-
Apomixis is a natural phenomenon in some flowering plants that results in clonal seeds through bypassing the normal meiosis and double fertilization[1]. In apomicts (the species able to propagate through apomixis), the seeds can develop from the embryo sac formed by somatic ovule cell (apospory) or megaspore mother cell (MMC) without meiotic division (diplospory)[1]. Apospory and diplospory are collectively referred to as gametophytic apomixis[1]. In Citrus and Fortunella, the nucellus or integument of ovule can develop into adventitious embryo (sporophytic apomixis)[2,3]. These asexual reproduction modes result in offspring that are genetically identical to the maternal parent plant (Fig. 1). Since the first documentation of apomixis in Alchornea ilicifolia by Smith in 1841[4], apomixis has now been found in more than 400 species, most of which are tropical and temperate plants[5]. However, no staple crop species have been found to propagate in the way of apomixis[1,6,7] .
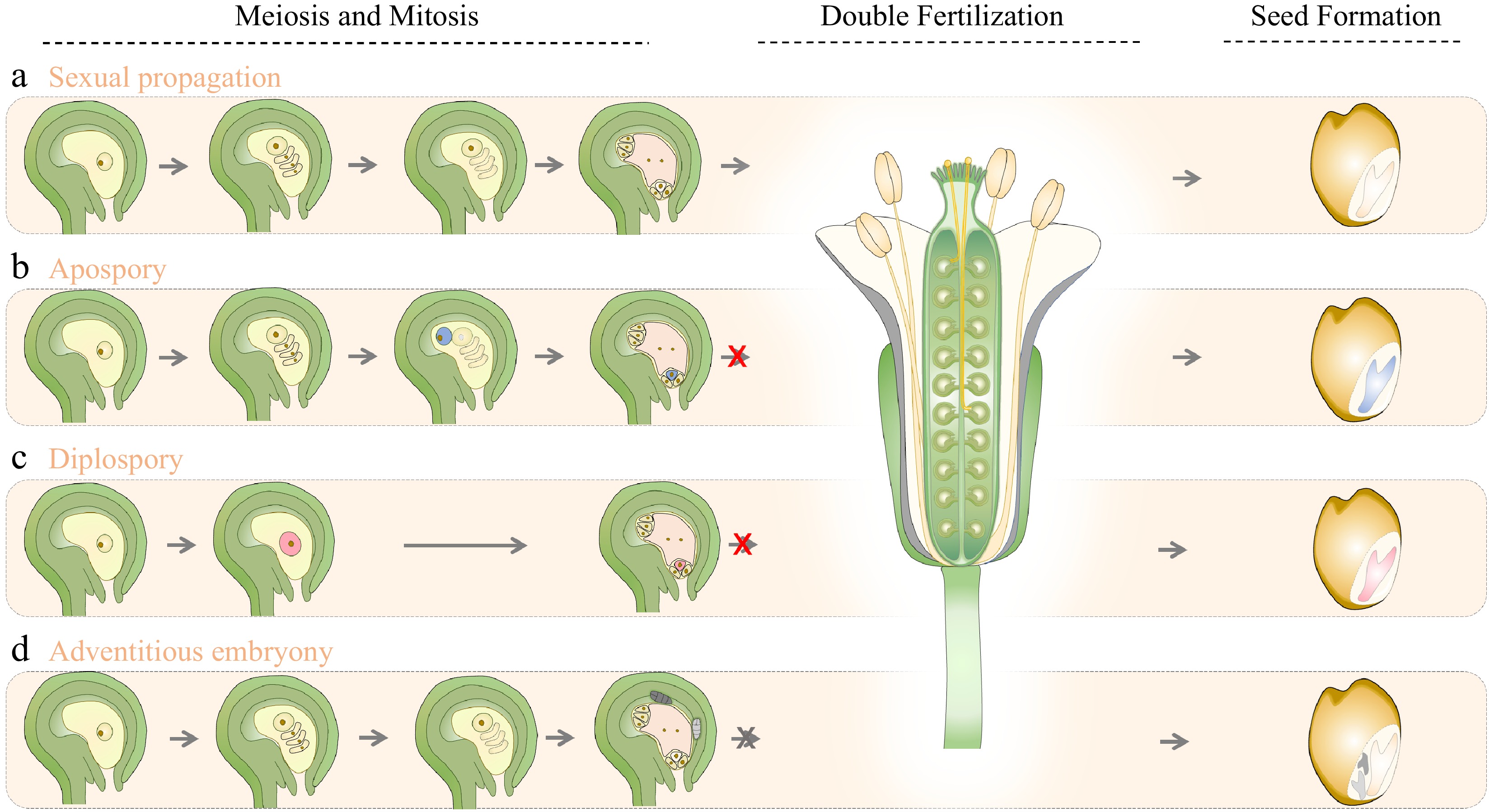
Figure 1.
Schematic representation of sexual and apomictic seed formation in flowering plants. Apomixis deviates from sexual propagation mainly in meiosis, mitosis and double fertilization, which are the primary developmental stages in forming seeds. This diagram depicts the (a) normal sexual reproduction process, and the asexual reproduction processes of apomixis, (b) including apospory, (c) diplospory, and (d) adventitious embryony in flowering plants. Arrows indicate the developmental sequences for seed production. Red crosses mark the bypass of double fertilization in gametophytic apomixis (apospory and diplospory). In gametophytic apomixis (b & c), diplospory produces diploid embryo sac through mitotic division from MMC (red), while the unreduced embryo sac in apospory initiates from a different ovule precursor cell (blue) that divides mitotically, and the unreduced egg cell (b & c) can further develop into embryo without gamete fusion. However, the endosperm formation of apospory and diplospory may need central cell fertilization. In (d) adventitious embryony, embryos initiate directly from somatic cells of the nucellus or integument that are adjacent to the embryo sac, and endosperm develops from the fertilized central cell.
Nowadays, heterosis is widely exploited in modern plant breeding[8], however, the trait segregation of superior hybrid seeds in the progeny greatly limits the agricultural application of hybrid vigor. The breeders need to perform laborious cross-breeding yearly to harvest the F1 hybrid seeds with the desired characteristics. Considering the clonal-propagation properties of apomixis, apomixis has long been the research hotspot, which is thought to be the ideal propagation manner in agricultural practice allowing the instant fixation of superior traits[9]. In this context, great efforts have been exerted in dissecting the underlying molecular mechanism of apomixis and introducing the apomixis to the staple crop species.
The introgression strategy was the early method used to introduce apomixis from the wild relatives to staple crops. However, attempts in developing apomictic maize through introgression of Tripsacum dactyloides to Zea mays has yielded few successes despite of the elaborate and laborious hybridizing and backcrossing[10]. Therefore, another strategy termed synthetic clonal reproduction (also called synthetic apomixis), has been proposed[11]. In synthetic apomixis, the normal meiosis is circumvented to produce unrecombined and unreduced diploid egg cells, and double fertilization is replaced by parthenogenetic development[11,12].
In the last decades, great advances have been made in understanding the molecular mechanisms of plant sexual reproduction, particularly the meiosis and double fertilization in plants[13−15]. Apomeiosis, as one key hallmark of apomixis, has been realized through combining several meiosis-specific gene mutations in Arabidopsis and rice[12,16]. Recently, synthetic apomixis has been engineered in rice by combining Mitosis instead of Meiosis (MiMe) with the mutation of a sperm-specific gene MATRILINEAL (MTL) or ectopic expression of paternal gene BABY BOOM1 (BBM1) in the egg cell, which enables the clonal reproduction of F1 hybrids through seeds and stable transmission of heterotic phenotypes over generations[17,18]. These revolutionary achievements in rice implies bright prospect in applying synthetic apomixis to other staple crops.
In this review, we will briefly summarize the characterized genes in meiosis, double fertilization and chromosome stability that could be engineered for synthetic apomixis. Moreover, we will discuss the potential opportunities and challenges in the future application of synthetic apomixis in crops.
-
Sexual reproduction is the ubiquitous and dominant propagation mode in angiosperms[15]. In higher plants, sexual reproduction refers to the formation of seeds from the zygote through the union of haploid male and female gametes[15]. To achieving this, sequential events, including meiosis-dependent haploid gametophyte formation, pollen-pistil interaction and the double fertilization, need to occur in an error-free manner[13−15].
Meiosis
-
Meiosis is a specialized cell division that halves the ploidy of the germ cells (Fig. 2a). During meiosis (meiosis I and II), the chromosomes of MMC undergoes one round of replication but two rounds of chromosome segregation. The segregation of paired homologous chromosomes during meiosis I and then the following segregation of sister chromatids during meiosis II result in the formation of four haploid cells after cytokinesis (Fig. 2a). The core process of meiosis is characterized by the involvement of many genes (for reviews see[13,19]). In this context, only the pathways and genes of meiotic recombination and segregation of chromosomes that are directly associated with synthetic apomixis will be focused.
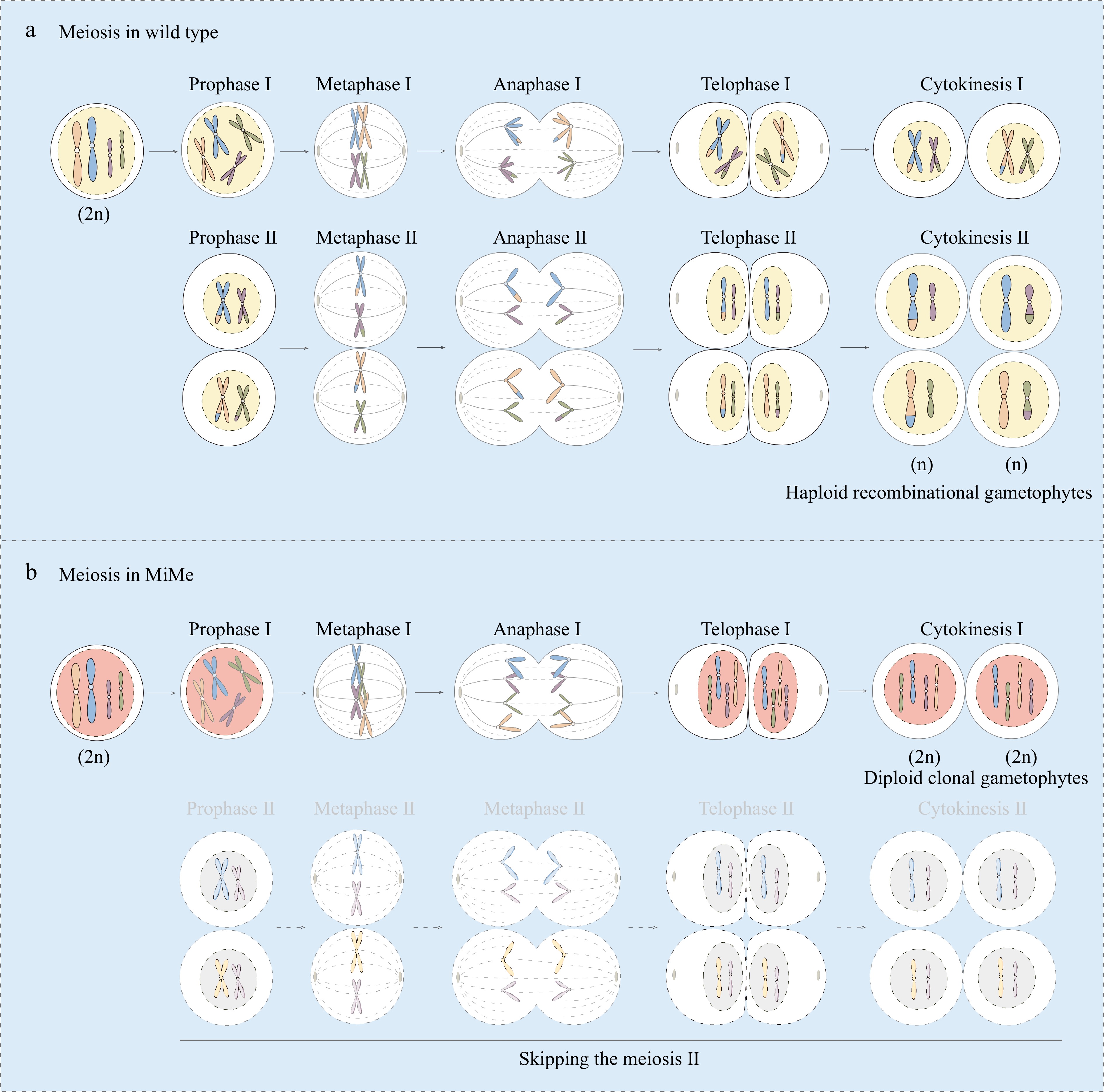
Figure 2.
Schematic representation of meiosis in wild type and MiMe plants. Meiosis in wild type plant is depicted in (a), and (b) exhibits the altered meiosis in MiMe plants. In normal meiosis, the chromosome undergoes one replication, but twice divisions (meiosis I and II). During meiotic prophase I, the homologous recombination takes places with exchange of genetical information between homologous chromosomes. After meiosis II, four haploid recombinational gametes are formed from one megasporocyte (a). However, only two diploid clonal gametes are formed from one megasporocyte in MiMe system without recombination event and meiosis II (b).
Meiotic recombination
-
The meiotic recombination is a highly complicated biological event that is characterized as a repair mechanism for programmed DNA double-strand-breaks (DSBs) at chromosomes during meiosis. Meiotic recombination initiates with the formation of DSBs, which is repaired by homologous recombination to form crossover (CO) or non-crossover (NCO) products[20].
Meiotic recombination is regulated by many conserved genes in a sequential way[20−23]. Sporulation-defective11 (SPO11) has been identified as the primary gene responsible for the formation of DSB in eukaryotes. SPO11 shares sequence similarity with the A subunit of Topoisomerase VI in Archaea[24]. The Arabidopsis genome harbors three homologs of SPO11, namely SPO11-1, SPO11-2, and SPO11-3, with SPO11-1 and SPO11-2 being involved in DSB formation, while SPO11-3 does not contribute to this process[25]. In rice, four homologs of SPO11 have been identified, with SPO11-1, SPO11-2, and SPO11-4 being implicated in DSB formation[26]. Moreover, one another meiotic topoisomerase VIB-like (MTOPVIB) protein that shows structural similarity with the Archaeal topo-VIB subunit, is also highly conserved in plants in regulating the DSB formation[27]. Similar with topoisomerase VI that functions in A2B2 heterotetramer formation, AtSPO11-1 and its homologs AtSPO11-2 in Arabidopsis are thought to form complex with MTOPVIB to catalyze the linkage of 5'-phosphotyrosine to induce DSBs[25].
In Saccharomyces cerevisiae, function of SPO11 requires various proteins for the generation of DSBs, including RAD50, XRS2, REC102, SKI8/REC103, REC104, REC114, MER2, MEI4, and MRE[19]. In contrast to SPO11, these proteins exhibit limited conservation at the sequence level among different kingdoms. Moreover, even when the protein sequences are conserved, functional divergence is frequently observed. For instance, the meiotic role of SKI8/REC103 is conserved in S. cerevisiae and several fungi[19,28]. However, the homologs of SKI8/REC103 in Arabidopsis do not exhibit any meiotic functionality[28]. Although RAD50 plays a crucial role in DSB formation in S. cerevisiae, the orthologs of RAD50 in Saccharomyces pombe are not necessary for the generation of DSBs[29]. Moreover, MRE11 is essential for DSB processing but not for the initiation of SPO11-dependent meiotic DNA breaks in Arabidopsis[30]. Additionally, diverse DSB formation-related genes have also been identified in different species via genetic screening, like PUTATIVE RECOMBINATION INITIATION DEFECT 1, 2 and 3 (PRD1, 2 and 3), and DSB forming (DFO) in Arabidopsis[31−35], and HOMOLOGOUS PAIRING ABERRATION IN RICE MEIOSIS1 (PAIR1), also called OsPRD3, Central Region Component 1 (CRC1), P31comet, and SOLO DANCERS (SDS) in rice[36−39]. Mutants impaired in any of these core recombinant-specific genes display defects in DSB formation, indicating their indispensable role in meiotic recombination[13].
After creation of DSBs by the SPO11 dimer, the nuclease MRE11 recruits RAD50 and XRS2/NBS1 to form MRX/MRN complexes, which associate with DSBs in eukaryotes[13,40]. In the presence of SAE2/COM1, MRX/MRN complexes process DSBs via endonucleolytic cleavage to produce asymmetrically spaced nicks flanking DSBs and exonucleolytic resection, yielding 3′-OH single-stranded DNA (ssDNA) ends[41]. Meanwhile, SPO11 is removed from the chromosome[42]. Replication Protein A (RPA) complexes bind to these ssDNA ends, which are then replaced by RAD51 and DMC1 to form nucleofilaments[43]. Subsequently, RAD51 and DMC1 targets the nucleofilaments for homology search and heteroduplex formation[43]. In most eukaryotes, there are two pathways involved in CO formation: ZMM pathway and non-ZMM pathway that form class I and II CO, respectively[13]. ZMM is the primary pathway, which consists of ZIP1, ZIP2, ZIP3, ZIP4 MER3, MSH4, and MSH5 in S. cerevisiae, and MSH4, MSH5, SHOC1, HEI10, and ZIP4 in Arabidopsis[13]. ZMM pathways account for more than 85% CO formation in eukaryotes. However, there are many other genes identified in yeast and plants to be implicated in CO formation, but not classified in ZMM pathways (for review see[13]). To date, less is known in regarding to class II CO formation. MUS81 is characterized in non-ZMM CO pathways in plants[44]. Lesion in MUS81 reduces CO frequency up to 10% in a wild-type background[44]. Collectively, COs generate genetic diversity in offspring, which is crucial for the survival and adaptation of a species[13]. The presence of the bivalent structure also allows homologous chromosomes to be equally separated during the first meiotic division[45]. When bivalent structure is absent between a pair of homologous chromosomes, unequilibrated segregation of the chromosomes may occur during the mid-prophase I to late prophase I of the first meiotic division[45]. However, meiotic recombination is detrimental to fix the superior traits in heterozygous hybrid seeds.
In addition to COs, NCOs also contribute to meiotic DSB repair. Unlike COs, NCOs involve the copying of a small patch of intact homologous chromosome to the broken chromosome without reciprocal exchange[13]. In Arabidopsis, several components inhibiting CO formation have been identified, which participate in various NCO pathways. Generally, the successful completion of recombination requires the involvement of topoisomerase IIIα and its associated protein, AtBLAP75/AtRMI1[46,47]. Mutations in these genes result in SPO11-dependent connections between homologous chromosomes during metaphase I, independent of ZMM proteins[46,47]. In addition to topoisomerases, the helicase FANCM, along with its cofactors MHF1 and MHF2, as well as the RECQ4 helicase (a homolog of BLOOM/Sgs1), also acts to suppress CO formation[13,48−50]. Notably, COs frequency increases dramatically in either fancm or recq4a/recq4b mutant lines, with these additional COs arising through the class II CO pathway[13,49,50]. Furthermore, FIDGTIN-L1 (FIGL1) represents an additional pathway that inhibits CO formation in a FANCM-independent manner[13]. Taken together, these findings suggest the existence of a delicate balance between NCOs and class II COs in plants.
Meiotic cell cycle progression
-
As mentioned above, meiosis includes two rounds of segregation of chromosomes. At the end of meiosis I, the interlink between homologous chromosomes is disrupted, and the sister chromatids of homologous chromosomes that are separately connected by kinetochores are faithfully oriented and steered to opposite poles[13]. To date, meiosis-specific Recombination-deficient 8 (REC8) is found to be the exclusive meiosis-specific cohesion subunit in plants[51]. The plants with lesion in REC8 exhibit abnormal meiosis, including defects in DSB repairment and bipolar orientation of sister chromatids during meiosis I[51]. In rice, REC8 is also reported to be necessary for chromosome axis building and bouquet formation, homologous chromosome pairing and recombination[52]. Other cohesion subunits, including sister-chromatid cohesion protein 3 (SCC3), Structural Maintenance of Chromosome 1 (SMC1) and SMC3 are shared by both meiosis and mitosis to be responsible for separation of sister chromatids[51,53]. Additionally, it has been observed that SHUGOSHIN1 (SGO1) and Bub1-related kinase 1 (BRK1) are implicated in the protection of centromeric cohesion[54,55]. Notably, the mutants of SCC3, SGO1, and BRK1 have been found to induce untimely separation of sister chromatids, as documented in previous studies[51,54,55].
Cyclin, cyclin-dependent kinases (CDKs) and the negative regulator anaphase-promoting complex (APC/C) constitute the conserved core regulatory module that ensures the faithful progression of the cell cycle for both meiosis and mitosis in eukaryotes[56−58]. During cell division, cyclin associates with CDKs to activate its kinase activity, which is indispensable for the entry and progression of metaphase[59]. The progression of meiosis is closely correlated with the nuanced regulation of the activity of the cyclin-CDK module. For instance, the onset of anaphase requires rapid degradation of cyclin by APC/C to suppress CDK activity[58]. In contrast, the initiation of meiosis II through interkinesis requires moderate inhibition of the activity of cyclin-CDK. Intriguingly, the dramatic suppression of cyclin-CDK leads to the termination of meiosis. This cell cycle machinery is also highly conserved in plants[13].
In Arabidopsis genome, several genes are associated with meiosis progression, including Tardy Asynchronous Meiosis (TAM), CDKs (CDKA;1, CDKB;1, CDKB1;2, CDKB2;1 and CDKB2;2), Three Division Mutation 1 (TDM1) and Omission of Second Division 1(OSD1)[60−62]. During the first meiotic division, TAM, as an A-type cyclin associates with CDKA;1 to phosphorylate TDM1 to suppress its activity[62]. TDM1 is an APC/C component that is necessary for exit from meiosis II[63]. Additionally, TDM1 is supposed to be functional during the whole meiosis since it is always highly induced along with meiosis, while TAM is expressed only at meiosis I[60]. OSD1 is responsible for the onset of meiosis II by regulating cyclin-CDK activity in both rice and Arabidopsis[12,16]. Mutations in any of these genes induce the formation of polyploid gametes, suggesting that they are of great importance in maintaining normal meiosis progression.
Double fertilization
-
After meiosis, the male and female gametophytes are formed after mitosis and cell differentiation[64,65]. Sperm cells encapsulated in the pollen are delivered to the ovule through the stigma and transmitting tissue to encounter egg cells for zygote formation[15]. A series of biological events, including pollen-stigma interaction, pollen tube germination and growth, pollen tube guidance, and pollen tube reception are carried out sequentially to ensure the successful selection and delivery of desired conspecific male gametes to egg cells[14]. Finally, double fertilization takes place in embryo sac when the two sperm cells unit with the egg cell and central cell to initiate the embryogenesis and endosperm development, respectively. To date, numerous advances have been made in deciphering the molecular underpinnings behind these processes (for review see[15,66,67]).
In angiosperms, double fertilization is a highly ordered and sophisticated process that involves multiple steps, including gamete attachment, sperm activation, membrane fusion, plasmogamy, and karyogamy[66,68]. After release into the embryo sac, immobile sperm cells are steered towards the site of gamete fusion and attach to the female gametes[69]. The GAMETE EXPRESSED 2 (GEX2) gene, which encodes a sperm-specific transmembrane protein in Arabidopsis, has been reported to mediate the gamete attachment[70]. The conserved filamin-repeat domain of GEX2 forms an immunoglobulin (Ig)-like fold that is essential for gamete attachment and is also shared by FUS1 in Chlamydomonas reinhardtii (Chlamydomonas)[71]. Sperm cells with a lesion in GEX2 show defects in attaching to the egg and central cells, and the resultant double-fertilization failure and single fertilization typically cause aborted seeds[70,71].
In the context of gamete fusion in flowering plants, little is known about the molecular mechanisms underlying the activation of sperm cells. In contrast, the signaling cascades initiated by gamete association prior to fusion have been more extensively studied in unicellular green algae and multicellular animals[72,73]. In Arabidopsis, upon the arrival of sperm cell, the membrane contact between the sperm and egg cells triggers the release of cysteine-rich EGG CELL 1 (EC1) by the egg cell through exocytosis to activate the sperm[74]. The secreted EC1 then triggers the translocation of the fusogen HAPLESS 2/GENERATIVE CELL SPECIFIC 1 (HAP2/GCS1) from endomembrane-associated sites to the surface of the sperm cell[74,75]. Furthermore, EC1 is reported to mediate the separation of the two physically linked sperm cells[76]. The positioning of sperm cells is also critical for the success of double fertilization. When the two sperms simultaneously attach to the egg cell, repositioning occurs with one sperm cell released and re-oriented towards the central cell and the other remains stagnant[77]. Proper adhesion of each sperm cell to one female gamete is then achieved[67,77].
After adhesion, HAP2/GCS1, as the essential fusogen, acts as the plasma membrane merger for the male and female gametes[75,78]. In Chlamydomonas, hap2 mutant gametes can interact with female gametes and stopped at a 10 nm distance between the opposing membranes, failing in the further plasma membrane fusion[79]. The presence of orthologs of HAP2 in all eukaryotic clades, except fungi, indicates that HAP2 is a highly conserved gamete fusion protein in eukaryotes[80]. Interestingly, the structure of Chlamydomonas HAP2 exhibits striking similarities to class II viral fusion proteins, with three extracellular domains, a single transmembrane domain, and a smaller intracellular domain[81]. These class II viral fusion proteins homotrimerize upon entry into host endosomes, and a conformational change facilitates the fusion of the host membrane and viral envelope[81]. Similarly, recent structural analyses of Arabidopsis HAP2 have shown that an apical amphipathic helix is required for membrane insertion in vitro and fusion with the egg and central cell in vivo[80]. These discoveries suggest a model for double fertilization in Arabidopsis, where HAP2 functions as a unilateral gamete fusion protein, mediating the fusion of one sperm cell with the egg plasma membrane and the other with the central cell, without requiring a binding protein on the surface of the female gametes. However, this model raises intriguing questions, such as how the activity of HAP2 is regulated in preventing the premature sperm cell fusion and what is the underlying mechanism that guide the two distinct fusion events between sperm and female gametes.
The absence of necessary genes for either gamete fusion or preventing polyspermy can result in haploid offspring in certain mutants, attributing to a double fertilization defect. Recent studies have identified two sperm cell-specific DOMAIN OF UNKOWN FUNCTION 679 membrane proteins (DMP8 and DMP9) that also facilitate gamete fusion, especially sperm-egg cell fusion[76]. The dmp8 dmp9 sperm cells are also defective in the secretion of HAP2 upon gamete activation by EC1[82]. Loss of DMP8/9 in Arabidopsis or the orthologs in maize, potato, tomato, Brassica napus, tobacco, watermelon, cabbage and Medicago truncatula causes maternal haploid offspring[83−91]. In a recent study, the identification of two aspartic endopeptidases, EGG CELL-SECRETED 1 (ECS1) and ECS2, has shed light on the intricate spatial and temporal control of gamete interactions in Arabidopsis[92]. These peptidases are secreted into the extracellular space from the egg cells to regulate the nuclear and plasma membrane fusion between the male and female gametes[93−96]. Interestingly, the generation of partially semigamous zygotes and then maternal haploid offspring containing only the genome of the egg has been observed in ecs1ecs2 double mutants in Arabidopsis and the rice esc1 mutant, through either selfing or hybridization[93].
In summary, the genetic alteration of some of the genes involved in double fertilization causes haploid production. Critical roles in gamete attachment and fusion are played by GEX2, HAP2/GCS1, DMP8/9, EC1 and ECS1/2 proteins, while only knockout of DMP8/9 and ECS1/2 have been tested and verified to induce haploid generation. This raises the possibility that the entry or physical adhesion of the sperm triggers certain zygotic programs, although the haploid generation occurs at a low frequency (less than 10%). Another commonality between DMP8/9 and ECS1/2 is that they both control preferential fertilization of the egg cell[76,94]. A more profound comprehension of the molecular mechanisms underlying double fertilization in angiosperms would facilitate the development of parthenogenesis.
-
Scientists have long sought to introduce apomixis into non-apomictic staple crops, as this could offer significant agronomic benefits. Two feasible routes for achieving this goal have been proposed: introgression and engineering synthetic apomixis. To date, researchers have achieved great success in engineering synthetic apomixis in Arabidopsis and rice using hybridization or genome-editing technology[17,18,97]. Synthetic apomixis involves three principles. First, meiosis, which normally produces haploid female gametophytes, must be circumvented (Fig. 2b). Second, embryogenesis must take place in the absence of nuclear fusion between male and female gametes or with the occurrence of genome elimination after gametes fusion (Fig. 3). Lastly, viable endosperm must develop via an autonomous or pseudogamous mechanism, or normal central cell fertilization. To bypass meiosis, Mitosis instead of Meiosis (MiMe) strategy can be employed to induce unreduced and nonrecombinant maternal gametes[12,16]. Parthenogenesis and elimination of the paternal genome are two approaches that can be used to create progenies with uniparental genome.

Figure 3.
Schematic representation of genome elimination and parthenogenesis. The haploid offspring can be induced through crossing plant carrying modified CENH3 gene with (a) wild type pollen, (b) mutating MTL or (c) DMP gene, or (d) nectopic expression of BBMs in the egg cell. In (a), genome elimination occurs in crosses between wild type pollen and CEHN3-based haploid inducer. The chromosome of haploid inducer is unstable during mitotic divisions of the early embryo, and will be lost to produce haploid plants, which are genetically identical to the genome of wild type parental gametophyte. (b) Depicts the haploid induction by chromosome fragmentation event. In mtl mutants, chromosomes undergo fragmentation with varied frequencies from microsporocyte to embryogenesis stage. During double fertilization, DMP is supposed to bridge the gamete fusion, however, mutating DMP results in gamete fusion failure and haploid production (c). Additionally, parthenogenesis can be induced in sexual plants by ectopic expression of BBMs in the egg cell (d).
Molecular underpinnings of mitosis instead of meiosis
-
The MiMe strategy involves mutating several selected core meiosis-related genes to disrupt chromosome recombination, induce separation of sister chromatids during meiosis I, and totally abolish the meiosis II[16]. Among these genes, SPO11-1, PRD1, PRD2, PRD3/PAIR1, MTOPVIB, DFO and P31comet, which play key roles in DSB formation during meiotic recombination, can be mutated to prevent recombination events in plants[98]. Premature sister chromatid segregation during the first meiotic division can be achieved by mutating the REC8 gene, which encodes the exclusive meiosis-specific cohesion subunit[98]. OSD1, TAM, and TDM1 are pivotal genes controlling meiotic cell cycle transition and lesions in any one of these three genes abolish the meiosis II[98]. Combination of mutations in single gene of each group have been proven successful in establishing MiMe in Arabidopsis and rice (for review see[98]).
Genome elimination
-
After successful induction of apomeiosis in sexual plants through MiMe strategy, the next hurdle towards clonal seeds is to circumvent the formation of zygote. The two ways to achieve this include genome elimination and parthenogenesis. The major difference between gametophyte apomixis and genome elimination in producing clonal seeds is whether fertilization is required. In genome elimination, either maternal or paternal chromosome set will be lost during early embryogenesis. To date, several strategies have been found to cause genome elimination, including CENH3-based crossing (Fig. 3a), interspecific outcrossing (Fig. 3b), and intraspecific hybridization.
CENH3 is a highly conserved centromeric histone H3 present in all eukaryotes[99]. It consists of a conserved C-terminal histone fold domain and a highly variable N-terminal tail. During cell cycle, CENH3 recruits proteins to assemble kinetochores at the centromere, where spindle fibers attach to guide accurate chromosome segregation in mitosis and meiosis[99]. Studies have shown that homozygous null mutants of Atcenh3-1 exhibit mid-global stage embryo development arrest[99]. Intriguingly, the embryo-lethal phenotype can be rescued by fusing CENH3 with green fluorescent protein (GFP), or by using GFP-tailswap, a variant of GFP-CENH3 with the N-terminal tail of CENH3 substituted with the N-terminal of histone H3.3[99]. When these rescued plants are used as male or female parents in crosses with wild type plants, haploid offspring with uniparental chromosome sets can be observed, transforming the plant into a flexible haploid induction system that can efficiently induce both maternal and paternal haploids, with the latter having an induction rate of over 20%[99]. The mechanism underlying this phenomenon is thought to be postzygotic incompatibility, where the parental chromosome set carrying the structurally altered CENH3 at its centromeres is mitotically unstable and is therefore left behind in early embryonic divisions[99]. Moreover, it has been proposed that mutations in CENH3 may impair chromatin loading, resulting in smaller centromeres that cannot compete with the larger centromeres of the crossing parent[100]. This size dimorphism of parental centromeres can cause early loss of chromosomes, resulting in haploid and aneuploid progenies[100,101]. These serendipitous discoveries have stimulated researches on the relationship between CENH3 and genome elimination.
Further studies have shown that orthologs of CENH3 from different species of Brassicaceae or even Zea mays can complement the embryo-lethal phenotype of Atcenh3-1 and generate haploid offspring when crossed with wild type plants[102]. In addition, genetic research has revealed that a single-amino-acid missense mutation in CENH3 is sufficient to mimic the effects of chimeric CENH3 or CENH3 from diverged species in inducing genome elimination[103]. However, despite the efforts of numerous researchers, the anticipated success has not been achieved in most crops, with only a few crops, such as wheat and maize, having successfully generated haploid induction systems based on CENH3, but with lower induction rates than those in Arabidopsis[101,104]. Excitingly, crosses between mutant lines expressing CENH3 variants and MiMe or dyad mutants produce clonal seeds in Arabidopsis[97].
In a recent study by Li et al., it was discovered that the GFP-tailswap haploid induction system is highly sensitive to environmental temperature in terms of both pollen viability and haploid induction ability, compared to wild-type plants[105]. Specifically, even slight increases in environmental temperature, from 22 to 25 °C, led to a near-total loss of GFP-tailswap pollen viability, while significantly enhancing haploid induction ability. Conversely, decreasing the temperature had the opposite effect[105]. These findings have important implications for the extension of CENH3-based haploid induction systems to other crops and provides valuable clues for future studies, emphasizing the need to consider the influence of environments. Moreover, the research sheds light on the fundamental biological issues of CENH3 in maintaining the precise separation control of chromosomes during cell division and the mechanism of haploid formation.
Outcross-related genome elimination
-
The phenomenon of uniparental genome elimination in interspecific outcrosses between monocots was first observed in 1970. Although the underlying mechanism is not yet fully understood, this phenomenon has been applied in agriculture to produce maternal haploid lines, such as triploid wheat, which is generated by crossing hexaploid wheat with maize, sorghum, or pearl millet[106,107]. Selective elimination of parental genomes can occur in interspecific hybridization, such as in the haploid offspring induced by the interspecific hybridization of barley (Hordeum vulgare L.)[108]. In crosses between wheat and pearl millet, the paternal genome of pearl millet is selectively eliminated[109], and a series of biological events, including paternal genome separation, chromosome structure rearrangement, micronucleus formation, and micronucleus degradation, are induced in this process[110]. In outcrosses between wheat and maize, a haploid wheat embryo forms, possibly due to the small genome size of maize and defective spindle attachment to the centromere[111], or defective gamete interactions. The formed wheat zygotic embryo gradually degrades after several divisions, and normal viable endosperm cannot form after successful fertilization of central cell due to post-zygotic cross barrier[111]. However, given the inadequate understanding of the underlying molecular mechanism, the use of outcross in producing clonal seeds of crops is still constrained.
Conspecific hybridization-related genome elimination
-
The use of Stock6-derived haploid inducer lines for intraspecific hybridization is a commonly employed technique in maize breeding, as established by Coe[112]. The haploid induction capability of Stock-6 is due to the mutations in ZmPLA1 and ZmDMP [87,113]. ZmPLA1, which encodes a maize phospholipase that localized on the endo-plasma membrane (endo-PM) that wraps the sperm cells within the cytoplasm of the pollen tube[114], was initially identified as qhir1, a quantitative trait locus[115], and is also known as MATRILINEAL (MTL)[116] and NOT LIKE DAD (NLD)[114]. Single nucleus sequencing of the zmpla1 haploid inducer lines revealed an unexpectedly high frequency of sperm DNA fragmentation (Fig. 3b), which has been suggested to be the cause of paternal genome loss following fertilization and the subsequent production of maternal haploids[117]. However, the mechanism triggering DNA fragmentation remains unknown. Recent comprehensive omics analysis has revealed a possible mechanism involving an ROS burst for haploid induction in zmpla1[7], suggesting that a ROS burst causes an imbalance in the redox state of the metabolome, leading to sperm DNA breakage. Once the constraints of the DNA-repair machinery are overcome, the breakage may extend from the centromeric regions to the entirety of the sperm genome, ultimately leading to fragmentation. Subsequently, continuous DNA fragmentation leads to the loss of the male genome and/or chromosomes with defective centromeres, ultimately culminating in haploid induction following fertilization (Fig. 3b). Furthermore, through the use of omics analysis, a unique peroxidase, ZmPOD65, has been identified as being specific to pollen and serving as a novel gene that regulates haploid induction[7]. This discovery serves to reinforce the relationship between reactive oxygen species and haploid induction in zmpla1.
MTL-based chromosome fragmentation has been utilized in generating haploid inducing lines in other staple crops. For instance, by mutating TaMTL in wheat using gene editing technique can create a haploid inducer line with a haploid induction rate (HIR) of up to 18.9%[118]. Haploid induction is also achieved in foxtail millet (Setaria italica) by gene editing of SiMTL[119]. In rice, the selfing progeny of mtl mutant includes haploids and doubled haploids with HIR of 4.44%. Moreover, simultaneously mutating OsMTL and three MiMe inducing genes REC8, PAIR1, and OSD1 can result in maternal clonal seeds, suggesting that the mechanism is likely to be conserved in plants[17]. However, expanding this method to Arabidopsis is hampered by the fact that the most relevant gene of MTL in Arabidopsis, AtPLP2, is only expressed in vegetative organs[120]. MTL-based genome elimination also relates to genome instability but occurs before fertilization. Therefore, searching for an endo-PM-specific phospholipase in dicots may be effective in haploid induction in dicot species. Intriguingly, recent studies indicate that the phospholipases expressed in pollen or gynoecium are also correlated to haploid induction. In Arabidopsis, null allele mutation of a gynoecium-expressed phospholipase All (pPLAIIγ) produces maternal haploid seeds, and the haploid induction in pplaIIγ is accompanied by the internalization of PIN1 at the plasma membrane of the basal funiculus[121]. In Zea mays, mutation line impaired in pollen-specific PHOSPHOLIPASE D3 (ZmPLD3) generates maternal haploids, which shows comparable haploid induction rate with mtl[122]. Additionally, mutating PLD3 in mtl background triples the haploid induction rate from 1.19% to 4.13%, indicating synergistic effect of PLD3 and MTL in haploid induction[122]. ZmPLD3 is highly conserved in other cereals, indicating the potential in developing PLD3-based haploid-inducer in other staple crops.
Parthenogenesis
-
After generating a diploid clonal gamete, autonomous transition to embryonic development can be achieved through ectopic expression of the transcription factor BABY BOOM (BBM) or its orthologs in the egg cell of monocots or dicots[18,123,124]. BBMs belong to the eudicot AINTEGUMENTA (euANT) family and play a crucial role in various processes, including embryogenesis, root development, and somatic embryogenesis[125]. In Pennisetum squamulatum, several BBM-like (BBML) genes located in the apospory-specific genomic region (ASGR) control the parthenogenesis of Pennisetum squamulatum[126,127]. PsASGR-BBMLs are orthologs of BnBBM, and their ectopic expression in the egg cell can induce parthenogenesis in tobacco, pearl millet, rice and maize[18,123,128−130]. Intriguingly, the expression of PsASGR-BBM1 driven by the Arabidopsis egg cell-specific AtDD45/EC1.2 promoter resulted in more autonomously developed egg cells in rice and maize than when driven by its native promoter[130]. However, this strategy was unsuccessful in Arabidopsis, and the native promoter of PsASGR-BBM1 has failed to be transcribed in Arabidopsis[130]. Recently, ectopic expression of BnBBM in the egg cell of Arabidopsis and the dicot crops Brassica napus and Solanum lycopersicon was suggested to initiate early embryogenesis in the absence of fertilization[124]. Moreover, haploid induction rates of the egg-cell-expressed BBM lines are even increased in crossing with dmp in the case of Arabidopsis and tomato, implying a synergistic effect between dmp and ectopic BBM expression on parthenogenesis[124].
Rice has four OsBBM homologous genes, among them, BBM1, BBM3, and BBM4, are exclusively expressed in sperm cells before fertilization[18]. Ectopic expression of OsBBMs in rice egg cell can initiate parthenogenesis and result in viable seeds when the endosperm develops after the normal fertilization of the central cell[18]. As shown in other species (for review see[131]), the ability of OsASGR-BBM1/OsBBM1 in initiating somatic embryogenesis before fertilization was also confirmed[132]. The successful engineering of synthetic apomixis in rice involves the mutation of three MiMe-causing genes, namely PAIR1, REC8, and OSD1 using the CRISPR/Cas9 technique, and the ectopic expression of OsBBM1 in the egg cell[18]. Nonetheless, this strategy is beset with low seed setting rates and low clonal seed ratio, which considerably limit its implementation in crops. Subsequent research aimed at improving this approach reported the enhancement of cloning seed proportion to as high as 90% in the hybrid rice variety BRS-CIRAD 302[133]. This was achieved through the combination of ectopic OsBBM1 expression in the egg cell and the mutation of PAIR1, REC8, and OSD1 genes through a single vector[133]. A new synthetic apomixis strategy for rice that combines the ectopic expression of BBM4 with the MiMe strategy has been developed for hybrid rice, resulting in the generation of Fixation of hybrids 2 (Fix2) plants that exhibit normal growth during the vegetative growth stage[134]. Additionally, the Fix2 plants displayed a high seed setting rate of 80.9%−86.1%, similar to that of normal hybrid rice at 82.1%−86.6%[134]. Although the seed setting rate of Fix2 strategy was not affected, the cloning seed proportion was quite low (1.7%)[134]. Thus, further research efforts are needed to develop a synthetic apomixis strategy for hybrid rice with high seed-setting rates and high cloning seed induction rates.
In addition to BBMs, a PARTHENOGENESIS (PAR) gene in apomictic dandelion has been cloned recently, which encodes a K2-2 zinc finger, ethylene-responsive element binding factor-associated amphiphilic repression (EAR)-domain protein[135]. Ectopic expression of PAR, under the control of AtEC1 promoter in sexual lettuce, induces haploid embryo formation in unfertilized embryo sac[135]. However, the haploid embryo undergoes abortion at later developmental stages due to the inadequate support from abnormal and nonsexual endosperm[135]. The recessive par gene are expressed in the pollen of sexual dandelion, and the two PAR homologs in Arabidopsis[135], DUO1 activated zinc finer 3 (DAZ3) and transcriptional repressor of EIN3-dependent ethylene-response 1 (TREE1), are among the highest expressed genes in Arabidopsis sperm cells[136]. Therefore, it is supposed that the function of PAR is to lift the autonomous inhibition on embryogenesis of unfertilized egg cell[135]. Overall, this study highlights the potential application of PAR homologs in inducing parthenogenesis in other dicots.
Endosperm formation in synthetic apomixis
-
As the last step, the formation of viable endosperm is critical for the success of synthetic apomixis. In most natural apomicts, endosperm develops from the fertilized central cell[137], however, endosperm can also form autonomously in some apomictic species[138−140]. It is worth noting that the bypassing of central cell fertilization is not essential for the induction of clonal seeds, and all reported instances of synthetic apomixis have relied on the normal fertilization of the central cell[17,18,97,133,134]. Nevertheless, the autonomous formation of endosperm represents the ultimate culmination towards the realization of fertilization-independent synthetic apomixis.
In Hieracium, the occurrence of autonomous endosperm development represents a qualitative trait governed by one single dominant locus[141]. Additionally, this trait is regulated by multiple other unknown gene(s)[141], implying a complex underlying mechanism for autonomous endosperm formation in natural apomictic species. While progress in elucidating the mechanisms of autonomous endosperm formation in natural apomicts has been sluggish, considerable knowledge have been amassed regarding the induction of autonomous endosperm formation in sexual plants. Notably, the pivotal role is played by Fertilization-Independent Seed (FIS) class genes, which encode the proteins involved in the Fertilization-Independent Seed-Polycomb Repressive Complex2 (FIS-PRC2) in embryo sac[142−147]. The FIS-PRC2 members include MEDEA (MEA), FIS2, FERTILIZATION INDEPENDENT ENDOSPERM (FIE), MULTICOPY SUPRESSOR OF IRA (MSI1), and BORGIA (BGA)[142−146]. Disruptions in these FIS genes induce autonomous endosperm formation in sexual Arabidopsis[142−145]. Moreover, the penetrance rate of autonomous endosperm formation, which estimates the percentage of ovules carrying these fis mutant, ranges from 41.2% to 92.4% in Arabidopsis[142]. However, fis mutants consistently exhibit abnormal endosperm development or embryo lethality[142−146,148]. To date, the orthologs of FIS genes have been cloned in many cereal plants, such as rice, maize and barley[149−154]. Encouragingly, mutations in many of these orthologs have been demonstrated to induce autonomous endosperm formation[149,150,154]. Further investigation reveals that the chromatin modifying complex FIS-PRC2 mediates the trimethylation of the histone H3 Lysine-27, thereby suppressing gene expression[155,156]. The causes underlying autonomous endosperm development in mutants of the PRC2 genes are likely attributed to the activation of the auxin biosynthesis pathway in the central cell of Arabidopsis and rice[155,157−160].
Interestingly, a recent study has shed light on the impact of mutations in two rice orthologs of FIE. These mutations have been found to abolish the suppression of asexual embryo development and autonomous endosperm formation prior to fertilization[161]. Moreover, it has been observed that the asexual embryo development is prompted by the autonomous endosperm, which also leads to the transcription of male-genome expressed genes such as OsBBM1, OsBBM2, and OsWUSHCEL-related homeobox 8/9 (OsWOX8/9) in the asexual embryo[161]. These findings suggest that PRC genes are involved in the silencing of male-genome expressed genes. Additionally, it is noteworthy that the syncytial phase and cellularization of endosperm development occur independently of embryo presence[162,163]. Considering these insights, it is tempting to speculate that combining MiMe with fis mutants could be a feasible approach towards achieving fertilization-independent synthetic apomixis.
-
Synthetic apomixis offers great opportunities for permanent fixation of hybrid vigor and genetic stability of staple crops. By bypassing meiosis and fertilization, synthetic apomixis can produce genetically identical offspring, eliminating the need for costly and time-consuming breeding efforts. Although much progresses have been achieved in the past two decades, the molecular understanding of apomixis and robust strategies for application of synthetic apomixis in major crops are still limited. Compared to the conserved MiMe strategy, the haploid induction appears to be the major hurdles to be resolved in this field. Moreover, MiMe mutations has only been studied in few species, whether it works in other crops still need investigation. In addition, achieving a basic understanding of the distinction between sexual and asexual reproduction remains a significant challenge.
Advancements in our understanding of gamete interaction and the activation of the zygotic genome have the potential to lead to the identification of new genes that can be leveraged for the engineering of synthetic apomixis in crops. In addition, new tools have emerged that can be used to screen for potential gene targets and to dissect gene function and regulation networks. For example, single-cell CRISPR screening techniques, such as DAP-seq, CROP-seq, CRISP-seq, or Perturb-seq[164], and CRISPR-Cas-based genetic and epigenetic manipulation of gene expression systems[165], as well as comprehensive omics analyses including transcriptome, metabolome, quantitative proteome, and protein modification analysis[7], are rapidly developing. These technologies have the potential to accelerate the development of synthetic apomixis, as they can be used to identify the desired genes related to haploid induction and MiMe in plants. The knowledge of the transcriptional regulation of genes also depends on an understanding of the cis-elements and chromatin-based regulations in the genome sequence.
There are also several challenges that must be addressed before synthetic apomixis can be successfully applied to staple crops. One major challenge is the complexity of the apomixis trait, which involves the coordination of multiple genetic pathways. Developing synthetic apomixis systems that can be efficiently transferred to target crops will require a thorough understanding of these pathways and how they interact with one another. Another challenge is the potential for unintended consequences, such as the loss of genetic diversity and the accumulation of deleterious mutations over time. To address these concerns, researchers must carefully monitor the performance of synthetic apomixis lines over multiple generations and develop strategies to maintain genetic diversity within breeding populations. In addition, there may be regulatory challenges associated with the adoption of synthetic apomixis in agriculture, for example, concerns about the safety and environmental impact of genetically modified crops, or doubt on the need for such technologies given the availability of alternative breeding strategies. Overall, while synthetic apomixis holds great promise for improving staple crops like maize and soybeans, there are a number of scientific and technical challenges that must be overcome before this technology can be widely adopted.
In summary, the successful engineering of synthetic apomixis in rice signifies the commencement of a novel agricultural revolution. However, the translation of this breakthrough to other major crops, such as maize and soybean, presents various limitations. Despite the potential difficulties and challenges, the development of synthetic apomixis has enormous benefits in the field of crop breeding, and it is imperative that continued efforts should be made to overcome the existing obstacles.
This study was supported by the Strategic Priority Research Program of the Chinese Academy of Science (XDA24020306) and the National Key Research and Development Program of China (2022YFF1003500).
-
The authors declare that they have no conflict of interest. Hongju Li and KejianWang are the Editorial-Board members of Seed Biology who were blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of these Editorial-Board members and their research groups.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press on behalf of Hainan Yazhou Bay Seed Laboratory. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Li S, Wang J, Jia S, Wang K, Li H. 2023. Synthetic apomixis: from genetic basis to agricultural application. Seed Biology 2:10 doi: 10.48130/SeedBio-2023-0010
Synthetic apomixis: from genetic basis to agricultural application
- Received: 19 May 2023
- Accepted: 04 July 2023
- Published online: 04 September 2023
Abstract: Apomixis in plants is a widely existing biological phenomenon, in which seeds are formed without egg cell and sperm uniting. Hybrid breeding exploits heterosis to obtain seeds with superior traits. However, segregation of traits in the offspring greatly limits the widespread use of hybrid vigor in agricultural production. Synthetic apomixis is considered a desired way of clonal propagation of heterozygous maternal parents, which bypasses laborious hybrid process. In recent years, with the increasing understanding of the molecular mechanisms of plant meiosis and double fertilization, scientists have introduced apomixis to rice and hybrid rice varieties by genetic engineering of genes that are involved in sexual reproduction. In this review article, we will summarize the recent research progress in the meiosis and double fertilization related to synthetic apomixis and provide perspectives on the potential application of synthetic apomixis in different crops and livestock pastures.
-
Key words:
- Synthetic /
- Apomixis /
- Genetic /
- Basis /
- Agricultural /
- Applications













