-
Humans have domesticated plants and animals since the Neolithic revolution around 13,000 years ago, which enabled the first sedentary agricultural societies and eventual development of human societies[1]. In domestication processes, morphological and physiological changes in plant and animal traits are chosen and developed through specific nurturing and breeding of wild species for the enhancement of specific beneficial traits[2]. While humans have had by far the highest success rate in domesticating species, in number and geographical area, domestication is in no way specific or limited to homo sapiens, as ants have been observed to domesticate aphids, viruses have been domesticated by parasitic wasps, and more[3,4]. Indeed, by looking at domestication as a type of co-evolution between two species, for reciprocal fitness increases and geographic spread, we can assume domestication to be a specific kind of mutualism, in which both species nurture one another for traits through multispecies interactions[5].
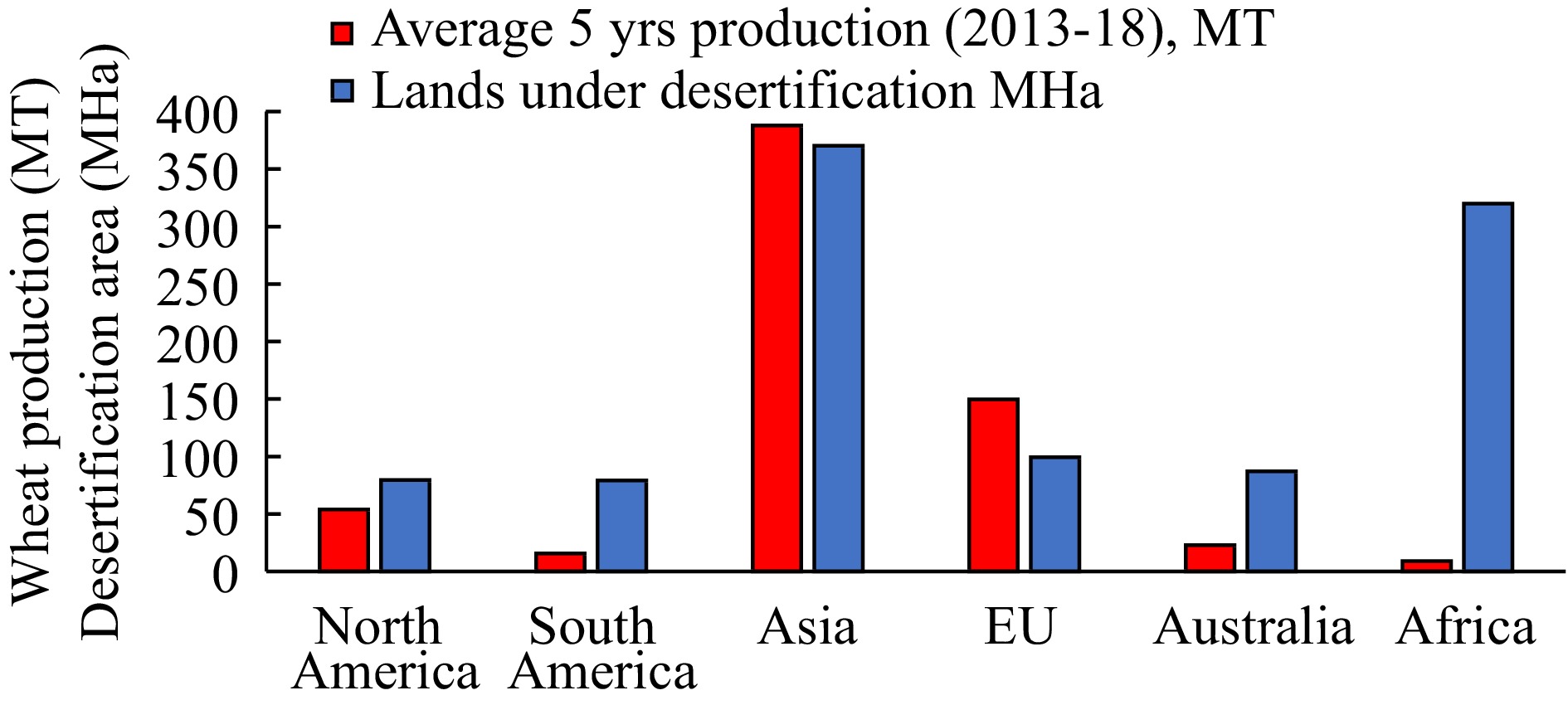
Grasses are among the most important staple crops for human use. Historically, wheat (triticum sp.) and barley (Hordeum vulgare) were the two first domesticated plants, followed by maize (Zea mays) and rice (Oryza sativa), with many more following for indirect consumption trough fodder, or industrial uses, like rye-grass (Lolium sp.) and cottons (Gossypim sp.) respectively[6]. Wheat, maize and rice are the three major cereal crops cultivated and consumed world wide, supplying food, feed and industrial raw materials for more than one-third of the world's population as both spring and winter crops[7,8]. Wheat is the second most produced staple crop, after maize (World food situation: FAO cereal supply and demand brief, 2016), while using the most land area compared to any other food crop, at 220.4 MHa (United Nations, 2016). Among the main producers, most are situated in areas under critical danger of desertification, according to UNEP's 1997 World Atlas of Desertification (2nd Edition), as can be seen in Fig. 1, and as such, the food production growth needed to feed the world population up to 2050 will come 90% from intensification of existing agricultural systems, and only 10% from expanding arable land.
-
In plants, domestication is usually associated with phenotypic traits like germination success rates, size, seed retention, root architecture, and other physiological and morphological traits, in functions of desired edibility and usage of specific plant organs[9−11]. From the plant's point of view, the development of such traits will depend on a shift in resource allocation, usually from stress tolerance, towards growth and reproduction[12,13].
The genetic suite of traits marking the divergence from a crop's wild relative is called 'domestication syndrome' and will take place biochemically as a conversion from the production of growth-related primary metabolites (common among all plants) to defense-related secondary metabolites that will be unique to the plant species, and/or to the event that induced their synthesis[14,15]. This exchange is critical since plant resources are limited and its defenses are metabolically expensive, carbon and nitrogen-wise[16]. The amplification of a defensive trait is usually generated through specific selection of individual genes and their neighboring regions (between 10−100 kb)[17] thus also possibly affecting random non-deleterious genes. Indeed, tradeoffs can also happen naturally due to the genetic links and placement of genes in chromosomes, as observed in Salicaceae seeds for example, which demonstrate a tradeoff between seed number and seed mass naturally, with seed mass strongly correlating with seed longevity[18]. This tradeoff can then easily turn into a genetic bottleneck in dry seasons since the species depends on plain floods for its survival. Thus, natural and artificial (anthropogenic) causes will eventually bring forth a reduction in gene diversity, as can be observed in the domesticated maize (Zea mays ssp. Mays), which demonstrates a loss of 38% in nucleotide diversity in comparison to its ancestor (Zea mays ssp. Parviglumis)[19], and in wheat that demonstrated 69% and 84% reduced diversity in bread and durum wheat respectively[20]. Many other cultivars, like corn, olives and sunflowers have been compared with their wild ancestors and show that the hypothesis of a trade-off between increased yields and defenses to be widely supported[12,21,22].
The domestication of wheat is especially well recorded due to the species importance to humans, with its first polyploidization event occurring around 500,000−150,000 years before the present, to form a new amphi-tetraploid species with 14 chromosome pairs named Triticum turgidum L. sp dicoccoides, which was in turn domesticated to form Triticum dicoccum – the direct ancestor of durum wheat. A second polyploidization occurred around the Neolithic (Agriculture) revolution ~10,000 years before present, between Triticum dicoccum and the wild diploid species Triticum tauschii to form the modern hexaploid Triticum aestivum – the 'bread wheat'[23], characterized by traits like reduced maturity shattering of spikelet, glume reduction, loss of seed dormancy, increased carbohydrates and decreased proteins and minerals in the seed's germplasm[24].
Additional phenotypical differences between domesticated and wild emmer have been observed to include increased shoot biomass in general, with higher total leaf area, and shoot fresh weight in particular, among the domesticated varieties in comparison to their wild counterparts[25]. Also, underground phenotypical differences to the plant's root architecture have also been observed to be substantial, with root biomass in general[26], and specific traits like primary and total root length, depth, width and dry weight in particular being observed to be higher in domesticated genotypes, compared to wild ones[25]. Differences were also observed in the exudation profile of the root's system among domesticated and wild genotypes, were despite the observation of general increase in exudation of organic compounds[27] in domesticated genotypes, specific compounds like poly alcohols, were found to be significantly higher in the wild emmer than in domesticated durum varieties[26].
It is important to state that along the plant's domesticated traits, its environment also had progressive changes, with human development and agrotechnical advances like increasing inputs of chemicals as fertilization and pesticides – leading to a decrease in both necessity and capacity of plants to self-support naturally[28], along direct changes to the soil physical, chemical, and microbial components[29]. Indeed, higher abundances of bacterial endophytes have been observed in wheat rhizosphere under low nutrients treatment[30], which along an increased amount of organic acids released into the rhizosphere in nutrient-poor plants[31], both support the theory that the domestication syndrome of wheat, as far as its effects on its rhizobiome, was driven by both genetics and human manipulation of agricultural soil.
-
The rhizosphere harbors a substantial number of microorganisms interacting with roots, with bacteria and fungi accounting for more than 90% of the total soil microbial biomass[32,33]. Fungi and bacteria in the soil interact with plants along a parasite-mutualist continuum, in which the microbes may harm or benefit its host as a function of the relative benefits and costs to each species[34]. Indeed, soil microorganisms play an essential part on plants' health and performance by positively or negatively manipulating their biochemistry, development and physiology[35].
Plant growth-promoting rhizobacteria (PGPRs), microorganisms that colonize the rhizosphere and roots, are known to improve both yields and tolerance of biotic and abiotic stresses in plants[36,37]. PGPRs can also help enhance plant growth through nitrogen fixation[38], hormonal secretions[39], specific antifungal and antibiotic activity[40−42], facilitation of essential minerals uptake[43,44] and induction of systemic resistance[45−48]. Specific strains have been found to help reduce the need for chemical fertilizers while maintaining commercially viable yields and grain quality[49] thus contributing to local and global environment by decreasing non-renewable resource dependence[50]. Furthermore, different studies demonstrate that there is also potential for exopolysaccharides (EPS) producing bacteria, like Bacillus subtilis and Azospirillum brasilense in water-stress amelioration[51].
Fungi also play a critical role in the rhizospheric microbial community, having a total soil microbial biomass ratio of fungi: bacteria ranging from 1:1 in agricultural soils and up to 1000:1 in coniferous forests[52], as intensity of soil management shows a high correlation to lower values, a phenomenon commonly thought to be caused by tillage and fertilization[53]. The Glomeromycota Arbuscular mycorrhiza fungi (AMF) for example, are considered a main player in maintaining soil carbon pools, as it can forms symbiotic relationships with nearly 90% of all plant species[54,55] by adding their hyphae to plant's roots, thus increasing the plant's water and mineral uptake network[56]. AMF will also expedite decomposition of organic matter, improving soil structure and nutrient carriage capacity[57]. Other fungi species have been found to increase grain yield, nitrogen uptake[58], alleviate biotic and abiotic stresses[59,60], promote growth through phosphorous solubility in soil[61] and increase seedling roots[62], among many other benefits.
As such, maintaining a beneficial and healthy rhizospheric community, should be in the plants' human caretakers best interests[63] as partially summarized for wheat in Table 1 (bacteria) and Table 2 (fungi) below.
Table 1. Known PGPR species for wheat.
Organism Benefit Source Azospirillum brasilensis Sp. 245 Growth rate. Water stress alleviation (Alvarez, Sueldo, and Barassi 2015) Azospirillum lipoferum Water stress alleviation (Agami, Ghramh, and Hashem 2017) Burkholderia phytofirmans Water use efficiency. Grain yield. Photosynthetic rates (Poupin 2015) Bacillus amyloliquefaciens Temperature stress alleviation (Tiwari et al. 2017) Azospirillum brasilense Sp245 Temperature stress alleviation (Hernaández-esquivel and Castro-mercado 2020) Pseudomonas Putida Temperature stress alleviation (Zulfikar Ali et al. 2011) Pseudomonas fluorescens Salt stress alleviation (Fathalla and El-Mageed 2020) Pantoea agglomerans Temperature stress alleviation (Cherif-Silini et al. 2019) Mycobacterium sp Temperature stress alleviation (Dilfuza Egamberdieva and Phylogeny 2014) Pseudomonas Putida Water stress alleviation (Mahmoudi et al. 2019) Pseudomonas extremorientalis Salinity tolerance (D. Egamberdieva 2011) Pseudomonas chlororaphis Salinity stress alleviation (Mahmoudi et al. 2019) Bacillus pumilus Salinity stress alleviation. Proline accumulation. P solubilization (Ansari, Ahmad, and Pichtel 2019) Hallobacillus sp. SL3 Root length. Dry weight (Ramadoss et al. 2013) Enterobacter asburiae Number of tillers. Grain weight. Growth rates (Kang et al. 2015) Pseudomonas aureantiaca Increased seedling root growth (Dilfuza Egamberdieva 2009) Bacillus safensis Increase in root and shoot biomass, height of plants, yield, as well as increase in chlorophyll content (Chakraborty et al. 2013) Aeromonas hydrophila/caviae (strain MAS-765 Increased the dry matter yield of roots and shoots (Ashraf, Hasnain, and Berge 2004) Bacillus mojavensis Increase in root and shoot weight, chlorophyll content, and nutrient uptake under salt stress (Pourbabaee AA, Bahmani E, Alikhani HA 2016) Lactobacillus plantarum Increased PGPR abundance (Agnolucci et al. 2019) Stenotrophomonas rhizophila Biotic stress resistance (Liu et al. 2021) Curtobacterium flaccumfaciens Growth promotor under drought (Hone et al. 2021) Table 2. Known beneficial fungi species for wheat.
Organism Benefits Source Morchella snyderi Increased root systems, biotic and abiotic stress alleviation (Ridout and Newcombe 2016) Penicillium sp. Abiotic stress alleviation Rhizophagus irregularis Nutrient uptake, growth, and yield (Li et al. 2018) Penicillum expansum Growth promotors trough P solubility in soil (Xiao et al. 2009) Mucor ramosissimus Candida krissii Azospirillum lipoferum Grain yield increase, nitrogen uptake (Gaur 1988) Trichoderma sp. Systemic resistance, mycotoxin suppression, seed germination rate increase (Basinska-Barczak Aneta 2020) (Basinska-Barczak Aneta 2020) (Nawrocka and Małolepsza 2013) Funneliformis mosseae Increased nutrient content, lower free radicals and increased root area under salt stress (Links et al. 2014) Penicillium olivicolor Increased seedling root (Khokhar et al. 2013) Sebacina Vermifera Increased biomass along resistance against biotic and abiotic stresses. (Ray and Craven 2016) Chaetomium sp. Biotic stress alleviation (Blaszczyk, Salamon, and Katarzyna 2021) Gloms etunicatum
Glomus intraradicesIncreased micro and macronutrients uptake in seedlings (Mardukhi et al. 2011) Aspergillus niger Catalase activity, nitrifier (Ripa et al. 2019) Aspergilus flavus -
In the past decade, with increasing understanding of the importance of the relationship between plants and their soil microbiome, the shaping of a plant's rhizobiome has come under study[64]. The 'Rhizospheric effect' (RE), defined as the chemical, biological and physical changes in the immediate vicinity of plants roots in the soil, and including the apoplastic spaces inside roots (the 'endorhizosphere'), is created through root exudates, decomposition of organic matter and rhizodeposition[65]. These will modify the soil environment in a way that will benefit the plant through recruitment of specific microorganisms, thus forming a distinct micro-environment and microbiome living within it[65,66].
It is estimated that plants allocate around 10% of all their fixed carbon (reaching up to 30%−40% of total fixed carbon in seedlings[67]) to compounds intended to be exuded to the rhizosphere[68]. These exudates can contain protons (H+), oxygen, water and inorganic acids, but mainly consist of primary and secondary metabolites, like amino acids, carbohydrates, organic acids, flavonoids, glucosinolates, hormones and etc.[69]. These compounds have shown to mediate interactions between the plant and its surroundings, from the immediate physical characteristics of the soil to other organisms like other plants, fungi, microbes insects or even herbivores[60,70−73]. Specific root exudates such as strigolactones and flavonoids for example, have been observed to play critical roles in communication between plants and rhizospheric microbes, from attracting beneficial PGPRs and mycorrhizal fungi, to staving off parasites[74].
The microbial composition of the rhizosphere has been observed to develop through both recruitment of a specific subset from the existing community in the soil[75], and through the seed's microbiome, inherited from its maternal tissues[76,77]. While there are various factors to the strength of the RE (and thus, the distinction of the new microbiome and rhizospheric physical properties from the bulk soil), the seed microbiome and its genotype have been observed to have a greater effect than the soil existing microbial community[77,78].
-
Although not all plants have been observed to maintain a distinct rhizobiome from the bulk soil, like in the case of rice and Arabidopsis[79,80], among those that do, it has been found that the rhizobiomes of domesticated crops have shown to differ significantly from their wild relatives[81,82].
Shifts in bacterial abundance and community composition are common in many domesticated interactions across evolutionary kingdoms. For example, a higher abundance of Bacteroidetes were found in the gut of hunter-gatherers' individuals of rural regions than in their modern, 'westernized' counterparts[83]. In plants, a general negative effect on the capacity for the formation of new symbiotic associations with PGPRs and mycorrhiza has been observed[29] in addition to decreased ability to benefit from existing mycorrhizal presence[84] in domesticated cultivars in comparison to their wild counterparts. Meta-analysis of root microbiome compositions consistently shows an enrichment in Bacteroidetes in wild relatives, while the predominant families in domesticated varieties are mainly Proteobacteria and Actinobacteria[11,85,86]. The exact mode of interaction between Bacteroidetes and roots is still unknown, but a cautious assumption on the dependency on root exudates can be argued based on the phylum's recognized capacity to degrade complex organic polymers[87].
It is also relevant to observe that in addition to the exudates' direct effects on the rhizobiome, there is also the indirect effect of the complexity and extent of root's presence effects on physical soil properties like pH[88], carbon content[89] and compaction[90], which cannot be ignored and has yet to be further studied and fully understood.
Root architecture, as it reflects closely the rhizospheric microbial architecture[65] must also be considered in addition to their exudates profile when comparing domesticated and wild plant varieties. Wild bean for example, which is known for its higher drought tolerance in comparison to its domesticated relative[91] demonstrates higher specific root length (SRL, i.e., root length per unit of root dry mass) and lower root density[92]. High SRL has been associated with higher efficiency of water search and uptake of nutrients in low water and nutrients soils, but the specific biochemistry of this correlation has not been clarified up to now. In wheat we see that wild relatives like Agropyron elongatum and T. turgidum spp. Dicoccoides demonstrate improved water stress adaptation in comparison to modern cultivars, seemingly through increased root biomass[93,94] and that in general, modern cultivars consistently demonstrate smaller shoot systems than their wild and older counterparts[95].
As a possible result of both architecture, exudates and probably other unknown parameters (such as carbon partitioning in the plant) differing between modern and wild wheat, we see also that in general, modern domesticated tetraploid and hexaploid (durum and bread accordingly) genotypes demonstrate a lower MD (mycorrhizal dependence – the degree of dependence on mycorrhizal symbiosis for maximum growth and yield) than their wild ancestors counterparts like T. tauschii[96]. Likewise, it is relevant to mention that domestication effects are still in development as observed by Hetrick et al., where it is described how varieties developed after 1975 had lower MD than those developed earlier[97].
-
A correlation can then be suggested, between the observed loss of genetic variability in domesticated plants in general and grasses in particular, and the limited capacity for interaction and recruitment of beneficial microorganisms, along other morphological and physiological domestication syndrome traits. Indeed, as wild crops are consistently more tolerant to stresses than their domesticated counterparts[98−100], and the extensive observed benefits of microorganisms' presence in tolerance of biotic and abiotic stresses[40,101,102], we can carefully assume that a part of the wild cultivar's durability to biotic and abiotic stresses is due to the biome it has recruited from the soil and inherited through maternal tissues in the seed. Indeed, wild genotypes microbiomes are proven to be consistently more diverse than their domesticated counterparts[92,103,104].
While a number of successfully developed commercial field crops inoculant exist based on popular and well-characterized species and strains of both fungi and bacteria, these products usually have unpredictable and unexpected lower performance in the field, due to the complexity of parameters in vivo and in situ between the product microbes, host and environment[99,105]. The gap between the expected positive effect of an inoculant, and its efficacy in the field, highlights the possibility that the use of microbial products, should be tailor-made for specific crops, climates, soils and geography, most of which would be solved by searching for specific microbes from the crop's wild relative's rhizobiome.
Latest studies elucidating specific differences in seed biomes between domesticated and wild genotypes, have shown various Pseudomonas ssp. (known to include several species beneficial in regard to general health, productivity, and even bio-control in plants)[27], and some Enterobacteria (known to include common PGPR species) species to appear in higher abundance in wild species seed metagenomes than in domesticated ones[106]. Also in rice, studies have found that despite the fact that both domesticated and wild species contained methylotropic and methanogenic archaea, some specific beneficial methanotrophs (Methylococcaceae and Methylocystaceae) had a higher affinity for the wild rice species[107] and could be further studied for their possible potential in domesticated crops.
These differences between various domesticated and wild genotypes represent a significant untapped potential for discovery of novel beneficial strains for agricultural crops, in a world focusing on the challenges of feeding a growing world population under changing climates.
Moreover, as domesticated cultivars have lost their communication and recruitment capacity, it is up to agriculture's human caretakers to take up the call and reestablish the beneficial dynamics between plants and its original biome.
This work has been partly financed by the Goldinger Trust, The Irving Goldman Foundation inc.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Blaschkauer M, Rachmilevitch S. 2023. Domestication in wheat affects its rhizobiome recruitment capacity: a review. Grass Research 3:5 doi: 10.48130/GR-2023-0005
Domestication in wheat affects its rhizobiome recruitment capacity: a review
- Received: 03 October 2022
- Accepted: 04 April 2023
- Published online: 24 April 2023
Abstract: Human domestication of grasses has been pivotal to human civilization as a main caloric source, however this has come at the expense of decreased genetic diversity. As plants evolved alongside a plethora of microorganisms, some of them critical to plant growth and health, domesticated plants demonstrate consistently changed rhizobiomes, along with lowered tolerance to stress. In the last few decades, the interest in specific beneficial microorganisms to staple crops has been growing gradually, due to improved high-output data techniques, extensive research, and rising concerns on the production of enough food for a growing world population undergoing world climate change. Here, we review how wheat domestication trade-off effects may have impacted the recruitment of an ideal rhizobiome assembly, describe known wheat-specific beneficial species of both fungi and bacteria, and propose the exploration of wild relatives and indigenous species for identification and reinstatement of beneficial microbial interactions that may have been lost through the effects of domestication.
-
Key words:
- Microbiome /
- Domestication syndrome /
- Rhizospheric effect /
- Rhizobiome /
- Wheat