-
Dianthus caryophyllus L. (carnation) is one of the most important floricultural crops in the world market. Double flower (DF) has been the key breeding target for carnations in the last century, resulting in the long dominant position of DF cultivars such as 'Master', 'Francesco' and 'Karen Rouge' in the carnation market. However, aesthetic habits change, especially for the younger generation. For instance recently, new cultivars with semi-double flowers (SDF) have become a new trend of carnation preference in China. Obviously, aesthetic diversity brings challenges but also provides opportunities for the commercial market.
In the last few decades, the molecular mechanism regulating flower development has been well studied in the model plant Arabidopsis, and the knowledge has also been transferred in part to ornamental plants[1,2]. In general, the A-, B- and C-class homeotic genes regulate the floral organ formation in a dependent or cooperative way[3,4]. The A-class genes control sepal formation alone and petal formation together with B-class genes; C-class genes determine the carpel fate alone and stamen formation together with the B-class genes[4,5]. In Arabidopsis, the function loss of the C-class gene AG can promote the over-accumulation of the A-class genes (AP1 and AP2), resulting in the homeotic conversion of stamens into petals[6,7]. In turn, the over-accumulation of AP2 can reduce the expression level of AG and lead to the same homeotic conversion of stamens into petals[8]. Similarly, the AG function leads to a petal number increase in Prunus lannesiana, Camellia japonica, Tricyrtis macranthopsis and roses[9−12]. And the mutation in miR172 target site causes the elevation of AP2 expression, resulting in the formation of double flowers in Prunus persica, roses, carnation, peonies, and camellias[13−16]. According to these studies, the DF phenotype is regulated basically by two genetic pathways, either through the loss of AG function or via miR172-mediated target-deficient of AP2[17,18].
The molecular mechanism underlying petal accumulation was studied also in carnation and the miR172 target-deficient of AP2 was uncovered[16]. However, the regulatory pathway of AG remains unknown. In this study, we cloned the two AG orthologs (DcAGa and DcAGb) and DcAP2 from one single flower (SF) and three SDF cultivars. Sequence polymorphisms causing amino acid changes were identified in DcAGa and DcAGb between SF and SDF cultivars while no difference was detected in the binding site of DcAP2. Further expression analysis indicated that these mutations caused the down-regulation of AGa and AGb. Our results suggest that the DcAG genes are associated with the semi-double flower phenotype as reported in other ornamental plants.
-
The single flower (SF) cultivar, 'Peach Party', typically has five sepals, five petals and 10 stamens. All three semi-double flower (SDF) cultivars possess five sepals but varying numbers of petals and stamens (Fig. 1, Table 1). The petal numbers of 'Pink Star', 'Purple Star' and 'Oscar' are 29.00 ± 3.30, 29.20 ± 3.33 and 26.00 ± 2.79, respectively, four times higher than the SF carnation while the stamen numbers of them are reduced by half, which are 5.20 ± 2.30, 4.90 ± 3.14 and 4.30 ± 2.06, respectively. There are also chimeric petals and stamens observed in the semi-double flower cultivars, including sharp petals similar to stamen and stamens with abnormal anthers, implying the homeotic transition from stamen into petals of the SDF cultivars.

Figure 1.
Characteristics of single and semi-double flower carnations. (a) 'Peach Party' (single flower, pink), (b) 'Pink Star' (semi-double flower, pink), (c) 'Purple Star' (semi-double flower, purple), (d) 'Oscar' (semi-double flower, red). Scale bars = 1 cm.
Table 1. Statistics of floral organs of the four cultivars.
Flower type Accessions Sepal
numberPetal
numberStamen
numberSingle Peach Party 5.00 ± 0.00 5.00 ± 0.00 10.00 ± 0.00 Semi-double Pink Star 5.00 ± 0.00 29.00 ± 3.30 5.20 ± 2.30 Semi-double Purple Star 5.00 ± 0.00 29.20 ± 3.33 4.90 ± 3.14 Semi-Double Oscar 5.00 ± 0.00 26.00 ± 2.79 4.30 ± 2.06 Sequence analysis of AGa, AGb and AP2 in SF and SDF carnations
-
It has been reported that the A- and C-class genes co-control the double flower phenotype of carnations. To understand the semi-double flower phenotype formation of this ornamental plant, we first cloned C-class genes from the SF cultivar and three other SDF cultivars. Multiple sequence alignments reveal that there are amino acid substitutions among the two types of cultivars in AGa and AGb. For AGa, the three SDF cultivars share two of the same amino acid changes (A62V, P73L) in the MADS-box domain. And 'Oscar' owns three more amino acid insertions, two at the end of the MADS-box domain and one in the K-box domain. 'Pink star' has another amino acid change (K167R) and an amino acid insertion in the K-box domain (Fig. 2a, Supplemental Fig. S1). 'Oscar' displays specific amino acid changes in the MADS-box domain (T34A) and K-box domain (Q110H) of AGb and shares three of the same amino acid changes with 'Pink star' and 'Purple star' in the K-box domain (K152I, V194A) and AG motif region (E236D) (Fig. 2b, Supplemenatal Fig. S2).
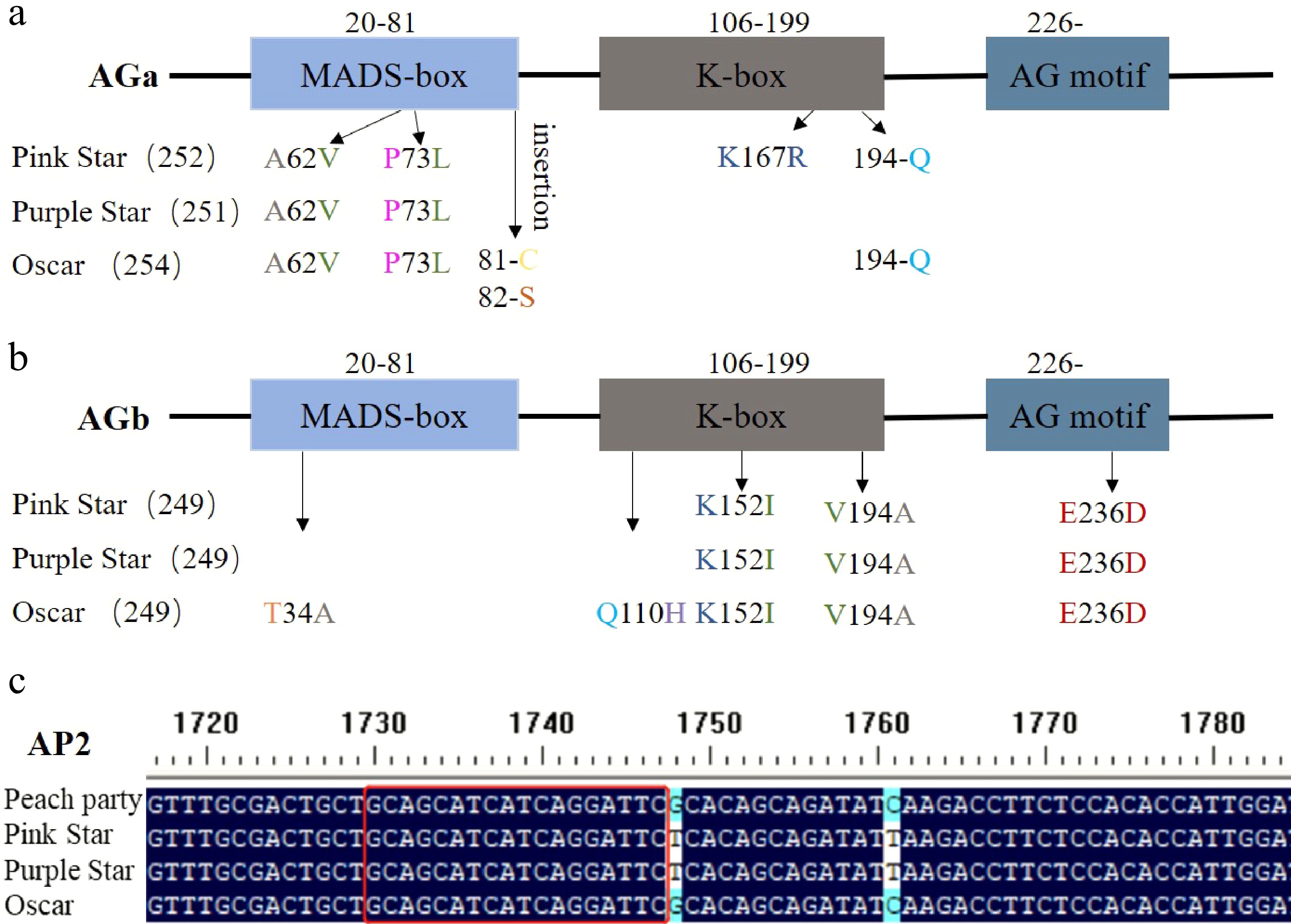
Figure 2.
Sequence polymorphisms of A-class and C-class genes in SF and SDF cultivars. (a) Amino acid substitutions in AGa. (b) Amino acid substitutions in AGb. (c) Sequence covering miR172 bind site of AP2.
We also analyzed sequences of the miR172 binding site of the A-class gene AP2. Our results show that there is no mutation in the miR172 binding site of AP2 isolated either from the SDF cultivars or the SF cultivar (Fig. 2c).
Expression analysis of ABCE model genes
-
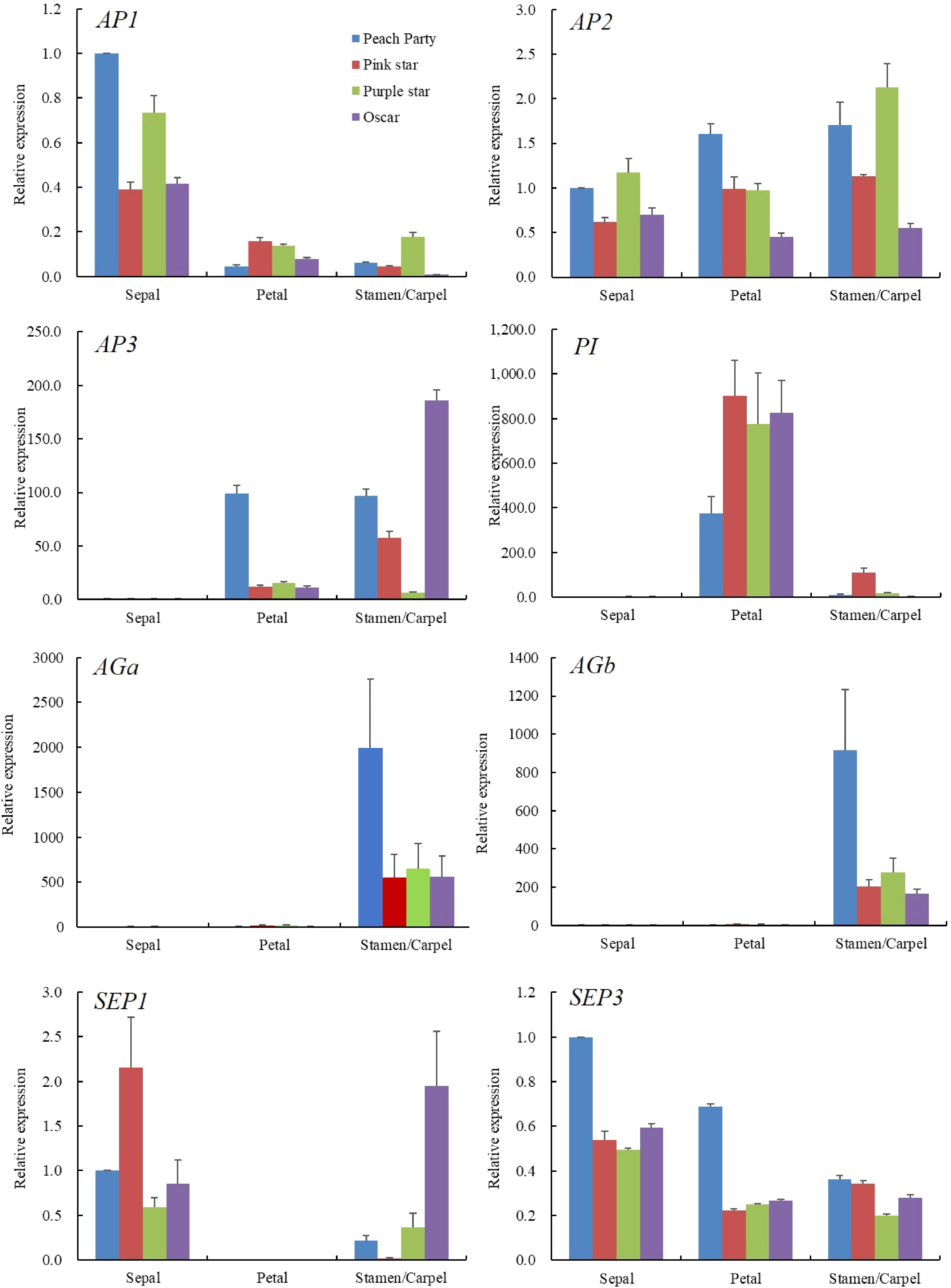
To investigate the relationship between the expression pattern of ABCE model genes and the SDF phenotype, we analyzed the expression levels of A-class (AP1 and AP2), B-class (AP3 and PI), C-class (AGa and AGb) and E-class (SP1 and SP3) genes in the sepal, petal and stamen/carpel of the four cultivars (Fig. 3). Firstly, we analysed the relative expression level of AGa, AGb and AP2, since the antagonistic expression between A- and C-class genes affects the homeotic conversion of petal and stamen. Our results show that AGa and AGb expressed significantly highly in the stamen/carpel and their expression levels in the stamen/carpel are reduced in the SDF cultivars compared with the SF cultivar. AP1 displayed a higher expression level in the sepal than that of other floral organs, while AP2 expressed consistently in all flower organs. Moreover, the expression level of AP1 was slightly reduced in the sepal but increased in the petal of the SDF cultivars. In contrast, AP2 expression is slightly increased in the petal of SDF cultivars. This is in line with the antagonism between class A genes and class C genes, indicating AG genes might play a major role in the formation of semi-double flower in carnations.
B-class genes, AP3 and PI, displayed higher expression levels in the petal and stamen/carpel than that in the sepal. However, theses two genes show opposite expression patterns in the SF and SDF cultivars. The expression level of AP3 is reduced significantly in the petal of the SDF cultivars while the expression level of PI is increased in the same organs. Similarly expression patterns happen in the carpel/stamen of the different cultivars. As for the E-class gene, the expression pattern of SEP3 is relatively smooth in all floral organs between single and semi-double flower, while there was no expression of SEP1 detected in the petal. One may conclude from the expression patterns of these genes that they fit the ABCE model in the general trend, but there is specificity in the expression of certain genes in specific tissues, implying their specific functions in floral organ formation which requires further exploration.
-
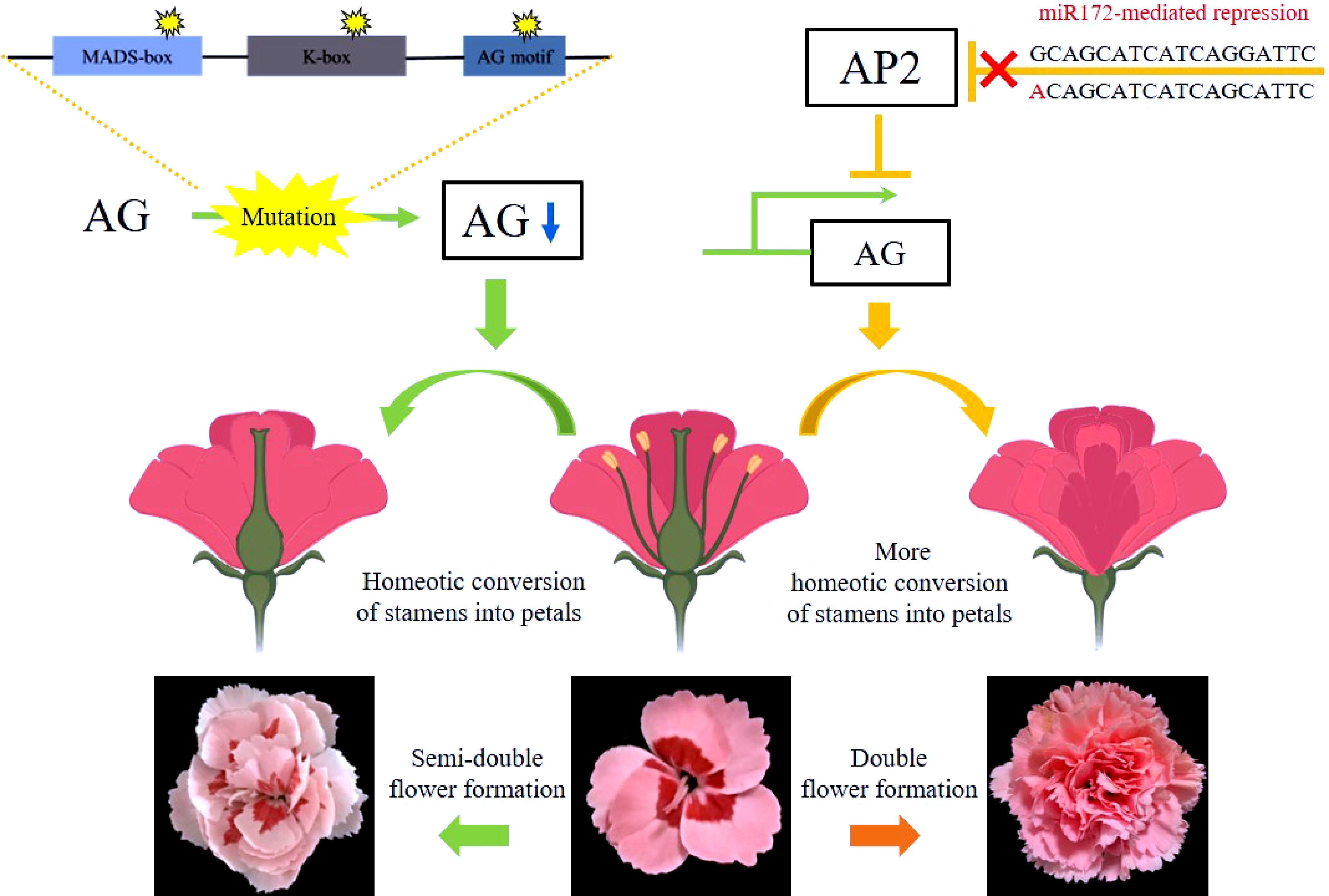
The genetic regulation networks governing the formation and subsequent development of each floral organ have been raised and supplemented in the past few decades, forming a well-known theory, namely the ABCDE model which suits most species[19]. AG, belonging to the C-class, is a key regulator of flower development. It specifies the fate of floral organs, whose mutation leads to the DF trait. Observation from scanning electronic microscopy shows that the Arabidopsis ag-1 mutant was defected in stamen development and displays extra petal formation[20]. Similarly, AG alleles with deletions in the exon were isolated from two Matthiola incana double flower cultivars. The deletions in the AG coding sequence also caused defects in its expression[21]. Studies in Magnolia stellate and Prunus lannesiana revealed that alternative transcriptional splicing of AG which lead to exon skipping also caused petal accumulation[12,22]. Here, we show that there are mutations in two DcAG orthologous, AGa and AGb of SDF carnation cultivars. A previous study on carnation DF trait demonstrated that there was no AG sequence polymorphism detected between SF and DF cultivars[23]. Based on our data, taken together with published data in carnations, we suggest the following model for the flower formation in carnation (Fig. 4), and predict that the DF phenotype is caused by the mutation of the micro172-binding site of AP2[15] whereas the mutation in AG is associated with the formation of the SDF trait.
The expression of AG is spatially controlled and restricted in the third and fourth whorl. Ectopic expressing AG controlled with CLV3 promoters in the outer whorl lead to the formation of carpelloid sepals and reduced petal number in Arabidopsis[24]. Expressed AG in the second whorl with the promoter of AP3 resulted in stamen replacement of petals, even causing the failure of the second whorl development in Arabidopsis[25]. Our data shows that AGa and AGb express primarily in the third and fourth whorl of either SF or SDF carnation flowers but barely express in the first and second whorl. The expression levels of these two genes drop in SDF cultivars, constantly with the observation that the stamen number of them are reduced compared with SF cultivar.
The double flower is more attractive, especially in ornamental plants. Nevertheless, with the expansion of aesthetic variance, people sometimes appreciate semi-double flowers more. Breeders never stops gaining novel double flowers while researchers continue to investigate the mechanism underlying double flower traits. Our results provide insights into the semi-double flower carnation cultivars, which would facilitate carnation breeding.
-
Four commercial carnation cultivars, 'Peach Party' (single flower, SF, pink), 'Pink Star' (semi-double flower, SDF, pink), 'Purple Star' (SDF, purple) and 'Oscar' (SDF, red), were used in this study (Fig. 1). The plants of four cultivars with different flower phenotypes were provided by JinPin Yunke (Yunnan) Seedling Co., Ltd. (Kunming, China).
cDNA and DNA preparation
-
Total RNA was extracted from 200 mg stamen/carpel mixture for each cultivar using EasyPure® Plant RNA Kit (TransGen, Beijing, China). cDNA was then synthesized using TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen, Beijing, China). The 20 μl reaction system contained 1 μg total RNA, 1 μl Random Primer, 10 μl 2*TS Reaction Mix, 1 μl RT Enzyme Mix, 1 μl gDNA Remover, and variable RNase-free Water. At first, a sufficient volume of RNA and the Random Primer was added to RNase-free Water. The mixture was then cultured at 65 °C for 5 min with a subsequent 2-min ice-bath. Afterwards, the other components were added and mixed, and the mixture was incubated at 25 °C for 10 min, 42 °C for 30 min and 85 °C for 5 s. The final product was then stored at −20 °C for further experiments.
Genomic DNA was prepared using the EDTA method. Leaf material (200 mg) was collected for each cultivar. After grinding, 800 μl DNA extraction buffer [per 100 ml including 35 ml ddH2O, 10 ml Tris-HCl (1 M, pH7.5), 10 ml NaCl (5 M), 10 ml EDTA (0.5 M) and 35 ml SDS (2%)] was added to the sample and the mixture was incubated at 65 °C for 30 min. It was then centrifuged at 12,000 rpm for 10 min and the supernatant was transferred to a new 1.5 ml tube and mixed with an equal volume of isopropanol. After mixing, the sample was centrifuged at 12,000 rpm for 10 min and the supernatant was then removed. The precipitate was washed with 70% ethanol twice and then dried. Finally, the DNA was dissolved in 50 μl ddH2O and stored at −20 °C.
PCR amplification and sequence analysis
-
The CDS or genomic DNA of the genes studied in this research was amplified by KOD one mix. The 10 μl reaction system contained 8.8 μl KOD one mix, 0.4 μl forward primer, 0.4 μl reverse primer and 0.4 μl template. Primers used in the experiments are listed in Table 2. The PCR mixture was then incubated at 98 °C for 3 min; followed by 35 cycles at 98 °C for 30 s, Tm (depending on the primers) for 30 s, 68 °C for 1 min; with an extension at 68 °C for 5 min. The PCR products were separated by 1% agarose gel and the expected bands were cut and purified by EasyPure® Quick Gel Extraction Kit. The purified PCR products were then constructed onto pEASY®-Blunt Simple Cloning Vector and transformed into Trans1-T1 competent cells. Positive clones selected by relative antibiotics were confirmed by PCR and sent for sequencing. The sequences were analyzed by CLC seqviewer and Snapgene.
Table 2. Primer sequence for gene cloning and expression analysis used in this study.
Gene Application Forward primer Reverse primer AGa cloning ATGGAATTTTCAAGCCAAATAACTAGG CCAAACACCTCTTCAACTTGTTTGA AGb cloning ATGGAGTTTTCAAGCCAAATTAC AAACTCCTCTCCAACTTGTGTAA AP2 cloning TGGTACGCCTGATGAAACGAA TGCCCCCTAATGGTTTCCAC DcUbq3–7 RT-qPCR GTTGTTGGTTTCAGGGCTGGTTTG CTACGGTAATTGAGAATTCACACCGAAATG AGa RT-qPCR ATGCTAATCATAGCGTGAAGG GTTGGCTTCGGCAACAGA AGb RT-qPCR CCTCAAGCCAAAGGAAGCTA ACCCATTTCTTCTCTTGCAG AP1 RT-qPCR TAGGTCAAGATTTGGATACGCT ATCTAATGTGTTTGAGGCCG AP2 RT-qPCR CGCGTATGGGTCAATTTCT AATTAGTAACCGCATCCTTCC AP3 RT-qPCR GTCTGCTCGCTCTCAGATT GTAAGTCGTGACACACGAT PI RT-qPCR CTTCGGTTGAAGAAATCCTAGA GGCTGAGATTTTCATGTTTTGC SEP1 RT-qPCR GCAGCAAACATGGGAAGG GTCAATGGGCTGGAAAAGAG SEP3 RT-qPCR TGATAGAAGCAAATCAAGCGAC GTGAAAGAAGACATGGTCTCC RT-qPCR analysis
-
Total RNA was extracted from the three independent parts: sepal, petal and stamen/carpel for each cultivar using EasyPure® Plant RNA Kit (TransGen). Real-time PCR was performed using LightCycler® 480 II Real-time PCR Instrument (Roche, Swiss) with 10 μl PCR reaction mixture that included 1 μl cDNA, 5 μl 2×PerfectStartTM Green qPCR SuperMix, 0.2 μl forward primer, 0.2 μl reverse primer and 3.6 μl nuclease-free water. Reactions were incubated in a 384-well optical plate (Roche, Swiss) at 94 °C for 30 s, followed by 45 cycles at 94 °C for 5 s, 60 °C for 30 s. Each sample was run in triplicate for analysis. At the end of the PCR cycles, melting curve analysis was performed to validate the specific generation of the expected PCR product. The expression levels of mRNAs were normalized to DcUbq3–7 and were calculated using the 2−ΔΔCᴛ method[26]. The gene-specific primers for qPCR of the target genes are shown in Table 2.
This research was funded by the Major Science and Technology Project of Yunnan Provincial Department of Science and Technology (202102AE090052), High-level Talent Introduction Program of Yunnan Province -Industrial Talent Special Project (YNQR-CYRC-2020-004), and the Green Food Brand—Build a Special Project (Floriculture) supported by Science and Technology (530000210000000013742). The author would like to thank Hejun Li, the production manager of JinPin Yunke (Yunnan) Seedling Co., Ltd., for kindly providing the plant materials.
-
The authors declare that they have no conflict of interest.
-
# These authors contributed equally: Chunlian Jin, Huaiting Geng
- Supplemental Fig. S1 Multiple alignment of AGa of the four cultivars.
- Supplemental Fig. S2 Multiple alignment of AGb of the four cultivars.
- Copyright: © 2022 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Jin C, Geng H, Qu S, Zhang D, Mo X, et al. 2022. AGAMOUS correlates with the semi-double flower trait in carnation. Ornamental Plant Research 2:11 doi: 10.48130/OPR-2022-0011
AGAMOUS correlates with the semi-double flower trait in carnation
- Received: 31 March 2022
- Accepted: 06 July 2022
- Published online: 26 July 2022
Abstract: Flower type is the most valuable ornamental trait in floricultural plants, for which the ABCDE model was proposed to explain the identity of each floral organ in flowering plants. The C-class gene AGAMOUS (AG) is responsible for stamen formation and plays an essential role in the double flower phenotype. A previous study in carnation revealed that the mutation in the miR172 binding site of the A-class gene APETALA2 (AP2) leads to petal accumulation. And the expression level of AG was reduced significantly in the double flowers compared with that in the single flowers. However, there was no sequence polymorphism detected between AGs isolated from the double flowers and single flowers. Here, we performed AG analysis using single and semi-double flower carnations, and detected several mutations located in the crucial position like the MADS-box domain in the AGs of semi-double flower carnations while no changes were found at the miR172 binding site of AP2. As a result, the expression levels of AGs are reduced in the semi-double flower carnation, which could be caught by the loss function of AGs. Our data proves that AGs mutations are also associated with the semi-double flower formation in carnation, complementing the lack of research about AG-mutation-associated double flower formation in carnation.
-
Key words:
- Carnation /
- Semi-double flower /
- MADS-box /
- AGAMOUS /
- APETALA2