-
Areca palm grows widely in the tropical areas of Asia and East Africa although it is believed to originate in the Philippines and Malaysia. In South Asian countries, the areca nut has been used for chewing and medicinal purposes for more than 2,000 years. Religious and masticatory purposes make their significance more unique[1]. The economic importance of areca palm is highly significant, especially in China and India, because it provides a way of living for millions of people. The economically valuable part of the areca palm tree is fruit called areca nut or betel quid, having great popularity due to its chewing features[2]. In old Indian and Chinese prescripts, the medicines extracted from areca nut are used for the treatment of leprosy, fits, leukoderma, cough, obesity and worm's anemia. The inhibitory potential of areca tannins has been detected against reverse transcriptase enzymes[3]. Various extracts of areca nut seeds and other constituents encompass numerous pharmacological possessions. The traditional use of old medicines obtained from different medicinal plants have great importance, because it provides the foundation to learn the potential use of today's advanced synthetic medicines[3,4].
During the life cycle of an individual plant, various predators such as fungi, nematodes, bacteria, virus and various herbivores affect the growth and development. Plants defend themselves by synthesizing variety of chemical compounds, including secondary metabolites that play their role in plant adaptation against the biotic and abiotic stresses[5−7]. In a natural ecological system, the relation between the plants and the environmental biotic stresses is kept in harmony despite the intense competition. However, the harmonious balance could be broken if the area of individual plant species sharply increases resulting in decline of biodiversity, affecting the transmission of infectious diseases of plants[8,9].
-
The consumption of areca nut has been sharply increasing which has given the opportunities to expand plantation areas. It is estimated that the area in Hainan Province in China is over 200,000 hectares at present, which is under serious threat from different pathogens, such as bacteria, fungi[10,11], phytoplasmas[12,13], viruses[14−19], and insects, such as Tirathaba rufiven and Brontispa longissimi[20,21]. Among all of them, the YLD posed a serious threat to areca palm plantations in the world. The YLD was first observed in areca palm in India in 1914[22,23], and later in China in 1985[24], and in Sri Lanka in 2015[13], destroying areca palm plantations on a large scale.
As the name suggests, the most visible symptom of YLD is the yellowing of the leaf. However, the occurrence of YLD is usually combined with one or more other pathogen and/or abiotic stresses, which makes it difficult to describe the specific YLD symptoms objectively. That's why YLD symptoms are described by different authors with different assumptions and results[25−28]. According to the observation and analysis in our laboratory, fungal, bacterial and viral infections on areca palm usually cause significant leaf necrosis spots or streaks. For example, Diaporthe limonicola causing leaf spot disease on areca palm. Firstly, small yellow spots are developed, as the symptoms progressed, the middle of the lesions appeared black with distinct yellow halos[29]. Curvularia pseudobrachyspora causing leaf spots in areca palm appeared dark red lesions which gradually developed into spindly, dark brown spots. The internal area of these lesions had a dark brown edge and yellow halo, gradually turned black and then coalesced to form larger necrotic areas[11]. Areca palm necrotic ringspot disease (ANRSD) and areca palm necrotic spindle-spot virus (ANSSV) exhibited translucent spots (1−2 mm) in the growing spindle in top leaves and necrotic ringspot and spindle-spot symptoms in medium and bottom leaves, respectively[16,30]. Nutrient deficiency (an abiotic factor) can also cause a wide range of yellow leaf symptom on the whole crown without apparent green-yellow demarcation. The soil-borne disease causes rotten root on areca palm, also resulting in a wide range of leaf yellow symptoms, which is similar to physiological yellowing. The YLD symptoms described in the previous reports were obviously different due to the co-occurrence of other pathogens. For example, Menon[25] reported that the translucent spots (1−2 mm) in the growing spindle were the first visible signs of YLD; after that the necrotic streaks having brown color appeared, expand towards the lamina of unfolding leaves, but the images of the symptomatic leaf and symptoms examined by Menon are mostly similar to the symptoms of the ANRSD and the ANSSV[16,30]. Nayar & Seliskar reported that the necrotic streaks in the leaf spindle were the initial symptoms of YLD, running parallel from the frond tip towards the frond base, although sometimes the initial effects may also have be shown by the leaf whorls of different stages i.e., young, middle, or older whorls[26]. However, the described necrotic streaks also seem to be caused by co-infection with fungal pathogens. The YLD symptoms were first reported in Hainan, China, in 1986. According to previous studies, the yellowing starts from old leaves in the lowermost position and extends upward towards younger leaves. No known causal agent and no evident infection center were found; therefore, the disease was declared to be caused by potassium deficiency[24].
Although the YLD symptoms reported by different authors are various due to co-infection by other pathogens or impact by environmental factors, the typical YLD symptoms have been summarized[1,17,18]: The yellowing initially starts from leaflet tips of lowermost leaves or the mid-crown (Fig. 1a). The diseased leaves were observed with the apparent demarcation of infected yellow and normal green areas. Chlorosis (yellowing) expands in the direction of vascular tissues while the midribs remain green, resulting in the formation of yellow-green borders (Fig. 1b) that distinguished it from physiological yellowing (Fig. 1c). Later, the border on older leaves becomes indistinct, and the chlorosis extends to the lower and younger leaves. In advanced stages, new stunted leaves emerge, crown size decreased markedly, and narrowing of the crown width was observed permanently, resulting in 'bunchy top' symptom[1,17,18]. Later, as much as the disease advanced stage arrives, roots become rotted, the size of the nuts are reduced, the stem becomes spongy and friable, and finally breaks up at the top[17,18,23]. Jin et al. have classified YLD into two types: 'bunchy top type' and 'yellowing type'[31]. To our opinion, these two types are not separate diseases, but are the different stages of the YLD. However, it is unclear whether the YLD symptoms mentioned above are caused by different pathogens or one pathogen.

Figure 1.
The typical YLD symptoms on areca palm. (a) YLD symptoms initially starts from leaflet tips of lowermost leaves and the mid-crown; (b) chlorosis (yellowing) expands in the direction of vascular tissues while the midribs remain green, forming yellow-green borders; (c) yellowing symptoms due to nutrition deficiencies or rotten root.
YLD has serious deleterious effects on areca palm yield production, worldwide. Approximately, preliminary estimates put the plantation area of areca palm seriously damaged by YLD in Hainan province of China at over 80,000 ha (40%). In India, area of areca palm garden damaged by YLD had reached 10,000 ha in the 1970s[23], while in Sri Lanka 11,968 ha of areca palm plantations have been affected by YLD[13]. Due to the severity of the attack, the YLD decreased the yield of areca palm up to 50% worldwide.
-
Numerous yellow-type diseases of plants have been associated with phloem colonized phytoplasma. It is a fastidious, pleomorphic cell wall-less bacteria, transmitted through hemipteran (sap-sucking) insect vectors among plants and infects a variety of different quality foods, fodder, timber crops, and fibers hence causing huge crop losses[32−34]. In 1994, the term 'phytoplasma' was adopted by the Phytoplasma working team at the 10th Congress of the International Organization of Mycoplasmology to collectively denote this taxon of plant pathogens[33]. The genus 'Candidatus Phytoplasma' was proposed by IRPCM Phytoplasma/Spiroplasma working. Phytoplasmas have sizes variable from 200 to 800 nm, they are pleomorphic because of the lack of cell wall. Phytoplasmas couldn't be cultured in vitro, the association of phytoplasma with plant diseases was depended on indirect biological proof, such as electron microscopy observation, symptoms elimination after tetracycline treatments, insect and dodder transmission[33,35]. Nayar & Seliskar[26] first reported that phytoplasma were closely associated with YLD based electron microscopy, and later the etiology was proved by other researchers[12,28,36]. The phytoplasma could be found in the transverse section of the midribs of pinnae in infected areca palm leaves that appeared separately or in clusters, stick to the inner surface of sieve tubes of infected palms, while absent in healthy trees[13]. Phytoplasma was purified from YLD samples through percoll density gradient centrifugation[37], the polyclonal antibodies were prepared and then the direct antigen coated-enzyme linked immunosorbent assay (DAC-ELISA) was available for pathogen detection in infected tissues[38,39]. SYBR green-based RT-PCR, loop-mediated isothermal amplification (LAMP) and RT-LAMP techniques have been established for the purpose to detect the areca palm YLD phytoplasma using YLD phytoplasma sequence ((JN967905.1, JN967906.1, JN967904.1) and Coconut root wilt phytoplasma sequence (FJ794816.1, JX273772) as template for picking primers[34,40].
Phytoplasmas with highly similar 16S rRNA gene sequence could be associated with different symptoms in the same or in different plant host(s), and different phytoplasmas might show similar symptoms (Bertaccini et al. 2014)[41]. To classify the phytoplasmas more objectively, multilocus sequence typing (MLST) have been applied to phytoplasma identification[42,43]. YLD phytoplasma collected from India had shown 99 % sequences similarity of 16S rRNA genes to sugar cane white leaf phytoplasma and Napier grass stunt phytoplasma, the phylogenetic analysis clustered the YLD phytoplasma with members of 16SrXI, e.g. Bermuda grass white leaf and Rice yellow dwarf groups. Based on secA gene-based phylogenetic analysis, this phytoplasma was assumed as a member of 16SrXI-B subgroup[12,44]. While phytoplasma detected in YLD of areca palm in China was grouped in 16SrI-G and 16SrI-B subgroup, significantly different from the India YLD phytoplasma (Che et al., 2010[45]. In Sri Lanka, areca YLD is co-occurrent with Weligama coconut leaf wilt disease (WCLWD). The YLD phytoplasma shared 100% nucleotide identity of secA gene sequences with the WCLWD phytoplasma and 99% similarity with 'Candidatus Phytoplasma cynodontis' strain. The phytoplasma was identified as a member of 16SrXIV subgroup associated with both YLD and WCLWD[13]. Abeysinghe et al. proposed that the confused classification might be due to the relatively poor resolution of 16S rRNA sequence among phytoplasmas strains. Then they amplified ca. 1 kb fragment of the leucyl transfer RNA synthetase (leuS) gene along with the secA gene to classify these phytoplasmas. Based on this data, 16SrXI and 16SrXIV phytoplasmas from coconut and areca palm in southern India and Sri Lanka were grouped in the same 16SrXI[46].
The guidelines for naming a new phytoplasma was recently revised[47,48]. The newly revised guidelines enhanced the threshold of 16SrRNA gene identity from 97.5% to 98.65%; The length of 16S rRNA gene sequence was extended from > 1,200 bp to > 1,500 bp. Furthermore, the whole genome-based average nucleotide identity (ANI) criterium was also proposed[47,48]. So far, 47 phytoplasma genomes (35 draft and 12 complete) involving 13 groups and 29 subgroups have been reported. The genome lengths the phytoplasmas range from 576 to 960 Kb[48−50]. However, no genome sequences of YLD phytoplasmas have been provided to date, the taxonomy of YLD phytoplasma is facing a huge challenge.
Palmaceae family is infected by a variety of ailments associated with phytoplasmas throughout the world, such as coconut yellow decline, coconut lethal yellowing, oil palm stunting disease, and WCLWD[51,52]. However, the transmission and validation by Koch's postulates of these diseases have not been well documented. Citrus Huanglongbing (HLB) is the most documented disease caused by wall-less Candidatus bacteria which induced devastating economic losses worldwide[53]. Candidatus Liberibacter asiaticus (Las) and C. Liberibacter africanus (Laf) are associated with Asian HLB and Africa HLB, respectively. The Asian citrus psyllid (Diaphorina citri) was identified as the insect vector of HLB[54]. Las has a 1.23-Mb genome which does not contain many necessary genes encoding the essential housekeeping pathways and metabolic pathways[55], revealing the mystery why it is difficult to cultivate in vitro. Like phytoplasma, Liberibacters are unculturable in vitro, but a modified Koch's postulate has been fulfilled to establish the status as the causal agents of HLB[56]. In contrast, the key etiological evidences supporting phytoplasmas as the causative agents of YLD are still lacking, although a lot of reports have shown the association. Proutista moesta (Westwood) was proposed to be the putative transmission vector of phytoplasma to areca palm[57], but the transmission of phytoplasma to areca palm has not been proved. Many YLD samples collected in India and China were not positive for phytoplasma by RT-PCR and RNA-seq analysis[19,58], it is therefore reasonable to suspected whether phytoplasmas are the etiological agent of YLD or not. The status of causal agent of phytoplasma for YLD still requires elucidation.
-
Areca palm velarivirus 1 (APV1) was first identified in YLD sample collected in China through small RNAs sequencing[14]. APV1 was later detected through a modified RNA-seq method, in which the ribosomal RNA was removed from the total RNA and poly d(T) primer was replaced by random hexamer primers for reverse transcription, enabling the transcription of prokaryotic and viral RNA without poly (A) tail[18]. Noteworthy, both methods have not discovered any phytoplasmas related genes in the Hainan YLD samples tested. The association of APV1 with areca palm YLD was determined by Digital Gene Expression (DGE) analysis and RT-PCR. APV1 was consistently detected in all YLD symptomatic samples from different locations of Hainan, China, by RT-PCR using APV1 specific primers. No sample was detected positive collected from non-epidemic areas, indicating the strong evidence for the association of APV1 with areca palm YLD[18]. Furthermore, the symptoms of YLD of areca palm show seasonal oscillation, which means that in winter or dry season when the temperature becomes low the YLD symptoms becomes more dominant and the whole plantations seem to be burnt, however, in rainy and warm seasons the disease symptoms becomes low or almost indiscernible. This phenomenon puts forward a possibility that areca YLD might be a physiological yellowing syndrome[24]. To reveal the mechanism underlying, Khan et al. analyzed the APV1 accumulation in the YLD samples in different seasons under natural condition and the samples under different temperatures in control condition through qRT-PCR and ELISA assays. APV1 titer were found to be higher in winter or under low temperature than in summer or under high temperature, revealed that the temperature is a key factor effecting the APV1 titer, which is closely associated with the YLD symptom severity[1]. This discovery strengthens the evidence of APV1 as the causal agent of YLD.
Closteroviridae constitutes of more than 30 plant viruses comprising four genera: Closterovirus, Ampelovirus, Crinivirus, and Velarivirus[59,60]. Viruses in this family have mono or bipartite positive-sense, single-stranded RNA genomes[59,61,62]. Based on comparison of the genome structure and phylogenetic analysis, APV1 was classified as a new member of the genus Velarivirus (family Closteroviridae)[15,48]. The genus Velarivirus derived from the Latin word velaris, which means belonging to a veil or curtain. The current members of the genus Velarivirus include areca palm velarivirus 1 (APV1, KR349464), grapevine leafroll- associated virus 7 (GLRaV-7, HE588185), little cherry virus 1 (LChV-1, Y10237), and cordyline virus 1 (CoV-1, HM588723), cordyline virus 2 (CoV-2, JQ599282), cordyline virus 3 (CoV-3, JQ599283), and cordyline virus 4 (CoV-4, JQ599284)[60,63−65].
APV1 has a typical flexuous and filamentous viral particle. APV1 genome encodes 11 open reading frames (ORFs)[18]. ORF 1a encodes a large protein with papain-like proteinase, methyltransferase and Helicase domains ORF 1b is predicted as an RNA-dependent RNA polymerase (RdRp) expressed by a frameshift of ORF 1a. ORF 2 encodes a 4 kDa hydrophobic protein. ORF 3 encodes a heat-shock protein 70 homolog (70 kDa), partially overlapping with ORF 4 which encodes a 21 kDa polypeptide. ORF5 encodes a 60 kDa protein. ORF 6 and ORF 7 encode the putative coat protein (CP) and the CP minor (CPm), respectively, whereas ORF 8, ORF 9 and ORF 10 encode three polypeptides with unknown functions. Phylogenetic analysis of complete genomes divided the 23 APV1 isolates into three phylogroups, of which phylogroup A is the most prevalent in Hainan. All APV1 isolates show high sequence conservation in seven ORFs (> 95% nt identity) at 3' terminal, whereas evident diversity (81%−87% nt identity) in three ORFs at 5' terminal[18,19].
Transmission vectors of APV1
-
The transmission of major plant viruses is utterly dependent on vectors. The arthropods, mostly Hemipterans are largely used by plant viruses as their transmission vectors[66]. The three genera of Closteroviruses are segregated based on the property of genome structures and the specificity of transmission vectors. Ampelovirus, Closterovirus and Crinivirus, are transmitted by mealybugs, aphids and whiteflies, respectively[59]. Velarivirus was ratified as a new genus of Closteroviridae in the 9th ICTV report, due to its distinct genome property and unknown insect vector[21,61,62,64,67]. Closteroviruses are classified based on phylogenetic analysis of conserved proteins, such as RNA-dependent RNA polymerase (RdRp), helicase (HEL), and HSP70, a tangible link has been founded among these conserved Closteroviruses proteins and the type of insect vectors responsible for their transmission[59,61]. Viruses belonging to the genus Velarivirus and genus Crinivirus have a close genetic relationship but due to their monopartite genomes and the lack of a known insect vector, the inclusion of genus Velarivirus into Crinivirus is not possible[60,65]. Our recent report first demonstrates that mealybugs are transmission vectors of Velarivirus. Both Pseudococcus cryptus and Ferrisia virgata mealybugs transmitted APV1 to areca palm seedlings and caused typical YLD symptoms (Fig. 2). On the one hand, the results provide a key etiological evidence of APV1 as the causal agent of YLD, on the other hand, an important question was raised why Velarivirus APV1 has closer genetic relationship with Crinivirus but has similar transmission vectors of Ampelovirus? that should be addressed in future research.
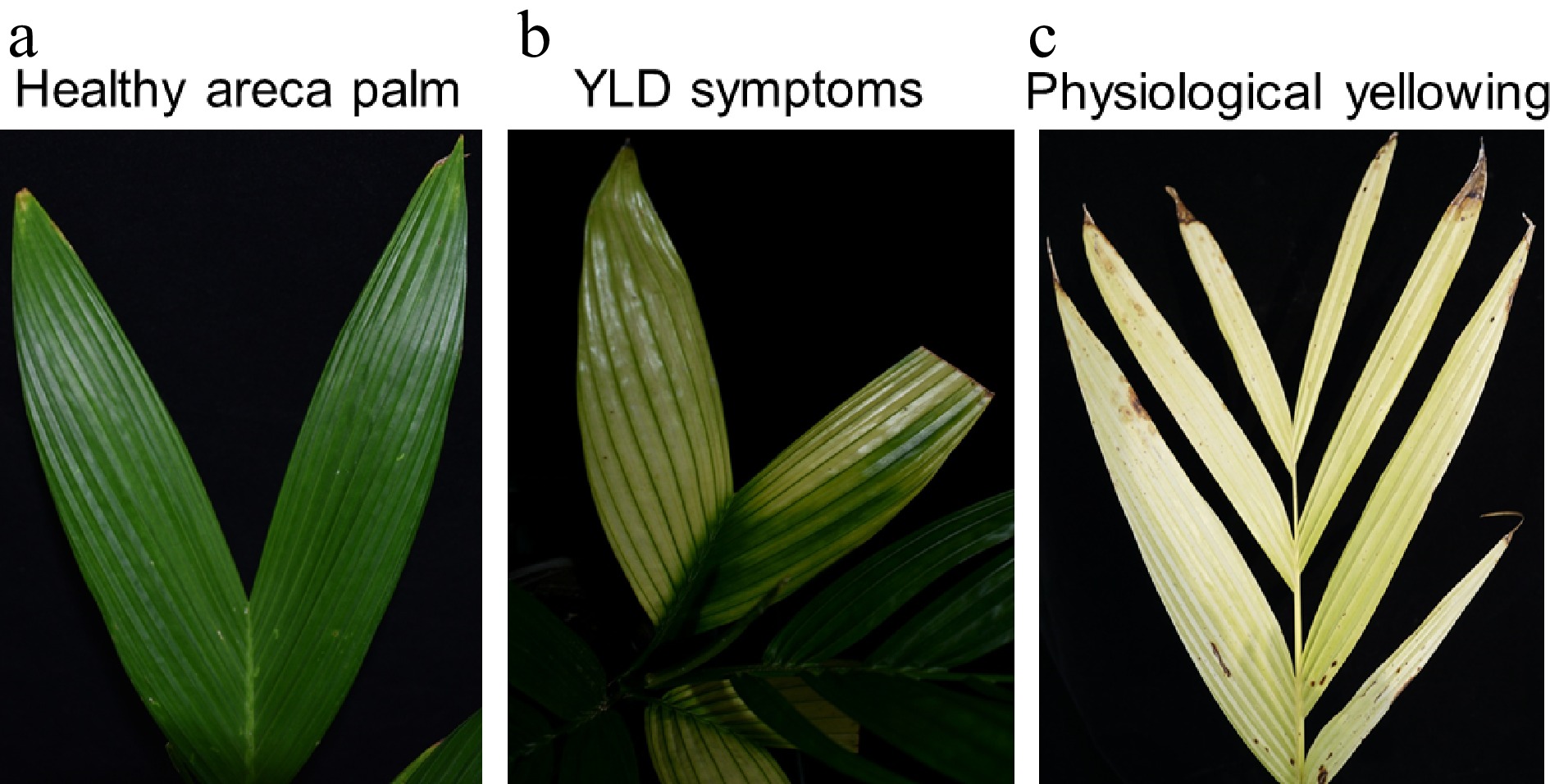
Figure 2.
APV1 transmitted by Ferrisia virgata mealybugs causes YLD symptoms on areca palm. (a) Healthy areca palm leaf, (b) YLD symptoms, (c) physiological yellowing.
The transmission of plant virus by Hemipteran vectors are defined based on many parameters, including acquisition access period (AAP), inoculation access period (IAP), retention time, trans-stadial passage, latent period, replication, and trans-ovarial. Transmission modes were separated into four types: non-persistent, semi-persistent, persistent-propagative and persistent-circulative[66]. Zhang et al.[17] revealed that the first instar mealybugs of Ferrisia virgata showed higher rate of transmission of APV1 than adult mealybugs. APV1 was detected in the stylet, foregut, midgut, and hindgut by immunofluorescence assays and immunocapture RT-PCR. APV1 was not transovarially transmitted. In sum, the transmission of APV1 by F. virgata occurs in a non-cyclic, semi-persistent manner[17]. The transmission mode of APV1 by P. cryptus is so far unknown.
Although the present work provides strong evidence of an association of APV1 with YLD, key etiological evidence, i.e. Koch's postulate, is needed to establish the causative status. Koch's postulates for viral pathogens are usually fulfilled by a modified way, e.g. construction and transformation of virus infectious clone into host plant[68,69]. The infectious clone of APV1 has constructed and successfully transformed into model plant N. benthamiana, but the attempts of either transformation of infectious clone or mechanical inoculation of APV1 virion into areca palms failed (data not shown). Mechanical transmission of a few Closteroviruses such as citrus tristeza virus (CTV), beet yellows virus (BYV), and grapevine leafroll-associated virus 2 (GLRaV-2) is available now[70], but no currently described velariviruses have been reported to be mechanically transmissible. Further, the genetic transformation of areca palm has not been reported. At the present stage, it seems feasible to fulfill the Koch's postulate through transmission of the virion generated by APV1 infectious clone in N. benthamiana into areca palms by the transmission vectors, although it is beset with difficulties.
Prevention of the APV1
-
Plant viruses cause severe declines in crop yields and even plant death worldwide. The preferred strategy for controlling plant viral diseases are resistance breeding, but conventional breeding is time-consuming and often difficult to succeed due to the lack of resistant germplasm. At present, genetic engineering techniques are the most effective way to control plant virus diseases[71]. In 1986, the first example of genetically engineered virus resistance (VR) was reported. The resistance to tobacco mosaic virus (TMV) was demonstrated by expression of the TMV coat protein gene in transgenic tobacco plants[72]. Since then, CP transgenic strategies have been widely applied to other plant viruses, such as Cucumber mosaic virus (CMV)[73], Potato virus Y (PVY)[74], Tobacco leaf roll virus (PLRV)[75], and so on, which have achieved good prevention and control effects, and become the most widely used plant antiviral strategy. At present, CP transgenic crops, such as CP gene antiviral zucchini[76], potato[77], papaya[78], and plum tree[79], have been approved for commercial use. Additionally, the artificial small RNAs (sRNAs), such as synthetic trans-acting small interfering RNAs (syn-tasiRNAs) and artificial microRNAs (amiRNAs), have been successfully used to confer antiviral resistance in plants[80−83]. Recently, CRISPR/Cas9 system has been proved to be used for plant virus disease prevention and control[84−86]. However, the genetic engineering techniques mentioned above are highly dependent on efficient genetic transformation which is still unavailable for areca palm.
In the areca palm YLD management program, although, there is no cutting-edge treatment of YLD to date, but one of the most important things is the timely detection of the disease. So far, various molecular and nano-technological techniques have been established for the timely detection of the disease. These techniques fulfil some important gaps and limitations in disease prevention and management program strategies. Additionally, management of YLD could be performed through control of the transmission vectors by applying variety of insecticidal medicinal spray, breeding and cultivation of resistant varieties, replacement of YLD plants with healthy plants, and plantation of the healthy seedlings away from the diseased pandemic areas. Noteworthy, the virus-induced gene silencing (VIGS) antiviral technology developed using latent viruses as vectors has achieved remarkable effects[87]. The VIGS vaccine vector constructed with apple latent spherical virus (ALSV) was effective in preventing and controlling three Tospoviruses after transient transformation in N. benthamiana and eustoma plants[87]. The same strategy could be applied for YLD control. Latent virus or mild isolate of viral pathogen infecting areca palm should be first identified and developed as VIGS vector and eventually as VIGS-based vaccines against APV1.
-
At present, YLD has greatly impacted the plantation of areca palm, but the causative pathogen of the disease is still disputed. Although a close association between the phytoplasma and YLD has been established, the causation of YLD by phytoplasma requires some key etiological evidences, e.g. transmission of phytoplasma into areca palm causing YLD symptoms by the vectors under controlled condition. The specific symptoms on areca palm caused by different subgroups of phytoplasmas require characterization. The draft genome sequence or even the complete genome sequence of different subgroups of phytoplasmas should have been determined. Nevertheless, the possibility of phytoplasma as causation of YLD has not been ruled out so far. The present work has provided strong evidence of an association of APV1 with YLD, Koch's postulate is needed to establish the causation. Then, a new question emerges, what are the difference between the symptoms caused by phytoplasmas, and by APV1? Only when this question was definitely answered, the management strategies applied for the control of YLD could be discussed.
The authors are thankful to China Scholarship Council (CSC), for giving us the opportunity to study in China. This work was supported by the earmarked fund for Hainan Agriculture Research System (Grant No:HNARS-1-G4) and the Project of Sanya Yazhou Bay Science and Technology City (Grant No:SCKJ-JYRC-2022-71).
-
The authors declare that they have no conflict of interest.
-
Received 4 April 2023; Accepted 9 June 2023; Published online 28 June 2023
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press on behalf of Hainan University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Khan LU, Zhao R, Wang H, Huang X. 2023. Recent advances of the causal agent of yellow leaf disease (YLD) on areca palm (Areca catechu L.). Tropical Plants 2:7 doi: 10.48130/TP-2023-0007
Recent advances of the causal agent of yellow leaf disease (YLD) on areca palm (Areca catechu L.)
- Received: 04 April 2023
- Accepted: 09 June 2023
- Published online: 28 June 2023
Abstract: Areca palm (Areca catechu L.), commonly known as betel palm, is a versatile perennial evergreen, a valuable economic crop in Southeast Asia. Its cultivation is seriously threatened by a destructive disease known as yellow leaf disease (YLD). Since the 1960s, the causal agent of the YLD was proposed to be phytoplasma, a pleomorphic wall-less, in vitro unculturable bacteria, which was supported by many reports from India, Sri Lanka, and China, but convincing etiological evidence is still lacking. Recently, the YLD was reported to be associated with a newly emerged novel areca palm velarivirus 1 (APV1) that belongs to the genus Velarivirus of the family Closteroviridae. Furthermore, APV1 could be transmitted by both Pseudococcus cryptus and Ferrisia virgata mealybugs to areca palm seedlings and caused typical YLD symptoms. A growing body of evidence suggests that APV1 is the causal agent of YLD, although Koch’s postulate has not been fulfilled. The conundrum of the causal agent of YLD is not yet solved. This review aims to compile and critically review all the published data concerning YLD as per our knowledge.
-
Key words:
- Areca palm /
- Yellow leaf disease /
- Phytoplasma /
- Areca palm velarivirus 1 /
- Closteroviridae.












