-
As sessile organisms, plants, unlike animals, cannot escape unfavorable growth environment, and are constantly affected by their natural environment with regards to their growth and development, including pests, diseases, drought, high temperature, low temperature, high salt, nutrient deficiency and so on[1,2]. In order to adapt to the environment, plants have evolved extremely complex response and defense mechanisms to avoid potential damage during their long-term evolution at the morphological, physiological, cellular and molecular levels[3,4]. With how plants cope with environmental stimuli, transcriptional regulation matters a lot[5,6].
In Arabidopsis, around 50 transcription factor families, accounting for approximately 2000 genes have been identified[7]. These transcription factors involve themselves in complex and dynamic networks, along with signal perception and transduction elements, which respond to biotic and abiotic stresses and regulate developmental processes. Among the transcription factor families, the WRKY proteins are a class of transcription factors unique to plants, and are named for their highly conserved WRKY motifs[8,9]. WRKY family members all contain 1–2 highly conserved WRKYGQK domains and a zinc finger motif at the C-terminal[10−12]. On the basis of the number of WRKY heptapeptide sequences and the structural characteristics of zinc finger motifs, WRKY proteins can be split into three classes: class I WRKY proteins contain two conserved WRKY heptapeptide sequences and one C2H2 (C-X4-5-C-X22-23-H-X-H) zinc finger structure; both class II and class III WRKY proteins contain only one WRKY heptapeptide sequence, and the difference is that the zinc finger structure of class II proteins is C2H2, while the zinc finger structure of class III WRKY proteins is C2HC (C-X7-C-X23-H-X-C)[10]. Most family members belong to class II WRKY proteins, which are further divided into five subgroups IIa-IIe based on their structural characteristics[9,10].
Transcription factors of WRKY family are induced by plant growth factors or environmental factors, and specifically recognize TTGAC sequence (W-box) and directly activate or inhibit the expression of target genes, which are widely involved in plant growth and development, defense responses, and abiotic stress responses[9,11,13]. A single WRKY transcription factor is often involved in the regulation of multiple biological processes, such as WRKY33, which acts as a key regulatory role in Botrytis resistance and is also involved in the establishment of salt, cold, submergence, and heat tolerance[14−21]. In addition, WRKY33 also affects root tissue differentiation and participates in developmental processes[22]. Previous research has reported that WRKY22 homologies mainly partake in plant disease resistance[23−27], while other biological processes are rarely reported.
Lily is an important flower crop and is loved by people all over the world. However, there are few research studies to date focusing on the abiotic stresses of lily. LpNAC17 from L. pumilum can enhance the tolerance to salt stress in tobacco by enhancing the expression of NtSOD, NtPOD, NtCAT, NtHAK1, NtPMA4, and NtSOS1[28]. The cut flower characteristics and growth traits under salt stress in 26 lily cultivars have also been explored[29]. With the exception of salt stress, the alkali tolerance and drought tolerance of transgenic tobacco were found to be reversely regulated by LpNAC6 from L. pumilum[30]. Since most lilies have poor thermotolerance, a large amount of published reports concentrate upon the mechanism underlying the heat stress response of lily; and various transcription factors from different families have been identified, such as LlHSFA3, LlHSP20, LlMYB305, LlWRKY39, LlERF110[31−37]. Our previous study identified a class IIe WRKY gene LlWRKY22 which was differentially expressed under heat stress in lily[38]. It positively regulates the thermotolerance of lily, but its role in other abiotic stresses is under-excavated[38]. Here, we analyzed the role of LlWRKY22 in other stresses with transgenic Arabidopsis plants. The results demonstrated that LlWRKY22 could not only be activated by heat, salt and osmotic stresses, but also promote the tolerance to these stresses when overexpressed in Arabidopsis. Furthermore, overexpression of LlWRKY22 also causes growth defects, accelerated flowering, and increased ABA sensitivity of transgenic plants.
-
The expression of LlWRKY22 in the leaves of lily 'White heaven' was detected with a 37 °C-heat stress treatment for different time periods. The results indicated that it was rapidly induced at the beginning of the treatment and sustained at a comparatively high level until it peaked at 12-h (Fig. 1a). In addition, the expression of LlWRKY22 was also activated by salt, mannitol, and ABA treatment (Fig. 1b).
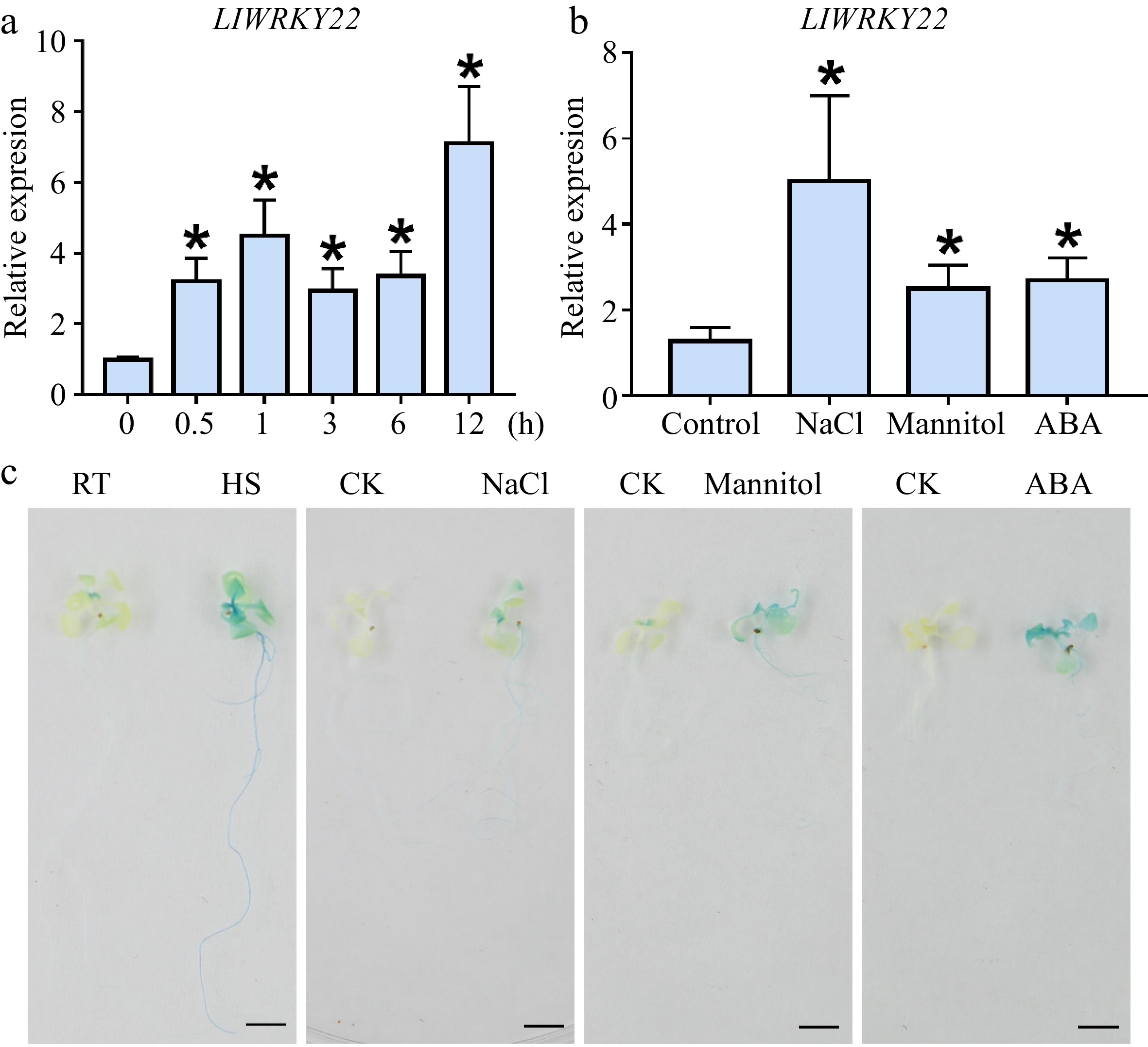
Figure 1.
The expression analysis of LlWRKY22 under different abiotic stresses. (a) Analysis of LlWRKY22 expression pattern in lily leaves after time-controlled exposure to 37 °C. (b) Detection of LlWRKY22 expression in lily under NaCl, mannitol, or ABA treatment. The tissue-cultured seedlings of lily were treated with water (CK), salt solution (NaCl, 200 mM), mannitol solution (300 mM), or ABA solution (10 μM) for 3 h, after which their leaves were collected for expression analysis. The data were normalized to the lily 18S rRNA, and the 2−ΔΔCᴛ method was used in the RT-qPCR analysis. Data are the mean (± SD) of three independent experiments (Student’s t-test, *P < 0.05). (c) Promoter activity in the LlWRKY22-pro::GUS transgenic Arabidopsis seedlings under different treatments. The 10-day-old seedlings were selected for the promoter activity analysis. RT, room temperature (22 °C), HS, heat stress (37 °C, 3 h). CK, control (sterile water). NaCl, NaCl solution (200 mM, 3 h). Mannitol, mannitol solution (400 mM, 3 h). ABA, ABA solution (10 μM, 3 h). Three replicates were conducted, and one representative picture is presented. Scale bar = 1 cm.
The promoter activity of LlWRKY22 under different abiotic stresses was also analyzed. The LlWRKY22 promoter had very weak activity under normal conditions in GUS reporter analysis using promoter-transgenic seedlings, but after HS treatment, its activity was largely increased in the leaves and roots (Fig. 1c). Additionally, the promoter activity of LlWRKY22 was also enhanced by salt, mannitol, and ABA treatments (Fig. 1c).
LlWRKY22 overexpression causes growth defects and ABA hypersensitivity in transgenic plants
-
To analyze the function of LlWRKY22 in vivo, we transformed LlWRKY22 into Arabidopsis under the control of 35S promoter. The lines OE-2, OE-4, and OE-5 were identified by RT-PCR and chosen for functional analysis (Supplemental Fig. S1). Under normal conditions, LlWRKY22 transgenic plants grew on MS medium for two weeks presenting a phenotype of significant growth defects with smaller rosettes and lower weights (Fig. 2a−c). After transplantation, LlWRKY22 transgenic lines, compared with wild-type plants, was dwarf-like, with plants being shorter and rosette leaves being smaller (Fig. 2d & e). These results implied that constitutive activation of LlWRKY22 might be harmful for plant growth under normal conditions. In addition, overexpression of LlWRKY22 in transgenic seedlings slightly but significantly promoted flowering (Fig. 2f); compared with wild-type seedlings, the LlWRKY22 transgenic plants required less rosette leaves and shorter growth time to flowering (Fig. 2g & h).
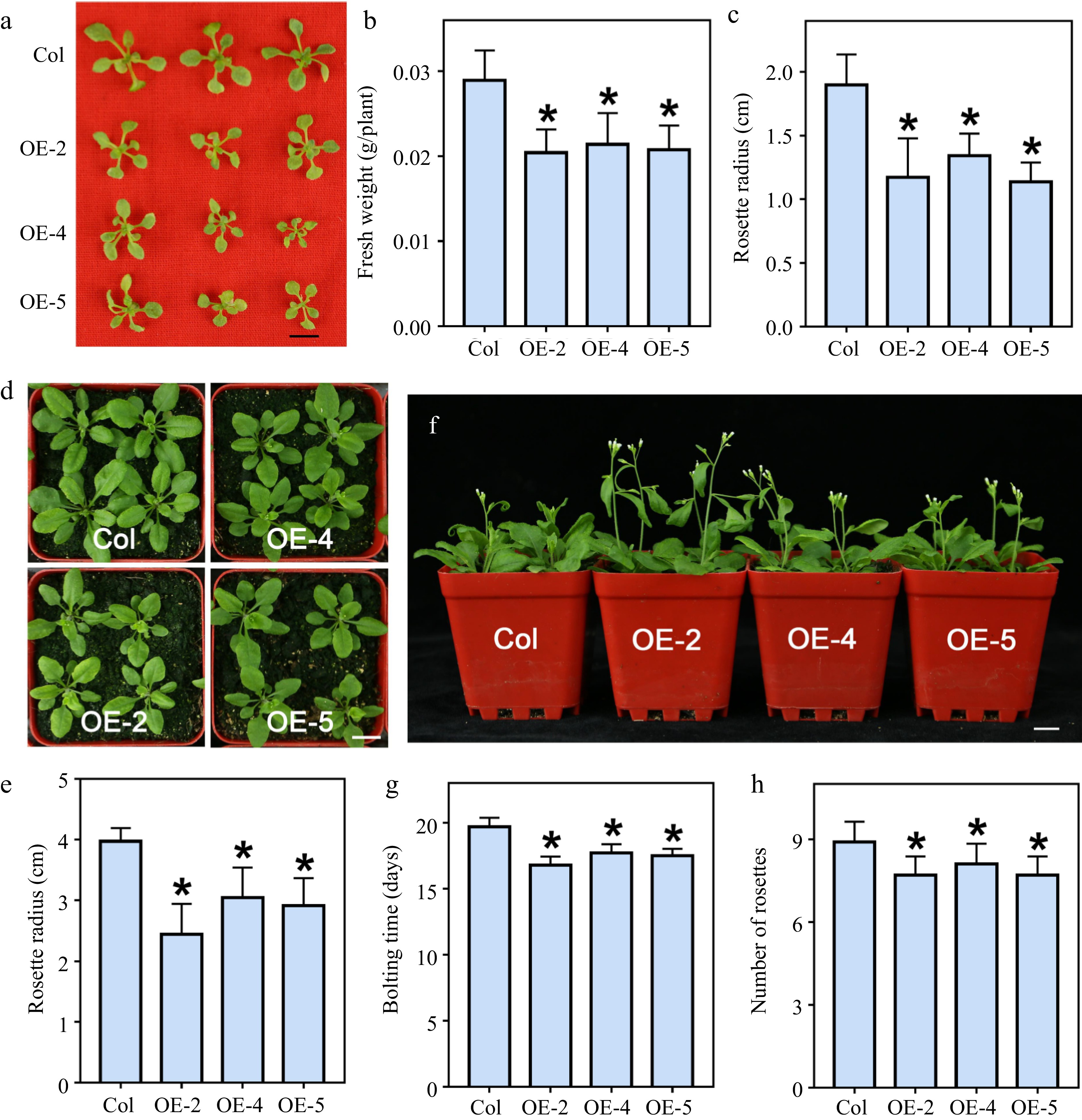
Figure 2.
The growth phenotypes of LlWRKY22 overexpression Arabidopsis plants. (a) The 3-week-old wild-type and transgenic seedlings grew on MS medium. Scale bar = 1 cm. (b) Fresh weight of three-week-old plants which grew on MS medium. Bars are means ± SD of the tested plants (n = 9). (c) Rosette radii of 3-week-old seedlings growing on MS medium. Bars are means ± SD of the tested plants (n = 9). (d) The seedlings growing on agar were transferred from agar plates to soil for 2 weeks. Scale bar = 1 cm. The representative picture is based on three replicates. (e) Rosette radii of 2-week-old plants which grew on the soil were counted. Bars are means ± SD of the tested plants (n = 12). Three replicates were performed, and one representative picture is shown. (f) Seedlings were planted on agar plates for 10 d and then transmitted to soil for 3 weeks. Scale bar = 1 cm. The representative picture is based on three replicates. (g) Bolting time of wild-type and transgenic seedlings. Bars are means ± SD of three independent experiments. (h) Rosette leaf number of the bolting plants. Bars are means ± SD of three replicates (n = 12, Student's t-test, * P < 0.05).
The WRKY22 homologies are reported to positively regulate ABA signaling in different plants[39,40]. We attempted to determine whether LlWRKY22 has a function similar to its homologies in ABA response (Fig. 3a). After 1.0 µM-ABA treatment, the seed germination rate of LlWRKY22 transgenic plants was lower than that of the wild-type plants after 6-days of growth (Fig. 3b & c). And the cotyledon green rate was also lower than that of the wild-type plants on the medium containing 0.5 µM ABA (Fig. 3d). To further determine ABA sensitivity at the late growth stage, we transferred the 5-day-old seedlings, which were germinated on MS medium, to a medium adding 5.0 µM ABA and grew for 12 d (Fig. 3e). It was illustrated that the root growth of the LlWRKY22 overexpression lines was largely restricted under ABA conditions compared to that of wild-type plants (Fig. 3f). The above results indicated LlWRKY22 overexpression led to ABA hypersensitivity of transgenic plants.
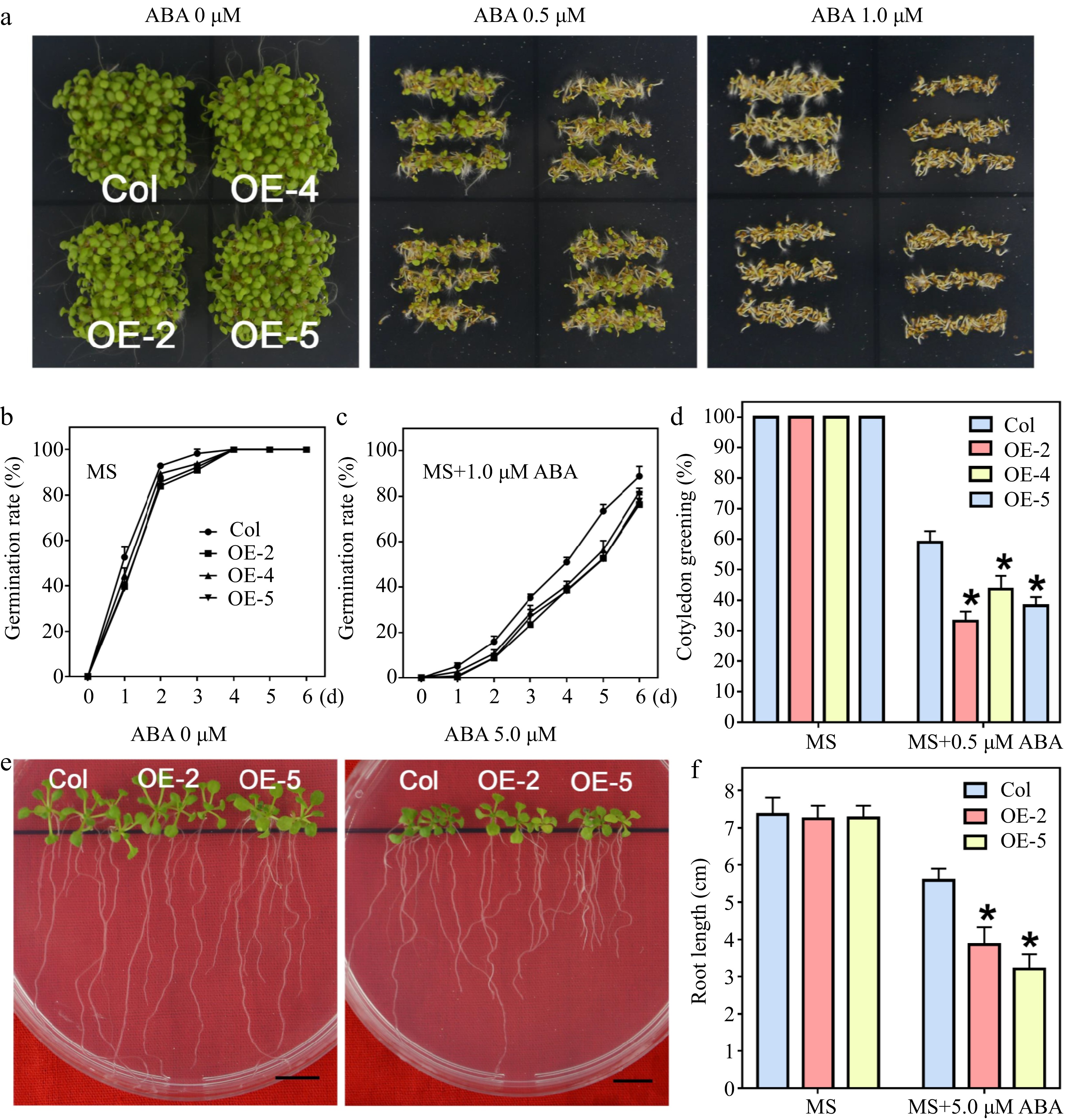
Figure 3.
Seed germination and seedling growth of LlWRKY22 overexpression Arabidopsis plants responding to ABA treatments. (a) The wild-type and transgenic Arabidopsis seeds were planted on ABA-supplemented MS medium for 6 d under light. Three replicates were performed and one representative image was exhibited. (b), (c) Seeds germinated on MS medium with or without 1.0 μM ABA and the germination rates were recorded daily. (d) Seeds were sown on MS medium with or without 0.5 μM ABA, and the percentage of cotyledon greening was recorded after 6 d. Data represent the means (± SD) of three replicates (Student's t-test, * P < 0.05). (e) The 5-day-old seedlings were used as materials and transplanted to MS medium with or without 5.0 μM ABA, and after 10 d of growth, the root lengths were photographed. Scale bar = 1 cm. (f) Root elongation of each plant was calculated, and the average value of 9 biological replicates for each line is shown. Bars are means ± SD of three replicates (n = 9, Student's t-test, * P < 0.05).
LlWRKY22 overexpression enhanced the thermotolerance of transgenic plants
-
For thermotolerance assay, we directly exposed the 5-day-old transgenic and wild-type seedlings to heat stress for 1 h at 45 °C and then put them at 22 °C to recover for 7 d (Fig. 4a). The three overexpression lines showed higher survival rates in comparison to the wild-type line, indicating that LlWRKY22 accumulation increased the thermotolerance of the transgenic plants (Fig. 4b). Next, we measured the expression levels of several heat-related genes in wild-type and transgenic seedlings. It was revealed that LlWRKY22 overexpression upregulated expression of abundant heat-related genes, consisting of AtDREB2A, AtDREB2B, AtDREB2C, AtJUB1, AtHSFA1a, AtHSFA1b, AtHSFA1d, AtHSFA1e, AtHSFA2, AtHSFA3, AtAPX2, AtHSP17.6, AtHSP22.0, AtHSP25.3, AtHsa32, and AtHSP101 (Fig. 4c; Supplemental Fig. S2). In particular, the expression of AtJUB1, AtDREB2A, AtDREB2B, and AtDREB2C was remarkedly elevated in the transgenic lines (Fig. 4c). Interestingly, the WRKY22 homolog of Arabidopsis, AtWRKY22, was also activated by LlWRKY22 overexpression (Fig. 4c). These results suggested that activation of these heat-related genes resulting from LlWRKY22 overexpression might contribute to the increased thermotolerance in transgenic plants.
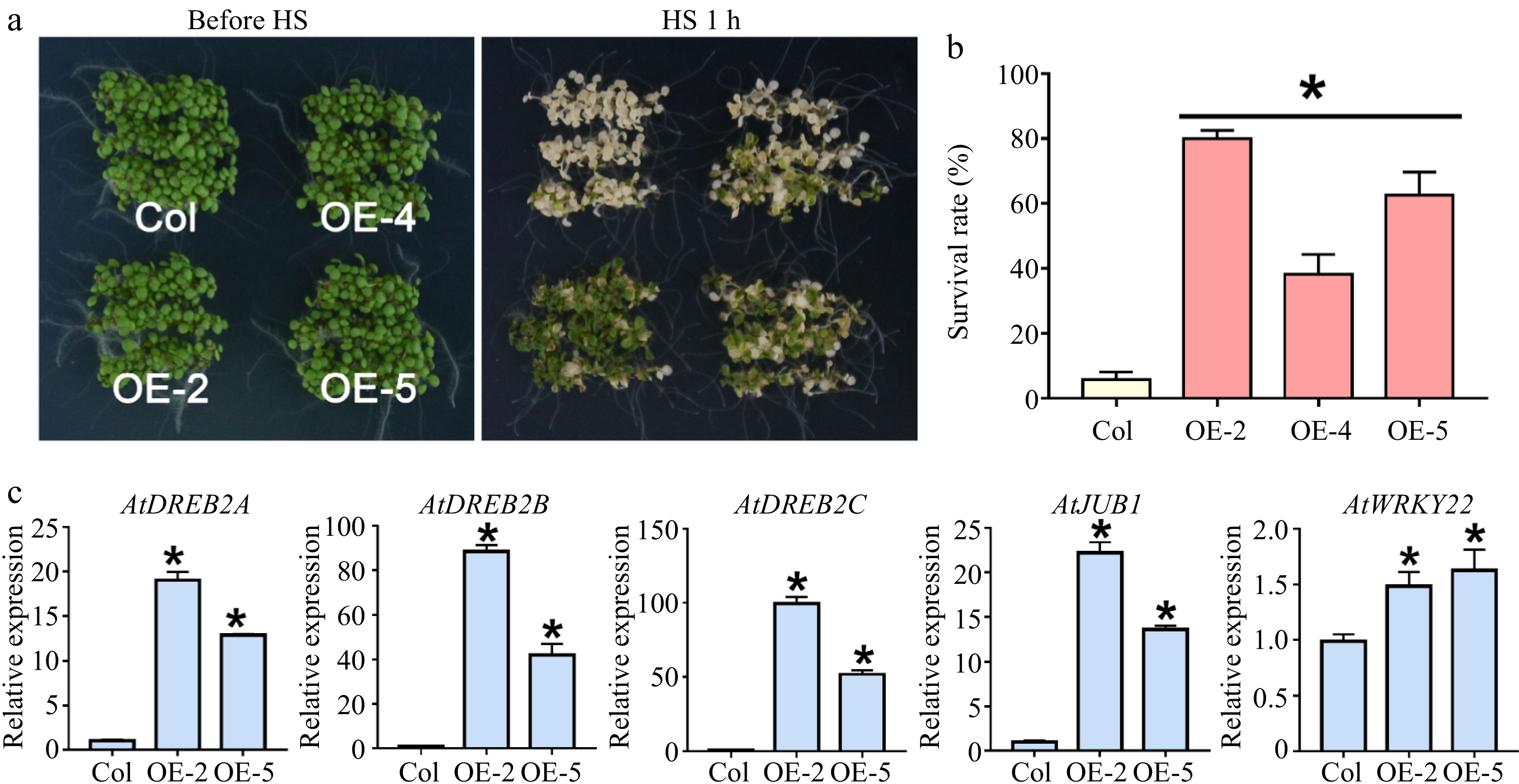
Figure 4.
Thermotolerance test of LlWRKY22 overexpressing Arabidopsis plants. (a) The thermotolerance of three LlWRKY22 overexpression lines (OE-2, OE-4, and OE-5) were examined. The phenotypes with 1-h 45 °C heat stress (HS) of wild-type and transgenic seedlings are exhibited. (b) The survival rate of wild type and transgenic lines was calculated after HS following by a 7-day recovery at 22 °C. Data are the mean ± SD of three replicates (Student's t-test, * P < 0.05). (c) Expression of AtDREB2A, AtDREB2B, AtDREB2C, AtJUB1, and AtWRKY22 under normal conditions was detected in transgenic Arabidopsis plants under normal conditions. Data are the mean ± SD of three replicates (Student's t-test, * P < 0.05).
LlWRKY22 overexpression enhanced the salt tolerance of transgenic plants
-
Under normal conditions, there was no difference in seed germination in the transgenic lines and wild type (Fig. 5a). Nevertheless, the 6-day cotyledon's green rate of all transgenic lines was higher than that of wild-type plants when the medium was added with 50 mM NaCl (Fig. 5b). When it was raised to a degree of 100 mM in NaCl concentration, the germination rate of the wild type was also markedly repressed compared to the overexpression lines (Fig. 5c). Furthermore, the root elongation in transgenic lines did not differ with the wild type at the post-germination stage when MS medium was not added with NaCl (Fig. 5d). However, root growth of the transgenic plants was significantly enhanced under salt stress compared with that of wild type (Fig. 5e). The above-mentioned results illustrated that LlWRKY22 overexpression in Arabidopsis could enhance the tolerance to salt stress, and that LlWRKY22 might positively regulate salt response.
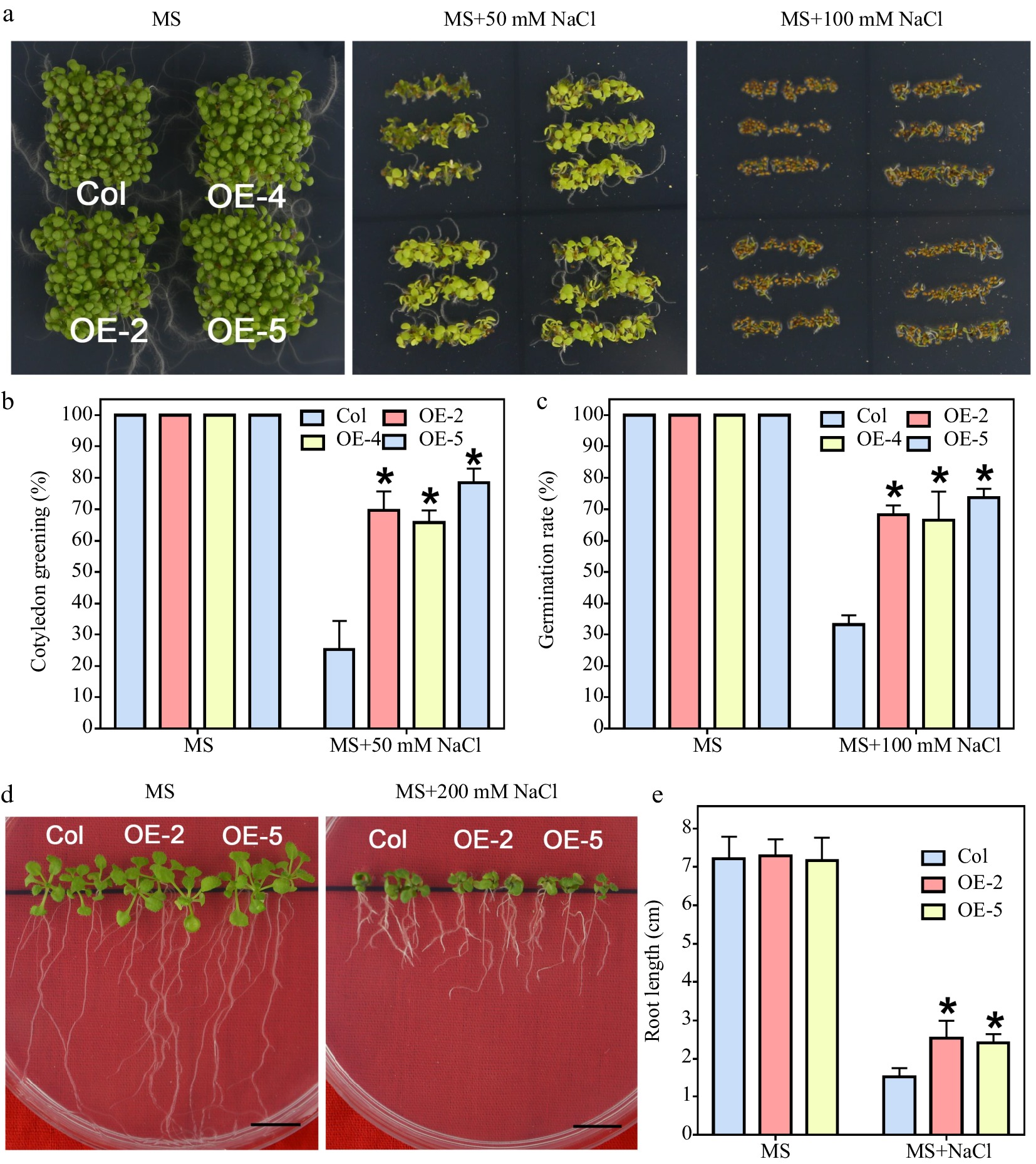
Figure 5.
Salt tolerance test of LlWRKY22 overexpression Arabidopsis plants. (a) Wild-type and transgenic Arabidopsis lines were cultivated and photographed on NaCl-supplemented MS medium for 5 d under light. Three independent experiments were performed and one representative picture is presented. (b), (c) The germination rate of seeds which germinated on the MS medium with 100 mM NaCl were recorded daily (MS medium without NaCl served as control). Data represent means ± SD of three biological replicates (Student's t-test, * P < 0.05). (d) The cotyledon greening rate of seeds which germinated on the MS medium with or without 50 mM NaCl was recorded after 6 d. Data represent means ± SD of three biological replicates (Student's t-test, * P < 0.05). (e) The 5-day-old seedlings were used as materials and grew on MS medium with or without 200 mM NaCl for 10 d, after which their root lengths were recorded and photographed. Scale bar = 1 cm. Bars are means ± SD of three replicates (n = 9, Student's t-test, * P < 0.05).
LlWRKY22 overexpression enhanced the mannitol tolerance of transgenic plants
-
Since LlWRKY22 expression was activated by mannitol treatment, the mannitol tolerance of the transgenic lines was also analyzed (Fig. 6a). The germination of wild-type and transgenic lines was almost the same without mannitol (Fig. 6a). When subjected to 200 mM mannitol treatment, the transgenic plants presented higher green rate of cotyledon (Fig. 6b), and the germination rate of transgenic lines was also elevated in comparison to that of wild-type plants with 400 mM mannitol (Fig. 6c). Obviously, the root growth of LlWRKY22 overexpression seedlings exceeded that of the wild type under 300 mM mannitol treatment (Fig. 6d & e). The results suggested that overexpression of LlWRKY22 elevated the tolerance to mannitol of transgenic Arabidopsis seedlings.
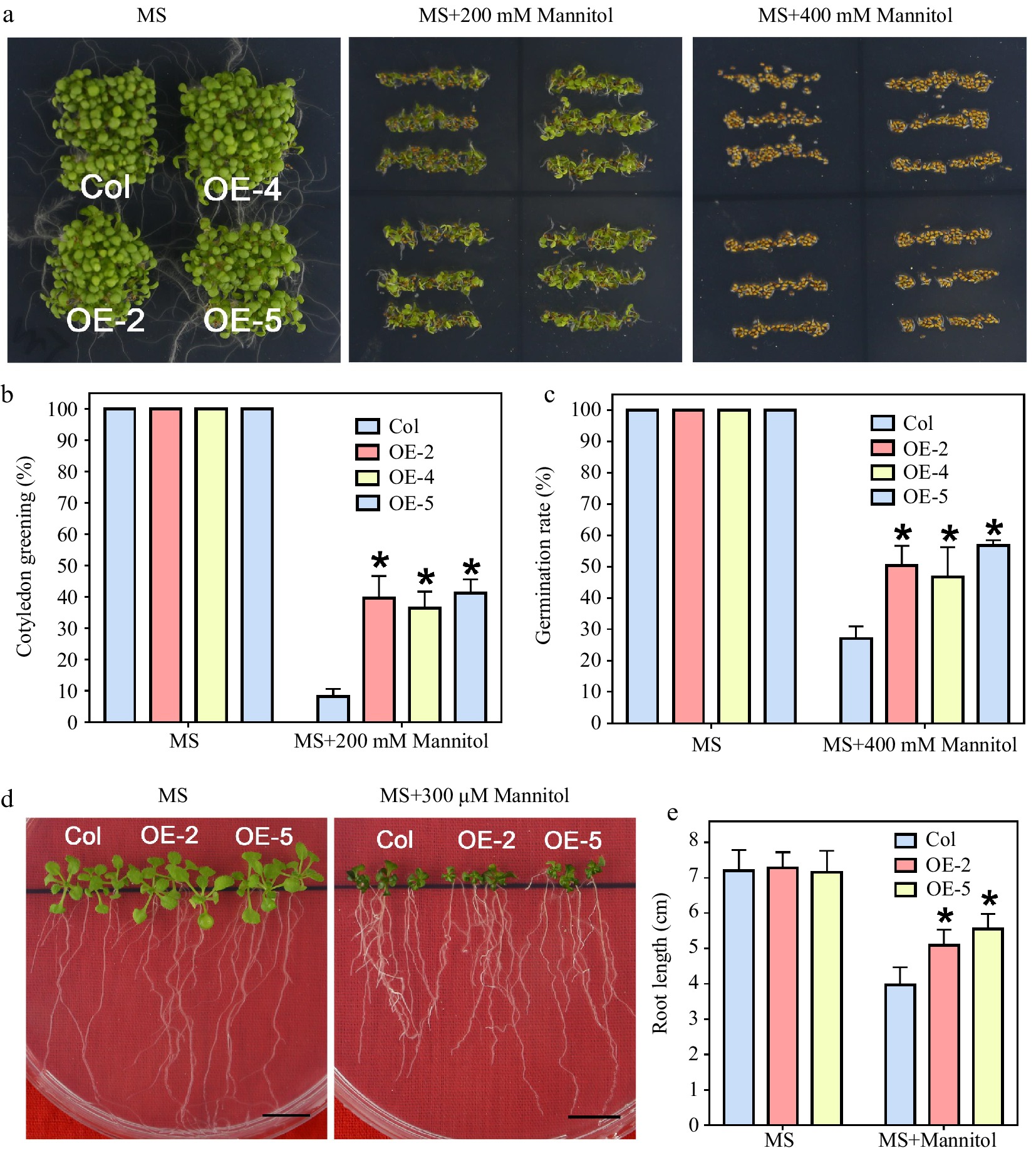
Figure 6.
Mannitol tolerance test of LlWRKY22 overexpression Arabidopsis plants. (a) Wild-type and transgenic Arabidopsis lines were planted under light for 5 d on mannitol-supplemented MS medium and the seed germination was photographed and recorded. Three independent experiments were conducted and one representative picture is shown. (b), (c) The germination rate of seeds which germinated on MS medium with 400 mM mannitol were recorded daily (MS medium without mannitol served as control). Data represent means ± SD of three biological replicates (Student's t-test, * P < 0.05). (d) The cotyledon greening rate of seeds which germinated for 6 d on the MS medium with 200 mM mannitol was recorded (MS medium without mannitol served as control). Data represent means ± SD of three biological replicates (Student's t-test, * P < 0.05). (e) Root lengths of 5-day-old seedlings were transplanted to MS medium with or without 300 mM mannitol. After 10 d, their root lengths were recorded and photographed. Scale bar = 1 cm. Bars are means ± SD of three replicates (n = 9, Student's t-test, * P < 0.05).
-
In Arabidopsis, AtWRKY22 is reported to take part in dark-induced leaf senescence, pathogen-triggered immunity, anti-aphid response, and oxidase stress response[24,39,41,42]. In rice, OsWRKY22 promotes Al-induced increases of the OsFRDL4 expression through directly binding W-box motif in the promoter of OsFRDL4, consequently strengthening Al-induced citrate secretion and Al tolerance[43]. Meanwhile, OsWRKY22 also responses to blast. The oswrky22 mutants exhibited an increased susceptibility to Magnaporthe oryzae, while overexpression of OsWRKY22 enhanced resistance to M. oryzae[23]. CsWRKY22 of citrus could enhance cell enlargement and CsLOB1 expression, thereby regulating resistance to Xanthomonas citri pathogen[25,26]. Additionally, MsWRKY22 of alfalfa was reported to directly bind MsWRKY11 promoter and activate its expression, thereby taking part in lignin biosynthesis, and drought tolerance[44]. VvWRKY22 interacts with VvSnRK1.1/VvSnRK1.2 (sucrose non-fermenting-1-related protein kinase 1) and regulates sugar accumulation in grape[40]. In peach fruit, through interacting with PpTGA1 physically, PpWRKY22 enhances the expression of several SA-responsive PR genes to partake in the tolerance to disease , and overexpression of PpWRKY22 confers enhanced resistance to Rhizopus stolonifera[27]. Our former work illustrated that LlWRKY22 is a differentially expressed gene in the heat-stress-transcriptome of lily leaves[38]. LlWRKY22 expression is continuingly activated by heat, whose protein is localized in the nucleus, shows transcriptional activation, and plays a positive role in thermotolerance of lily[38]. Not only than that, it can directly activate the expression of LlDREB2B and itself[38]. Although abiotic and biotic stimuli are most likely perceived by plants via stress-specific mechanisms and require differential plant responses, these studies imply that WRKY22 is a generalist performing specific functions in various biological processes. Nevertheless, no study to date has clarified function of WRKY22 in salt and osmotic tolerance. In this study, we found that LlWRKY22 was not only induced by heat but also responded to salt and mannitol stresses, whose promoter activity was also enhanced by salt and mannitol treatments (Fig. 1), supporting that it may partake in the establishment of salt and osmotic tolerance. Similar to the results in lily, overexpression of LlWRKY22 in Arabidopsis also showed enhanced tolerance to heat, and the expression of heat-responsive genes was also up-regulated (Fig. 4). In lily, it shows that LlWRKY22 can activate the expression of LlDREB2B along with itself through directly binding the tandem W-box element which was found in their promoters, thereby participating in the establishment of thermotolerance[38]. And in this study, we found that overexpression of LlWRKY22 could strongly enhance the expression of Arabidopsis DREB2-like genes, AtDREB2A, AtDREB2B, and AtDREB2C, and WRKY22 homolog AtWRKY22 (Fig. 4c). We supposed that the overexpression of LlWRKY22 in Arabidopsis could increase the endogenous expression level of AtWRKY22 by autoactivation; and after which, both up-regulated genes LlWRKY22 and AtWRKY22 were collaborated to enhance the expression of AtDREB-like genes probably by binding W-box elements. Therefore, it was speculated that the WRKY22-DREB2 regulation and WRKY22 autoactivation mechanisms through two tandem W-box copies are conserved between Arabidopsis and lily as well as in other plant species.
The DREB2-like transcription factors, AtDREB2A, AtDREB2B, and AtDREB2C, are extensively cognized as being related to various stress responses, particularly drought and heat tolerance[45−47]. And the salt and mannitol tolerances of transgenic Arabidopsis was identified. The results revealed that overexpression of LlWRKY22 raised the tolerance to salt and osmotic stresses in transgenic seedlings (Figs 5 & 6), indicating that LlWRKY22 positively regulated these stress responses, and among which the WRKY22-DREB2 regulatory module might make significant contributions. Through gene expression assay, it was also revealed that overexpression of LlWRKY22 stimulated the expression of AtJUB1 (Fig. 4c), which has been reported to be one of the core factors regulating multiple abiotic and biotic stresses[48,49]. Overexpression of JUB1 homology can improve heat, salt and drought tolerances in different plant species[48,50,51]. And JUB1 is a direct upstream regulator of DREB2A[50], suggesting that LlWRKY22 can also partake in the response to abiotic stress through the JUB1-DREB2A pathway. In addition, former findings have illustrated that the activation of stress-resistant genes, such as JUB1 and DREB2A, can lead to restricted growth and development of plants[46,52,53]. And we found that overexpression of LlWRKY22 showed growth defects also with small rosettes and low weights (Fig. 2).
In Arabidopsis, AtWRKY22 acts as a suppressor in JA and SA signaling, showing that AtWRKY22 has a role in both SA and JA signaling and partakes in transcriptional reprogramming in response to mechano-stimulation and aphid infestation[39]. Besides, aphid infestation also influences the expression of ABA-responsive genes in wrky22 mutant[39]. VvWRKY22 is induced by fructose and ABA, and it is illustrated that overexpression of VvWRKY22 decreases the content of sucrose, glucose together with fructose, and regulates the expression levels of sugar and ABA-related genes[40]. What's more, VvWRKY22 was revealed to interact with two significant kinases VvSnRK1.1 and VvSnRK1.2, which are involved in sugar metabolism and ABA signaling[40]. The wild-type Arabidopsis and two independent wrky22 mutants were treated with ABA, which promotes the closure of guard cells in wild-type leaves but not in both mutants, implying that the malfunction of WRKY22 greatly induces insensitivity to ABA[54]. The responsiveness to ABA response was limited, which was caused by the loss of the functional WRKY22 transcript in mutant shoot tips[54]. Our findings illustrated that ABA treatment activated the expression of LlWRKY22 (Fig. 1), whose overexpression would lead to enhanced ABA sensitivity in transgenic plants (Fig. 3), and the downstream DREB2-like genes were typically ABA-responsive genes (Fig. 4), suggesting that LlWRKY22 may partake in the ABA response.
Taken together, our results suggest that LlWRKY22 expression could be induced by various abiotic stresses, such as: heat, salt, osmotic stresses as well as ABA treatment; and its ectopic overexpression in Arabidopsis promoted the tolerances to those stresses and the sensitivity to ABA. Consequently, we speculated that the strengthened tolerances might be related to the increased ABA sensitivity in LlWRKY22 transgenic Arabidopsis plants.
-
The tissue-cultured Lilium longiflorum cv. 'White heaven' was used in the experiments. Sterile lily seedlings were cultivated on MS medium in a standard culture room. Arabidopsis thaliana (Col-0) seeds were sterilized with 0.1% NaClO and 0.01% TritonX-100, and then sown on MS medium, followed by dark conditions at 4 °C for 3 d. After germination, the seedlings were transferred from MS plates to plastic pots of a sterile rooting mixture under controlled conditions (22/16 °C, 16-h/8-h light/dark).
Cloning of LlWRKY22 from lily
-
We extracted total RNA from the heat-treated leaves by using an RNAprep Pure Kit (Tiangen, China). The leaves were sampled from the tissue-cultured 'White heaven' plants treated with 1-h heat stress at 37 °C. The cDNA was synthesized using a First Strand cDNA Synthesis Kit (R323-01, Vazyme, China). The open reading frame (ORF) of LlWRKY22 was isolated with special primers on the basis of the transcriptome data (Supplemental Table S1).
Abiotic stress treatments and gene expression assay of lily
-
The 2-week-old, healthy, tissue-cultured lily seedlings with similar sizes were chosen for the treatments and gene expression assay. To analyze the expression patterns under high temperature conditions, the selected plants were exposed to 37 °C for various durations (0, 0.5, 1, 3, 6, 12 h) in a temperature-controlled incubator (Liance, China). For salt and mannitol treatments, different time course and concentration gradients were pre-tested, and then we transferred plants from the growth medium to 200 mM NaCl, 300 mM mannitol, or 10 μM ABA solution and put them at 22 °C for 3 h, deionized water served as control. Immediately after treatment, leaves were frozen in liquid nitrogen. Total RNA was extracted as described above, and reverse transcription was performed with a HiScript II kit (Vazyme, China). Real-time quantitative PCR (RT-qPCR) was applied to detect the expression levels. The 18S rRNA of lily was applied as an internal control. The primers are listed in Supplemental Table S2 for the RT-qPCR assay.
Isolation of LlWRKY22 promoter from lily
-
Genomic DNA of lily leaves was extracted using a Plant Genprep DNA kit (Zomanbio, China). We isolated LlWRKY22 promoter through the Hi-tail PCR method[36,55]. The 1053-bp upstream fragment from the ATG of LlWRKY22 were cloned and identified from lily 'White heaven'.
Generation of LlWRKY22 transgenic Arabidopsis lines
-
The LlWRKY22 ORF was cloned and inserted into pCAMBIA1300 vector driven by 35S promoter. Promoter of LlWRKY22 was cloned into pCAMBIA1391 vector containing a GUS (β-glucuronidase) reporter gene. The recombinant vectors were transformed, respectively, into 5-week-old Arabidopsis plants using the floral-dip method. The transformed seeds were screened by adding 30 mg·L−1 hygromycin to MS medium. We used RT-PCR to identity all transgenic lines, and selected three T3-generation homozygous lines for gene functional analysis. The primers used for vector constructions are shown in Supplemental Table S3.
GUS activity assay of LlWRKY22 promoter transgenic plants
-
The LlWRKY22 transgenic seeds were sown on MS medium, and put in dark conditions at 4 °C for 3 d. After 5 d of germination, the seedlings were transplanted to vertically oriented MS medium for an additional 10 d. For heat stress, we put the seedlings at 37 °C for 3 h, and the untreated seedlings was used as a control. For NaCl, mannitol, and ABA treatments, the plants were submerged in 200 mM NaCl, 400 mM mannitol and 10 μM ABA solution for 3 h, and the controlled seedlings were cultured in sterile water. After treatment, the seedlings were collected and submerged in GUS staining solution for incubation at a temperature of 37 °C for 12 h. For the removal of chlorophyll, 70% ethanl was used.
Thermotolerance test of transgenic seedlings
-
The thermotolerance test was performed similar to that described in previous studies[34,37,56]. Briefly, seedlings were vernalized on MS medium and grown at 22 °C in a standard culture room as previously described. For heat stress, the 5-day-old seedlings were treated with 1-h 45 °C treatment, and we recorded the survival rate after a period of 7-day of recovery under normal growth conditions.
Abiotic stress treatments of transgenic seedlings
-
To investigate the effects on germination, different time courses and concentration gradients of salt, mannitol and ABA were pre-tested and appropriate treatments were selected. Wild-type and transgenic seeds were sown onto MS medium containing NaCl (0, 100, 200 mM), mannitol (0, 200, 400 mM), or ABA (0, 0.5, 1.0 μM), and their germination rates were recorded daily. To investigate the effects of NaCl, mannitol, and ABA on root growth, the 5-day-old seedlings were transferred to MS medium containing NaCl (200 mM), mannitol (300 mM), or ABA (5.0 μM). After 10 d of growth, a picture was taken and the root length of each plant was recorded.
Gene expression assay of transgenic Arabidopsis seedlings
-
Gene expression analysis was performed with 5-day-old transgenic and wild-type seedlings. Their RNA extractions were performed as described above. The expression level of detected genes was determined by RT-qPCR. AtActin2 was used as a normalization control. The primers used for RT-qPCR are listed in Supplemental Table S2.
Statistical analysis
-
GraphPad Prism 7.00 was used to analyze the experimental data and draw the diagrams. Data for P-value determinations and inference were analyzed by Student's t-test at a significant level of 0.05 or 0.01.
Data availability
-
The data underlying this article will be shared on reasonable request to the corresponding author.
This work was supported by the National Natural Science Foundation of China (31902055), the Fundamental Research Funds for the Central Universities (KYZZ2022004), the National Key R&D Program of China (2019YFD1000400), and the Natural Science Foundation of Jiangsu Province, China (BK20190532).
-
The authors declare that they have no conflict of interest.
- Supplemental Fig. S1 Molecular analysis of LlWRKY22 transgenic Arabidopsis lines. The 5-day-old seedlings were used to detect the expression of LlWRKY22 in transgenic Arabidopsis lines by RT-PCR. PCR of the endogenous control and tested gene was performed with 28 and 30 cycles, respectively. AtActin2 was used as an endogenous control.
- Supplementary Fig. S2 Expression analysis of heat-related genes in the wild-type and LlWRKY22 transgenic plants under normal conditions. Data are the mean ± SD of three replicates (Student’s t-test, * P < 0.05).
- Supplementary Table S1 Primers of LlWRKY22 isolation.
- Supplementary Table S2 Primers used for vector reconstruction.
- Supplementary Table S3 RT-qPCR primers.
- Copyright: © 2022 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Li T, Zhou T, Liang J, Zhang D, Teng N, et al. 2022. Overexpression of lily LlWRKY22 enhances multiple abiotic stress tolerances in transgenic Arabidopsis. Ornamental Plant Research 2:17 doi: 10.48130/OPR-2022-0017
Overexpression of lily LlWRKY22 enhances multiple abiotic stress tolerances in transgenic Arabidopsis
- Received: 05 September 2022
- Accepted: 30 September 2022
- Published online: 28 October 2022
Abstract: In our previous study, a heat-induced differentially expressed WRKY-IIe gene LlWRKY22 is isolated from lily (Lilium longiflorum), which acts as a positive role in thermotolerance, but whether it is involved in other stress responses is unknown. Here, the expression of LlWRKY22 was indicated to be positively influenced by heat, salt, or mannitol treatments, and its promoter activity was also enhanced after heat, salt, or mannitol treatments. In addition, LlWRKY22 responded to ABA treatment, which activated its expression and also increased the promoter activity. Overexpression of LlWRKY22 in Arabidopsis contributed to growth defects and early flowering. Simultaneously, compared with the wild type, the ABA sensitivity in transgenic lines was increased in both the germination stage and late growth stage. Further analysis showed that LlWRKY22 overexpression elevated the thermotolerance of transgenic plants and induced the expression of AtDREB2A, AtDREB2B, AtDREB2C, and AtJUB1. The salt and mannitol tolerances of the overexpression lines were also improved. Overall, our results illustrated that LlWRKY22 is affected by heat, salt, and osmotic stresses, and positively regulates heat, salt, and osmotic tolerances, which reveals that it acts as a generalist character responding to different abiotic stresses. And further to that, the regulatory pathway of LlWRKY22 also involves in ABA signaling.
-
Key words:
- Lilium longiflorum /
- LlWRKY22 /
- Abiotic stress /
- ABA /
- Thermotolerance













