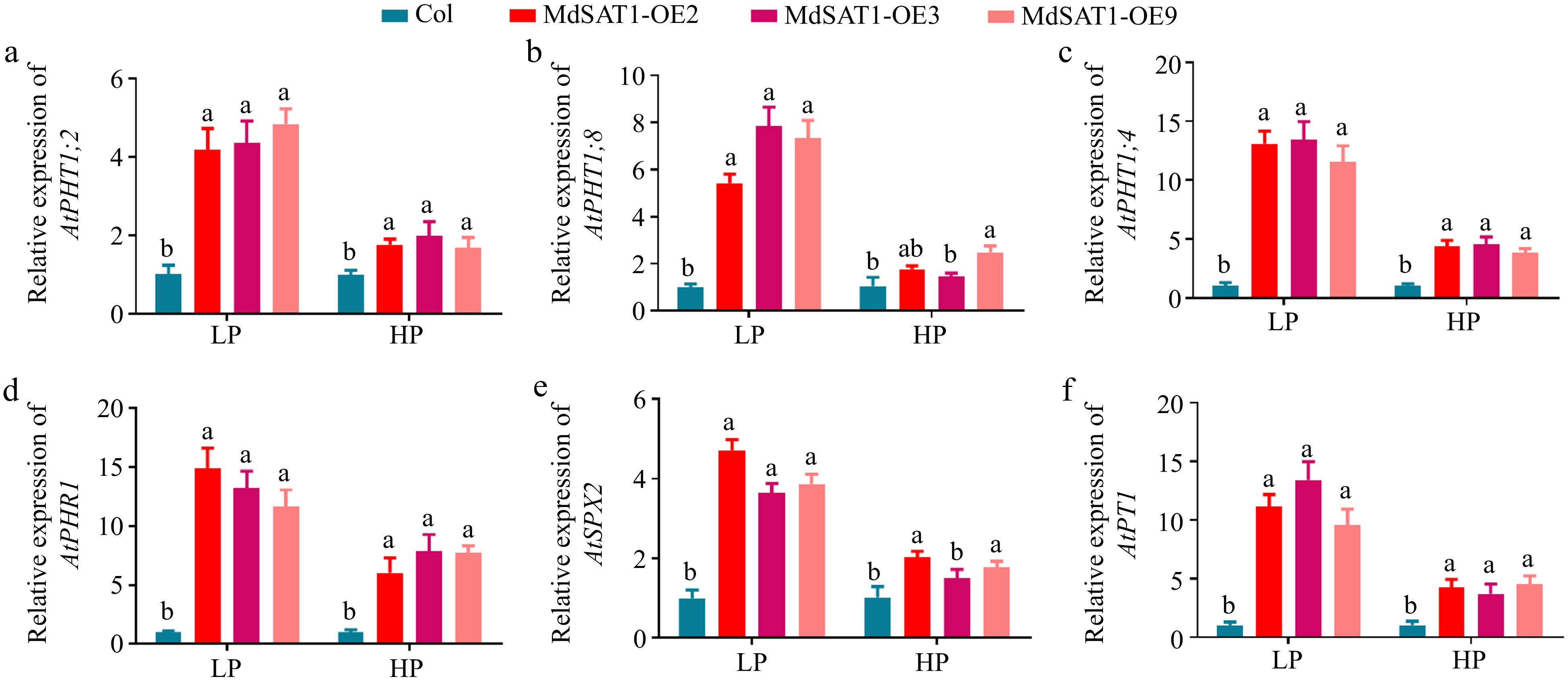
Figure 4.
MdSAT1 regulates the expression of Pi-responsive genes. MdSAT1-OE and Col Arabidopsis plants were grown for 3 weeks under LP (1.25 μM K2HPO4) or HP (1.25 mM K2HPO4) conditions. Plants grown at 23 °C/21 °C for day/night with 16L/8D, with an irradiance of 150 μmol·m−2·s−1 and relative humidity of 60%–70%. (a)–(f) Transcript levels of AtPHT1; 2, AtPHT1; 8, AtPHT1; 4, AtPHR1, AtSPX2, and AtPT1 in MdSAT1-OE and Col plants under LP conditions. Error bars represent standard deviations (n ≥ 3). Different letters above the bars indicate significantly different values (p < 0.05).
MdSAT1 promotes early flowering
-
The results show that the MdSAT1-OE lines grew faster and accumulated more Pi in vivo under Pi deficient conditions (Fig. 3a−c), and Pi had a flower-promoting effect[34]. MdSAT1-OE lines germinates 3-4 hours earlier than Col seedlings (Fig. 5b). The initiation of rosette leaves and flowering was significantly earlier in the MdSAT1-OE lines than in Col, and was more pronounced under the LP than the HP treatment (Fig. 5a, c & d). The MdSAT1-OE lines had significantly more rosette leaves than Col under the LP treatment (13.86/13.65/14.09:9.56), but no difference was detected under the HP treatment (23.98/22.58/22.09:20.98) (Fig. 5e). These indicate that overexpression of MdSAT1 advances the life process of Arabidopsis, including increasing the number of rosette leaves and early flowering, especially under Pi deficiency conditions. It is well known that flowering is regulated by flowering-related genes. The qRT-PCR analysis showed that the MdSAT1-OE lines had higher expression levels of flowering-promoting genes (AP1, FT, FUL, CO, and CAL) than Col, while the expression of the flowering-inhibiting gene FLC was lower than that in Col (Fig. 5f). Thus, we hypothesized that increased expression of the flowering-promoting genes in the MdSAT1-OE lines promotes flowering.
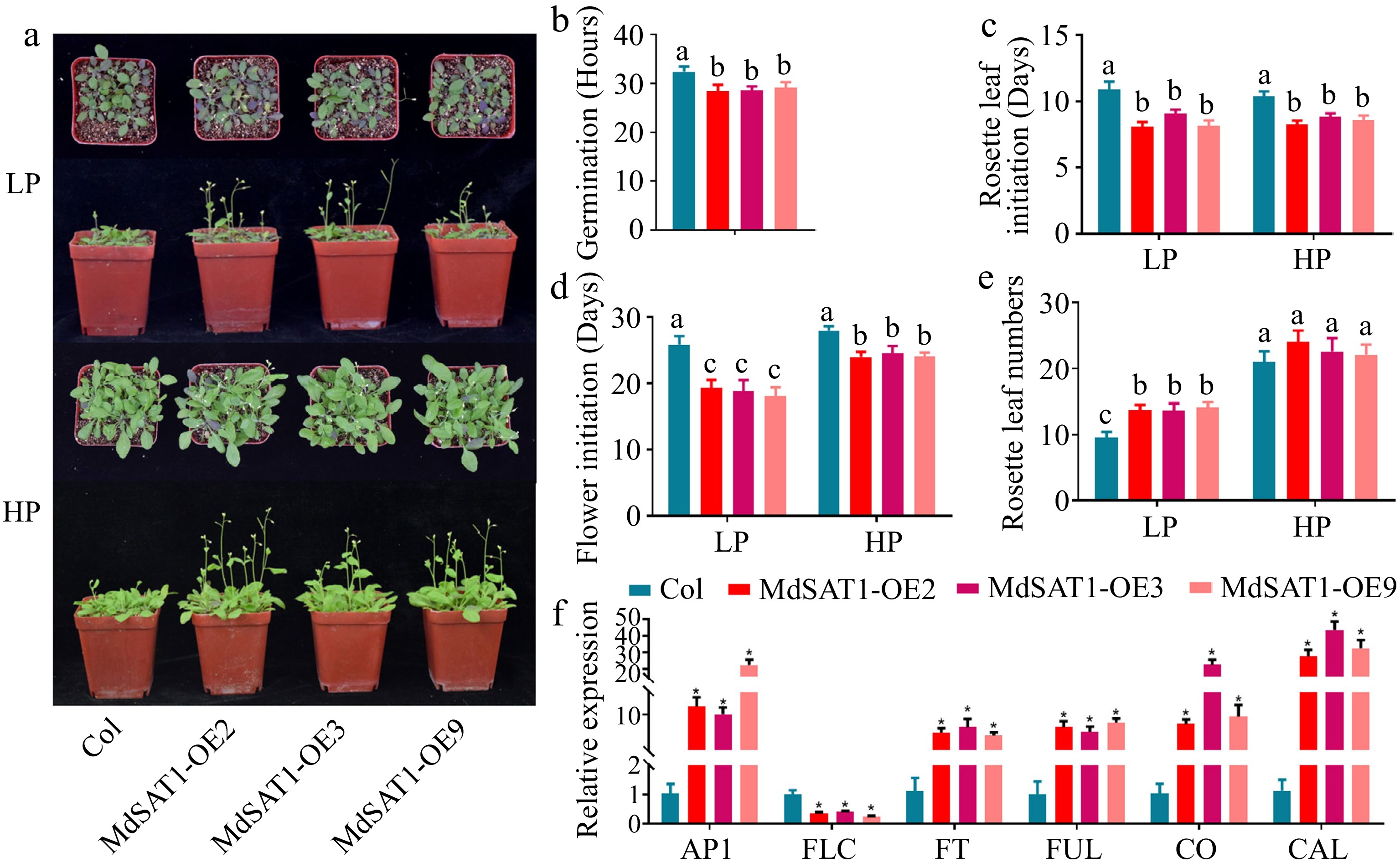
Figure 5.
MdSAT1 promotes early flowering. MdSAT1-OE and Col Arabidopsis plants were grown for 5 weeks under LP (1.25 μM K2HPO4) or HP (1.25 mM K2HPO4) conditions. Plants grown at 23 °C/21 °C for day/night with 16L/8D, with an irradiance of 150 μmol·m−2·s−1 and relative humidity of 60%−70%. (a) Morphological changes, (b) germination, (c) initiation of rosette leaves, (d) flower initiation, and (e) the number of rosette leaves are presented. (f) Transcript levels of AP1, FLC, FT, FUL, CO, and CAL in MdSAT1-OE and Col plants under LP conditions. Error bars represent standard deviations (n ≥ 3). * and different letters above the bars indicate significantly different values (p < 0.05).
MdSAT1 promotes Pi deficiency-induced leaf senescence
-
An important feature of senescence is the intensive transfer of nutrients (including Pi) from the vegetative organs to the developing seeds[34, 35]. Consequently, the senescence process of the MdSAT1-OE lines was faster than that of the Col, particularly in the LP treatment (Fig. 6a, b). Chlorophyll degradation leading to leaf chlorosis and increased ion leakage due to cell membrane fragmentation are important indicators of leaf senescence[36, 37]. Taken together the results suggest total chlorophyll content, Fv/Fm ratio, and ion leakage rate were consistent (Fig. 6c−e). These results suggest that MdSAT1 acts as a positive regulator of leaf senescence to maintain late reproductive growth in plants, particularly in response to Pi deficiency.
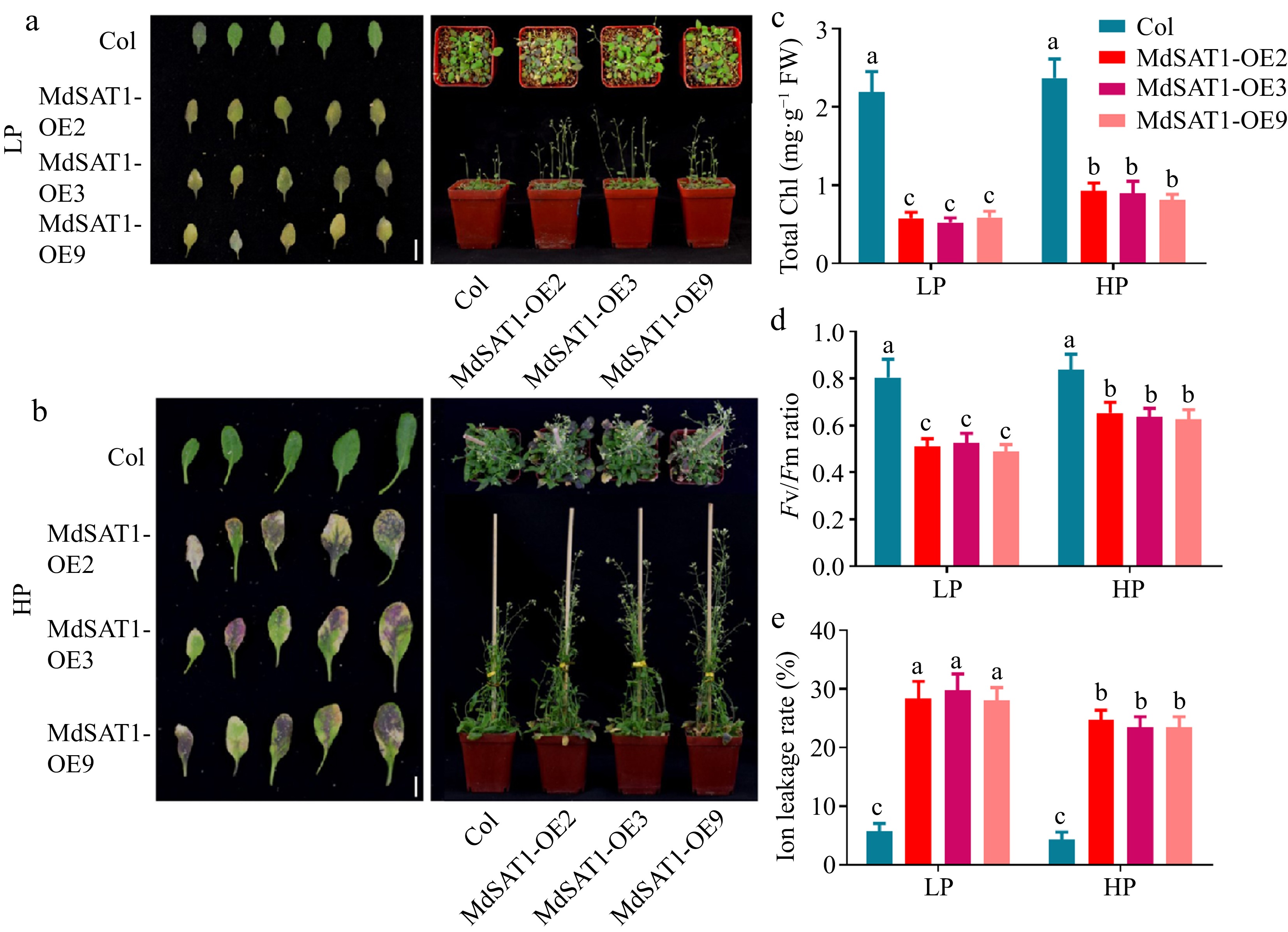
Figure 6.
MdSAT1 promotes leaf senescence. MdSAT1-OE and Col Arabidopsis plants were grown for 7 weeks under LP (1.25 μM K2HPO4) or HP (1.25 mM K2HPO4) conditions. Plants grown at 23 °C/21 °C for day/night with 16L/8D, with an irradiance of 150 μmol·m−2·s−1 and relative humidity of 60%−70%. (a), (b) Morphological changes, (c) total chlorophyll, (d) Fv/Fm ratio, and (e) ion leakage rate are presented. Error bars represent standard deviations (n ≥ 3). Different letters above the bars indicate significantly different values (p < 0.05).
-
Soluble phosphate is low in soil, which greatly limits its utilization by plants, and affects their growth and development[38,39]. Plants have evolved a set of responses to maintain Pi homeostasis, collectively known as PSRs[10]. Many transcription factors involved in PSRs have been identified after analyzing the Pi deficiency transcriptome, identifying low phosphate mutants, and comparing sequences between species[27]. The transcription factors involved in the response to Pi starvation have been experimentally confirmed and include PHR1[40], MYB62[25], ZAT6[30], bHLH32[29], and WRKY75[28]. Most PSR transcription factors are induced by Pi deficiency, such as the expression of ZAT6, WRKY75, and bHLH32 is induced by Pi deficiency in all parts of the plant; OsPTF1 expression is induced by Pi deficiency only in roots[28−30, 41]. In the present study, we determined that the bHLH transcription factor MdSAT1 was involved in the PSRs in apples, and MdSAT1 expression was significantly induced in roots by Pi deficiency (Fig. 1). Adaptive changes in root architecture are a major form of PSR occurring in plants. The root system of crops generally exhibits shorter primary roots and clustered roots to meet the nutritional needs of the ground under Pi deficiency. The roots stimulate lateral root vitality to promote the increase and lengthening of lateral roots, which take up more P[42]. Our results show that the MdSAT1-OE lines produced longer primary roots and more lateral roots in response to Pi deficiency when compared to Col (Fig. 2). This change in root architecture caused by MdSAT1 increased the contact area of the root system with the soil and increased the number of root tips. The root tip is an important P-uptake region that locally perceives exogenous Pi[13, 42]. All nutrient deficiencies stimulate the production of root hairs[8], yet MdSAT1 failed to produce root hairs under Pi deficiency (Supplemental Fig. S2). We hypothesized that MdSAT1 regulates root architecture to increase Pi uptake and allow the plants to be more adaptive under Pi deficiency. The regulation of root architecture by MdSAT1 was investigated in our previous work. MdSAT1 improves plant salt tolerance and reduces drought sensitivity by forming longer primary roots, but it is unable to respond to Cd, Cu, or ETH treatments by regulating the growth of primary roots[43]. MdSAT1 is expressed at the highest level at the beginning of the Arabidopsis lateral root primordial stage and promoted lateral root genesis and growth[33]. MdSAT1 also regulates the development of root hairs by regulating the transcript levels of root hair-related genes in an ammonium dosage-dependent manner[33]. These findings, together with the present results, show that MdSAT1 helps in adaptation to changes in the external environment by improving root architecture.
APase is an adaptable enzyme whose activity is influenced by the P supply status of the plant[44]. When the plant obtains a P starvation-induced signal, the root system secretes large quantities of APase into the inter-root zone to hydrolyze organic phosphorus in the growth medium to release
${\text{PO}_4^{3-}} $ Phosphorus fertilizer, also known as flower fertilizer, promotes the development of reproductive organs late in the growth cycle[34]. The flowering time of the MdSAT1-OE lines was earlier than that of Col and more pronounced in the Pi deficient environment (Fig. 5a–e). Flowering time is regulated by multiple pathways, which are ultimately controlled by flowering-related genes[48]. The expression levels of genes that promote flowering (AP1, FLC, FT, CO, and CAL) were significantly higher in the MdSAT1-OE lines compared with Col; the expression levels of gene that inhibit flowering (FUL) were significantly lower (Fig. 5f)[49]. Therefore, we hypothesized that MdSAT1 might affect the expression of flowering-related genes at the transcriptional level, thereby promoting flowering. Leaf nutrients are remobilized and relocated from dying leaves to seeds or other storage tissues during senescence, thereby contributing to the fitness and survival of the plant[35, 50−52]. Leaf chlorosis due to the degradation of chlorophyll is a characteristic of leaf senescence[36]. Pi plays a very important role in photosynthesis and chlorophyll synthesis[53]. Here, the results show that MdSAT1 in a Pi-deficient environment promoted senescence in old leaves, showing decreases in chlorophyll content and the Fv/Fm ratio (Fig. 6a–d). Plants are susceptible to rupture of cell membranes under unfavorable conditions, such as nutrient stress, resulting in an increase in ion leakage due to extravasation of the cytosol[37]. The MdSAT1-OE lines had the highest ion leakage rates under the LP treatment, indicating that the long-term Pi-deficient environment severely damaged the cell membranes of the MdSAT1-OE lines. In conclusion, we speculate that MdSAT1 shortens the plant growth period by reactivating nutrients in aging leaves to transfer to storage organs, such as seeds.
In summary, this study shows that the transcription factor MdSAT1 positively regulates Pi deficiency resistance and root architecture and is involved in the flowering and senescence processes of plants. The results of this study strengthen our understanding of transcriptional regulation in response to Pi deficiency in apples and provide a basis for future studies on the mechanisms by which MdSAT1 responds to other nutrient stressors.
-
In vitro shoot cultures of the 'Royal Gala' (Malus × domestica 'Gala') cultivar were subcultured at 1-month intervals on MS medium containing 0.1 mg·L−1 of gibberellin (GA), 0.5 mg·L−1 6-benzylaminopurine (6-BA), and 0.2 mg·L−1 1-naphthalene acetic acid (NAA), and rooted in MS medium containing 0.15 mg·L−1 NAA at 24 °C under a long-day photoperiod (16L : 8D)[54]. Uniformly growing apple seedlings were selected for Pi starvation under hydroponic conditions with ddH2O for 1 week and subjected to different Pi treatments. The different Pi concentrations the seedlings were treated with 0, 1.25, 25, 625, and 1,250 μM K2HPO4 for 15 d, supplemented with 2,500 μM K+ using the corresponding concentration of K2SO4. The different seedling treatment times were treated with 1.25 μM (low P, LP) and 1.25 mM K2HPO4 (high P, HP, represents the P concentration in MS medium) for 0, 1, 3, and 6 h and 1, 6, and 15 d. The LP concentration treatment group was supplemented with 2,500 μM K+ using the corresponding concentration of K2SO4. The sample were collected at day phase. Then, RNA was extracted from the apple seedlings with different Pi concentrations and treatment times, and detected by real-time-polymerase chain reaction (RT-PCR) analysis.
Arabidopsis seeds were disinfected in 75% alcohol and 2.8% sodium hypochlorite and sown on 1/2 MS medium solid culture plates (containing 1.5% sucrose and 0.8% agar powder, pH 5.9) at 4 °C in the dark for 4-d of stratification. The seeds were germinated and grown at 23 °C/21 °C for day/night with 16L/8D, with an irradiance of 150 μmol·m−2·s−1 and relative humidity of 60%−70%.
Two or three days after germination, the different genotypes of Arabidopsis (MdSAT1-OE, Col) seedlings were transplanted and grown on LP or HP modified basic nutrient solution (containing 1.0 mM CaCl2, 1.0 mM NaH2PO4 (LP: 1.25 μM; HP: 1.25 mM), 1.0 mM MgSO4, 0.1 mM FeNa2EDTA, 50 μM MnSO4·H2O, 50 μM H3BO3, 0.05 μM CuSO4·5H2O, 0.5 μM Na2MoO4·2H2O, 15 μM ZnSO4·7H2O, 2.5 μM KI, and 0.05 μM CoCl·6H2O, organic matter (2 μM C6H12O6·2H2O, 0.02 μM NC5H4COOH, 0.001 μM C12H17ClN4OS·HCl, 0.01 μM C8H11O3N·HCl, 0.1 μM NH2CN2·COOH, 1.5% sucrose and 0.8% agar powder, pH 5.9). The final K+ concentration was adjusted to be the same in both solutions by adding K2SO4. The root hairs were observed after 3 d on the treatment media, and the primary root and lateral root were observed after 7 d on the treatment media. The inorganic salts used in this article are provided by Tianjin Kaitong Chemical Reagent Co., Ltd.
The different types of Arabidopsis [MdSAT1-OE, Columbia (Col)] seedlings were transplanted to vermiculite 7 d after germination, irrigated with dH2O water, and watered weekly with a modified Hoagland's nutrient solution with either LP or HP, pH 5.9. The final K+ concentration was adjusted to be the same in both solutions by adding K2SO4. The details of the growing periods for the phenotypic observations, the gene expression analysis, and the physiological statistics are shown in the figure legends.
Transgenic materials
-
The MdSAT1 (NCBI ID: XM_029109715.1) open reading frame was cloned, and a 2 kb promoter fragment (NCBI ID: OU744551.1) was inserted upstream of the MdSAT1 transcription start site in the pRI-101 vector (Takara, Dalian, China) and the pCAMBIA1300 (Takara, Dalian, China) vector with the GUS reporter gene, respectively. Then transgenic Arabidopsis MdSAT1-OE (Supplemental Fig. S3) and ProMdSAT1::GUS were generated through Agrobacterium-mediated genetic transformation[43, 54,55]. The nutrient culture for screening homozygous lines is 1/2 MS medium solid culture plates (containing 50 mg/L kanamycin, 1.5% sucrose and 0.8% agar powder, pH 5.9), and the growth conditions is at 23 °C/21 °C for day/night with 16L/8D, with an irradiance of 150 μmol·m−2·s−1 and relative humidity of 60%−70%. After germination, the Arabidopsis seedlings were transplanted and grown in vermiculite with Hoagland's nutrient, homozygous transgenic Arabidopsis plants were obtained at the third generation (T3) (Shandong Agricultural University, China).
RNA extraction
-
The Omni Plant RNA Kit (tDNase I) was used to extract the plant RNA (Tiangen, Beijing, China). The extracted RNA was stored in tissue RNA preservation solution (RNAfollow M6100 New Cell & Molecular Biotech) for protection and stored in an ultra-low temperature refrigerator for gene expression analysis.
Quantitative RT-PCR analysis of gene expression
-
The cDNA required for quantitative PCR was synthesized using the PrimeScript First Chain cDNA Synthesis Kit (Takara, Dalian, China). The synthesized products were used as templates for quantitative RT-PCR to detect the expression levels of selected genes. Each template contained three biological replicates and executed the program of pre-denaturation at 95 °C for 10 min, denaturation at 95 °C for 15 s, annealing at 56 °C for 15 s, extension at 65 °C for 10 s, and 40 cycles with a StepOne Plus Real-Time PCR System[56]. Apple 18S rRNA (AT1G13180, F: CGTGACCTTACTGATTAC; R: TTCTCCTTGATGTCTCTT) and Arabidopsis actin rRNA (DQ341382, F: ACACGGGGAGGTAGTGACAA; R: CCTCCAATGGATCCTCGTTA) were used as the loading controls. The PCR analysis was performed using the specific primer sequences listed in Supplemental Table S1, and relative gene expression was calculated using the 2−ΔΔCᴛ method[57].
Physiological measurements
Pi content
-
The Pi content was extracted from MdSAT1-OE and Col Arabidopsis after treatment in LP or HP vermiculite for 3 weeks, using a method described previously[58]. The pre-weighed fresh samples were dissolved in 1 mL of 1% concentration and repeatedly frozen and thawed. Next, 100 μL of the extract was mixed with 200 μL of ultrapure water and 700 μL of mix buffer [0.42% NH4MoO4, 2.86% (v/v) H2SO4, and 10% (w/v) ascorbic acid in a 6:1 ratio, pH 3.1] at 37 °C for 1 h. Then, Pi content was determined at A820 according to a standard curve[58].
Acid phosphatase (APase) activity assay
-
All of the extraction steps were carried out on ice. Root samples of MdSAT1-OE and Col Arabidopsis were powdered in liquid nitrogen and transferred to an extraction buffer (10 mmol·L−1 sodium acetate, pH 5.6). The homogenized samples were centrifuged twice at 12,000 × g and 4 °C for 30 min, and the supernatants of each time were used to measure APase enzyme activity. The enzyme activity measurements were performed at 37 °C for 30 min in 100 mmol·L−1 sodium acetate buffer (pH 5.6) containing 5 mmol·L−1 of ρ-nitrophenyl phosphate (ρNPP). To determine the enzyme activity, we constructed a standard curve using known concentrations of
${\text{PO}_4^{3-}} $ Quantification of chlorophyll content, the Fv/Fm ratio, and ion leakage rate
-
The following experimental materials were extracted from MdSAT1-OE and Col Arabidopsis after treatment in LP or HP vermiculite for 7 weeks.
Total chlorophyll content was measured as described by Fitter et al[59].
Fv/Fm ratios were analyzed according to the manufacturer's instructions (OS-30p instrument, OptiSciences) as described previously[60].
Ion leakage rates were measured as described previously[61].
GUS staining
-
Arabidopsis (ProMdSAT1::GUS) seedlings [10 d old after growing on 1/2 MS medium (containing 1.5% sucrose and 0.8% agar powder, pH 5.9)] solid culture plates) were watered with ddH2O for 3 d, then treated for 1 h with 0, 1.25, 25, 625, and 1,250 μM K2HPO4, supplemented K+ to 2.5 mM with the corresponding concentration of K2SO4 and pH adjusted to 5.9, immersed in GUS staining solution for 1 h in the dark, and photographed. The GUS staining buffer was prepared according to Feng et al.[62].
Root system analysis
-
The primary roots of Arabidopsis were photographed and observed using a body vision microscope. The number and length of the root hairs in the 4-mm area 2 mm above the root tip were counted using Digimizer software.
Statistical analysis
-
Three biological replicates with three parallel experiments were used to analysis the data in this study. All data were statistically analyzed by two-way analysis of variance and Tukey's multi-comparison test in SAS software version 8.1 (SAS Institute, Cary, NC, USA). A p-value < 0.05 was considered significant. The results are expressed as means with corresponding standard errors.
-
The authors confirm contribution to the paper as follows: experiments design: Wang X, Gao W; research performed: Li T, Feng Z, Yang Y , Li M and Li G; data analysis: Li T, Feng Z and You C; manuscript preparation: Li T, Wang X. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analysed during this study are included in this published article.
This work was supported by the National Natural Science Foundation of China, 31972378 (Xiaofei Wang), China Agriculture Research System of MOF and MARA, CARS-27 (Xiaofei Wang), and Shandong Province Key R&D Program, 2022TZXD008-02 (Xiaofei Wang). We sincerely thank our team leader Dr. Yujin Hao, who will be remembered for his great achievement and for the support and help in our work.
-
The authors declare that they have no conflict of interest.
- Supplemental Fig. S1 Dynamic changes of ProMdSAT1::GUS activity under different phosphate concentrations.
- Supplemental Fig. S2 MdSAT1 regulates root hair development and growth.
- Supplemental Fig. S3 Relative expression of the homologs of AtSAT1.
- Supplemental Table S1 Primers for qRT-PCR.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Li T, Feng Z, Yang Y, Li M, Li G, et al. 2023. Functional identification of the bHLH transcription factor MdSAT1 in the phosphate deficiency response. Fruit Research 3:26 doi: 10.48130/FruRes-2023-0026
Functional identification of the bHLH transcription factor MdSAT1 in the phosphate deficiency response
- Received: 25 June 2023
- Accepted: 26 July 2023
- Published online: 27 October 2023
Abstract: Inorganic phosphate (Pi) starvation severely affects the normal growth and development of plants. In this study, MdSAT1, a Pi-responsive bHLH transcription factor, was isolated from apples. Ectopic expression of MdAST1 in Arabidopsis increased the number of lateral roots and root tips, as well as the transcript levels of genes related to Pi uptake and transport; thus, improving Pi utilization in response to a Pi deficiency. Ectopic expression of MdSAT1 significantly accelerated flowering and leaf senescence in Arabidopsis under a Pi deficiency. Taken together, the present study provides a basis for an in-depth investigation of the mechanisms of MdSAT1 on apple Pi uptake and utilization as well as plant growth and development.
-
Key words:
- Flowering /
- MdSAT1 /
- Phosphate deficiency /
- Root growth /
- Leaf senescence













