-
Tropical crops mainly include industrial raw materials such as Hevea brasiliensis, Manihot esculenta Crantz and Elaeis guineensis Jacq., tropical fruits such as Passiflora caerulea L., Litchi chinensis Sonn., Mangifera indica L. and Saccharum officinarum L. and spice drinks such as Coffea arabica L., Cinnamomum cassia (L.) and Illiciumverum Hook. f., which are important strategic resources and daily consumer goods. At present, remarkable achievements have been made in the development of the tropical crop industry. Advantageous production areas for major crops have taken shape and relevant industrial systems have been continuously improved, playing an important role in ensuring national defense and economic security, meeting people's living needs, developing non-food biomass energy and increasing farmers' income.
In recent years, the genomes of a number of tropical species have been uncovered, for example, Passiflora caerulea L.[1], Canna indica 'Edulis' and Canna indica L.[2], Litchi chinensis Sonn.[3], Cocos nucifera L.[4], etc. Genome and related research results provide valuable resources for the breeding and improvement of many tropical species. At the same time, with the output of a large number of multi-omics data, specific databases for different families have been developed, such as SapBase and ArecaceaeMDB[5,6]. At present, there is a shortage of databases for tropical crops. TropGeneDB[7] is currently the only database related to tropical crops; however, it lacks important crops such as Manihot esculenta Crantz, Ananas comosus, and Mangifera indica L.. There is also a shortage of analytical and visual tools for breeders to utilize.
-
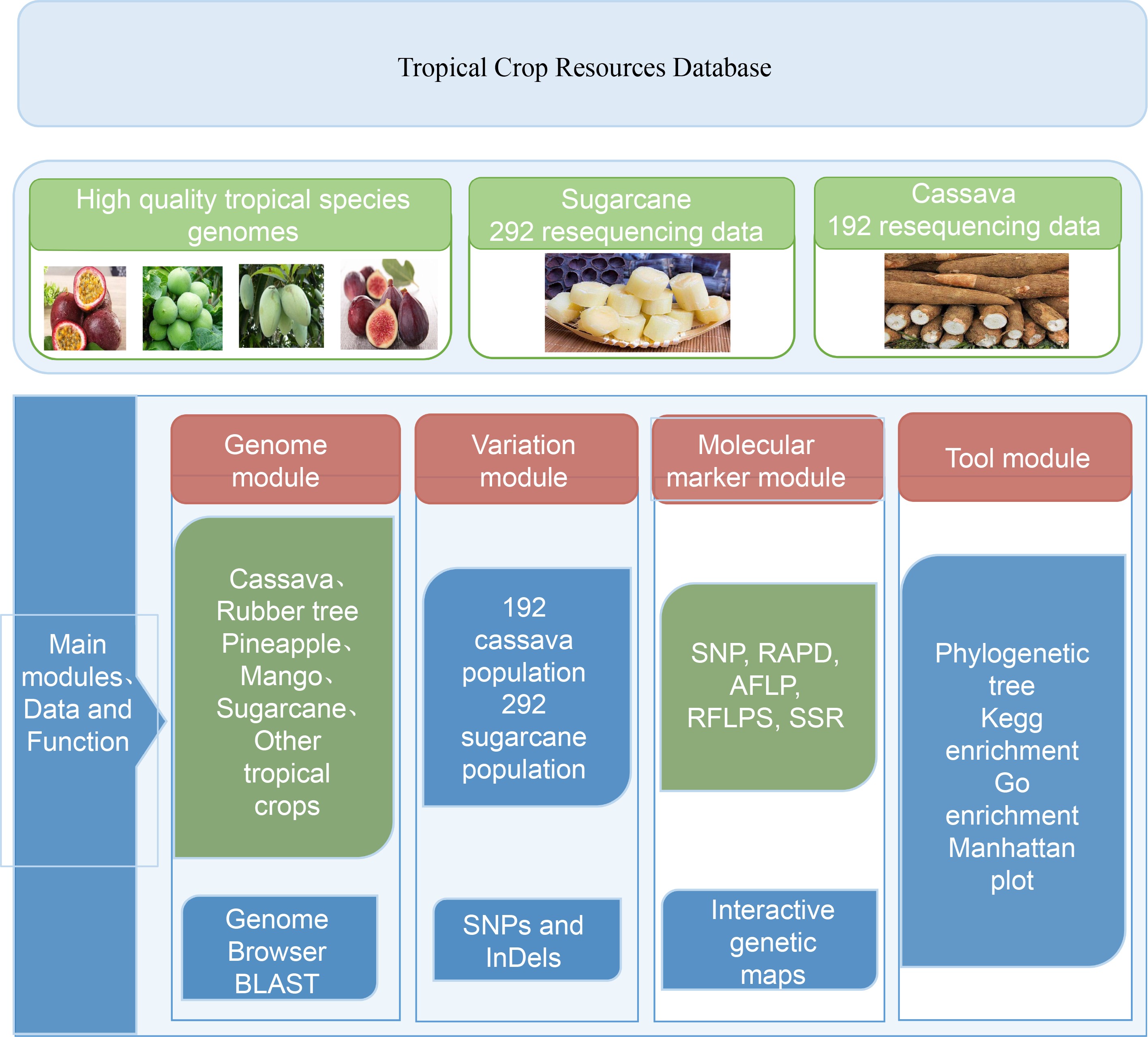
A total of 192 Manihot esculenta Crantz population resequencing data and 292 Saccharum officinarum L. population resequencing data were collected, and a number of high quality tropical species genomes and annotated data were collected from public databases as the basis for the establishment of this database.
Methods
-
The softwore Linux MongoDB Nginx and PHP development environment HTML5 CSS JavaScript JBrowse 1.16.11 SequenceServer R language R packages such as ggplot2, ggtree and treeio.
The classic Linux, MongoDB, Nginx and PHP development environments are deployed on the Ubuntu 20.04 operating system. The MongoDB database management system manages and stores all variation data (SNPs and InDels) and genotype data. The web front-end uses HTML5, CSS and JavaScript. HTML5 is mainly used for the layout of the page, CSS is used to control the style of the layout of the page, and JavaScript is responsible for implementing some interactive functions. Visualize the genome using JBrowse 1.16.11. SequenceServer is used for BLAST comparison. The rest of the data presentation and visualization is done in R language. It has been tested in browsers such as Firefox, Google Chrome and Internet Explorer.
-
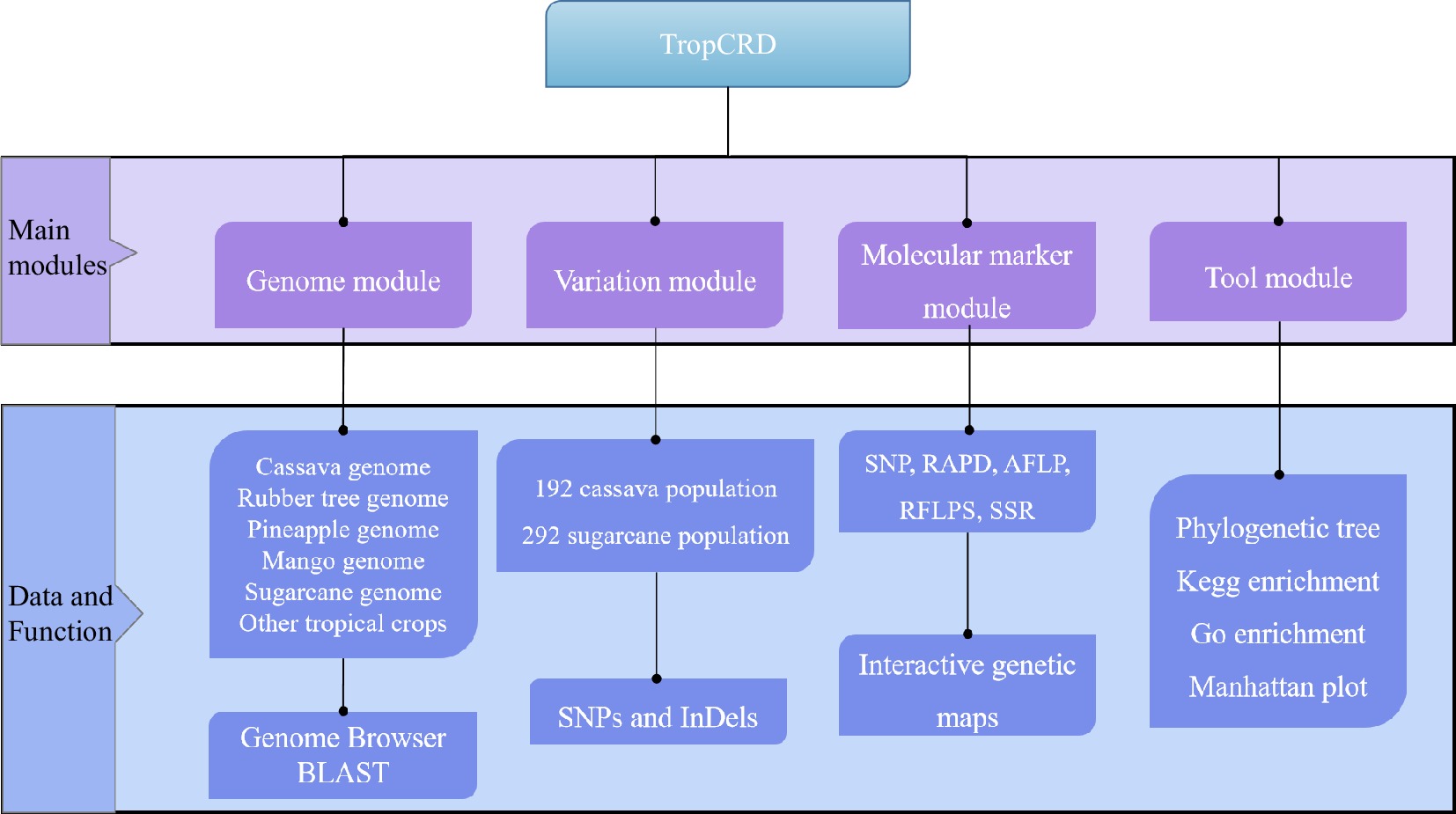
We have created TropCRD to provide data and technical support for tropical crop breeding. TropCRD was created to store genetic, molecular and phenotypic data on tropical crop species, with the variation information, molecular markers, quantitative trait loci, genetic maps, genetic diversity and phenotypic diversity studies data. At present, the database has included a total of five tropical species genomes, 12,850 QTL markers, 484 germplasm resources of tropical species, 75,396 SNPs and InDels, and 220,090 genes annotated. The genetic linkage map of the three species was constructed (Table 1).
Table 1. Overview of genetic, molecular and phenotypic data in TropCRD.
Species QTLs Germplasms SNPs Genes Genetic maps Manihot esculenta Crantz 4,501 192 30,427 33,030 1 Saccharum officinarum L. — 292 44,969 112,787 — Ananas comosus — — — 31,585 — Mangifera indica L. 1,409 — — — 1 Hevea brasiliensis 6,940 — — 42,688 1 The TropCRD is based on tropical crops and committed to building a platform serving tropical crop breeding, displaying various types of data from multiple dimensions, providing a more comprehensive display platform for data display of omics research, and providing a variety of new ideas and strategies for related in-depth research (Fig. 1).
Database software
-
TropCRD uses the classic Linux, MongoDB, Nginx and PHP development environment. It is deployed on the Ubuntu 20.04 operating system. All variation data (SNPs and InDels) and genotype data in the database are managed and stored by the MongoDB database management system. Its web front end uses HTML5, CSS and JavaScript. HTML5 is mainly used for page layout, CSS is used to control the style of page layout, and JavaScript is responsible for the implementation of some interactive functions. We used JBrowse 1.16.11 to visualize the genome. SequenceServer is used for BLAST comparison. The Echarts visualization library was used to present the genetic map, while the rest of the data presentation and visualization were handled by R language, which has been tested in browsers including Firefox, Google Chrome and Internet Explorer.
Genome module
-
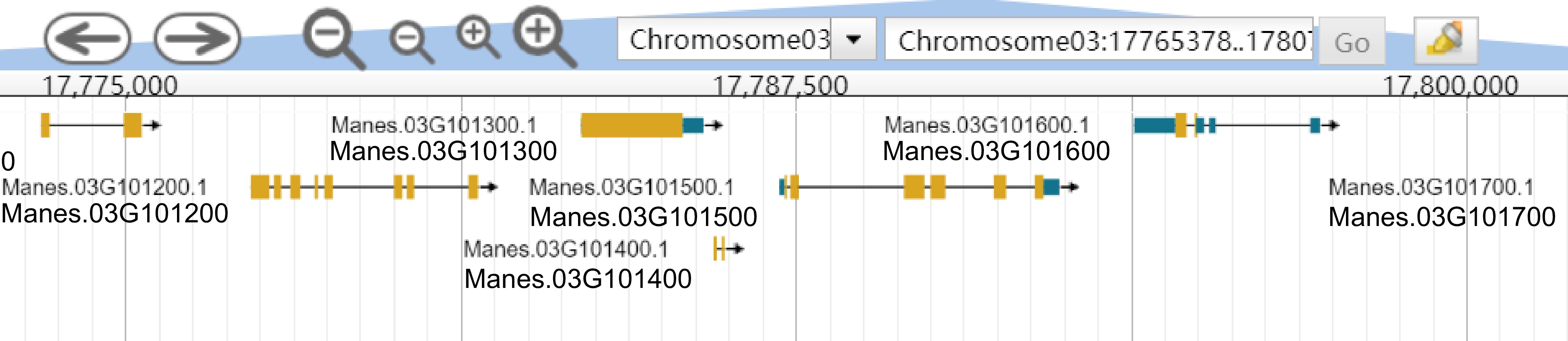
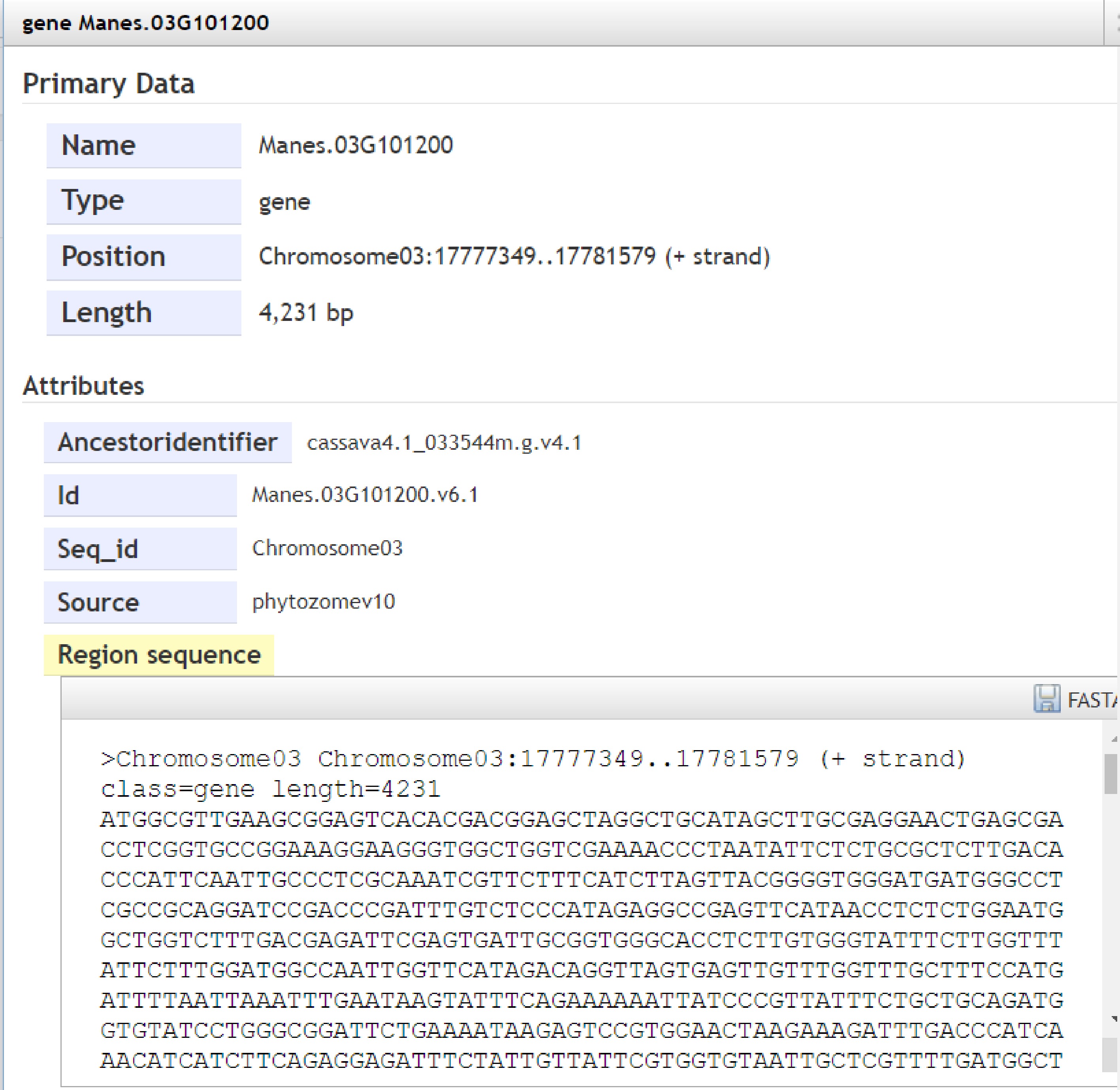
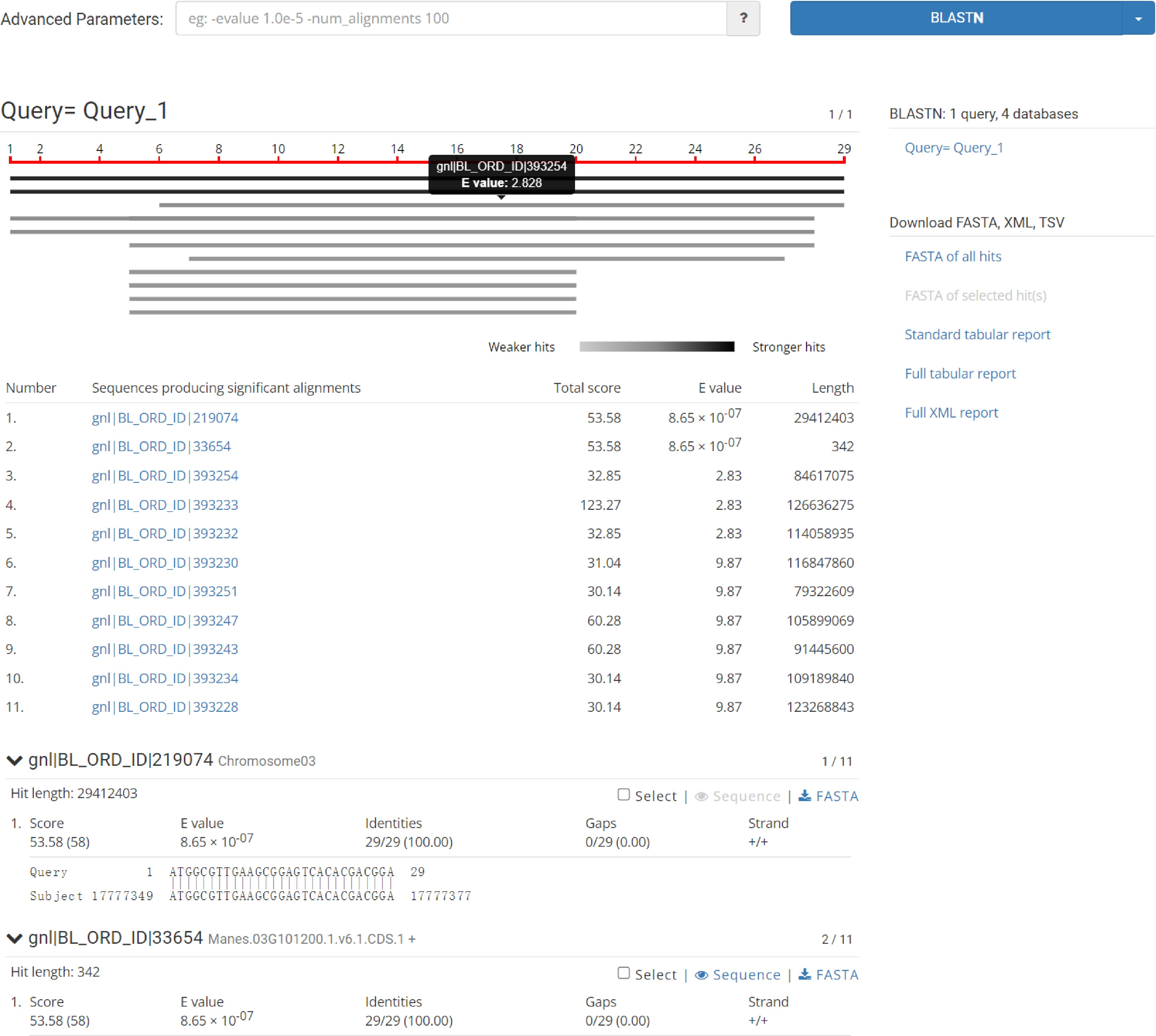
The Genome module provides users with the genomes of tropical species such as Manihot esculenta Crantz, Hevea brasiliensis, Saccharum officinarum L., Ananas comosus, Mangifera indica L., Jatropha curcas L. and Pennisetum sinese Roxb. The module adopts JBrowse software. Users can view genome sequence, gene, transcript sequence information and view annotation files, information about variation data, and complete visualizations in the Genome browser. For example, the user selects chromosome 3: 17,775,000 ~ 17,785,000 bp to browse the genome region (Fig. 2) and clicks 'Manes.0G101200' to display the type, location, length, and specific sequence information of the gene (Fig. 3). Support sequence alignment, can select multiple databases for comparison and analysis at the same time, compare the homology information between the output sequence, help judge the source of the input sequence or the evolutionary relationship with the known sequence (Fig. 4).
Molecular marker module
-
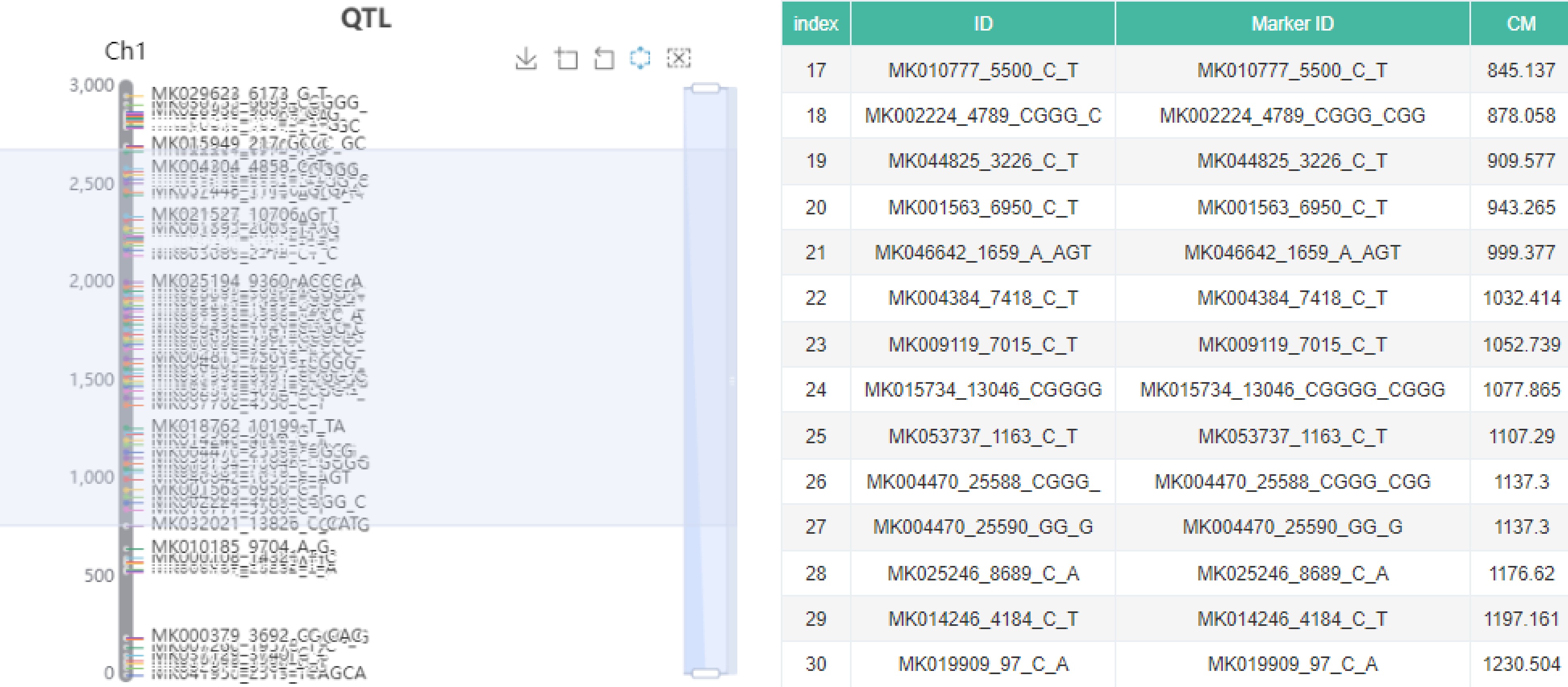
The molecular marker module included 37,852 markers of Manihot esculenta Crantz, Hevea brasiliensis, Ananas comosus and Mangifera indica L., including SNP, SSR, EST SSR, SCAR, CAPS, ISSR, RAPD, AFLP and RFLPS. Based on published articles[8−17], interactive genetic maps are constructed and markers are arranged on each chrome. Users can select the genetic map region they are interested in according to their own needs, obtain the marker ID, genetic map location, genome location and other information of all markers in the region (Fig. 5).
Variation module
-
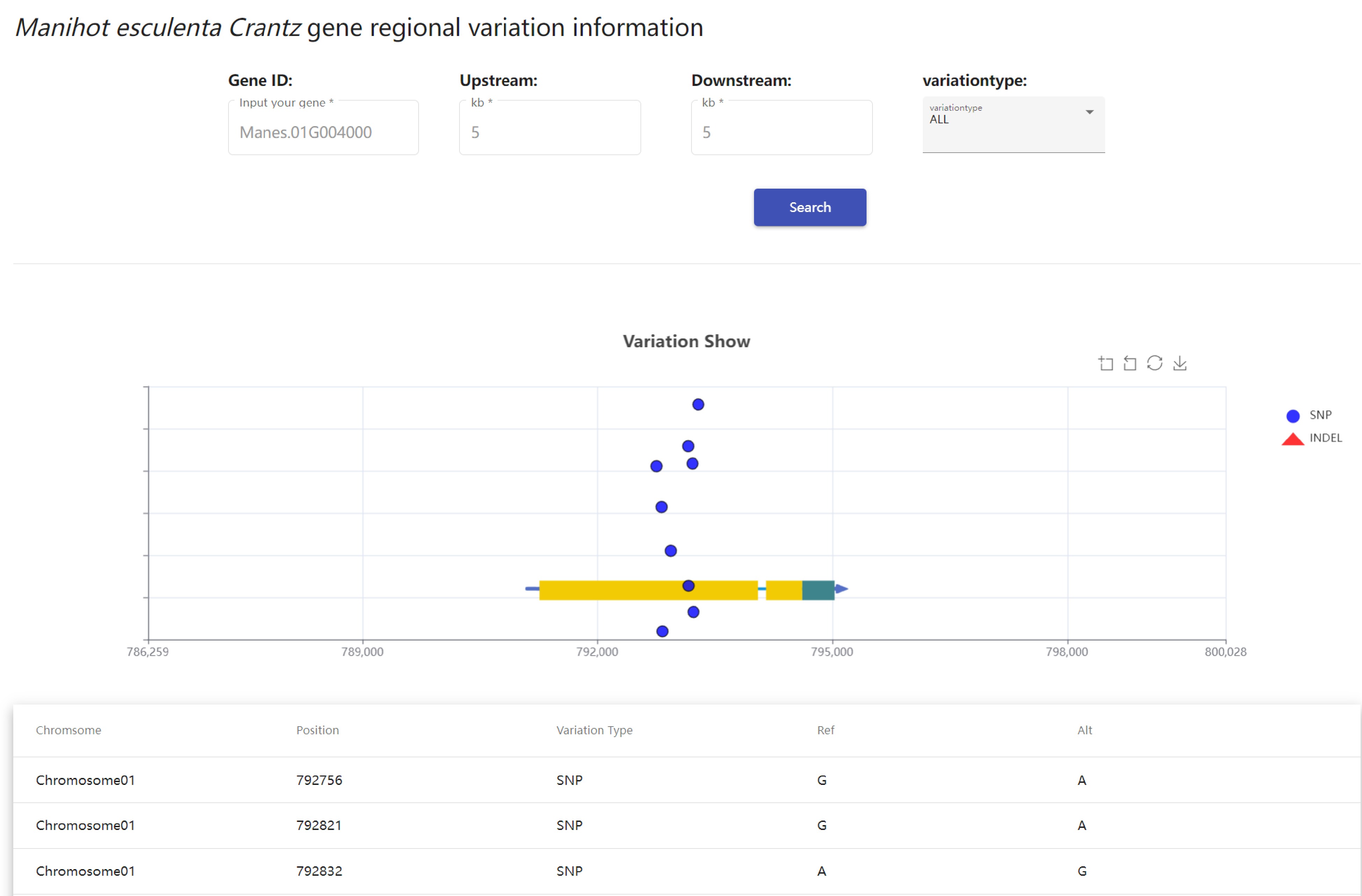
At present, in the tropical fruit genome database variation module, resequencing variation data of multiple species populations are included, and users can choose the species they are interested in. This module has realized the retrieval function of Manihot esculenta Crantz and Saccharum officinarum L., including 192 Manihot esculenta Crantz materials and 292 Saccharum officinarum L. samples with resequencing data. A total of 75,396 SNP and InDel sites were obtained through the processing of resequencing data. Among them,Saccharum officinarum L. had 44,969 sites and Manihot esculenta Crantz had 30,427 sites. Users can retrieve SNPs sites by gene ID, input gene ID, select upstream and downstream ranges and variation types, and variation information will be displayed in the form of charts (Fig. 6).
Tools module
-
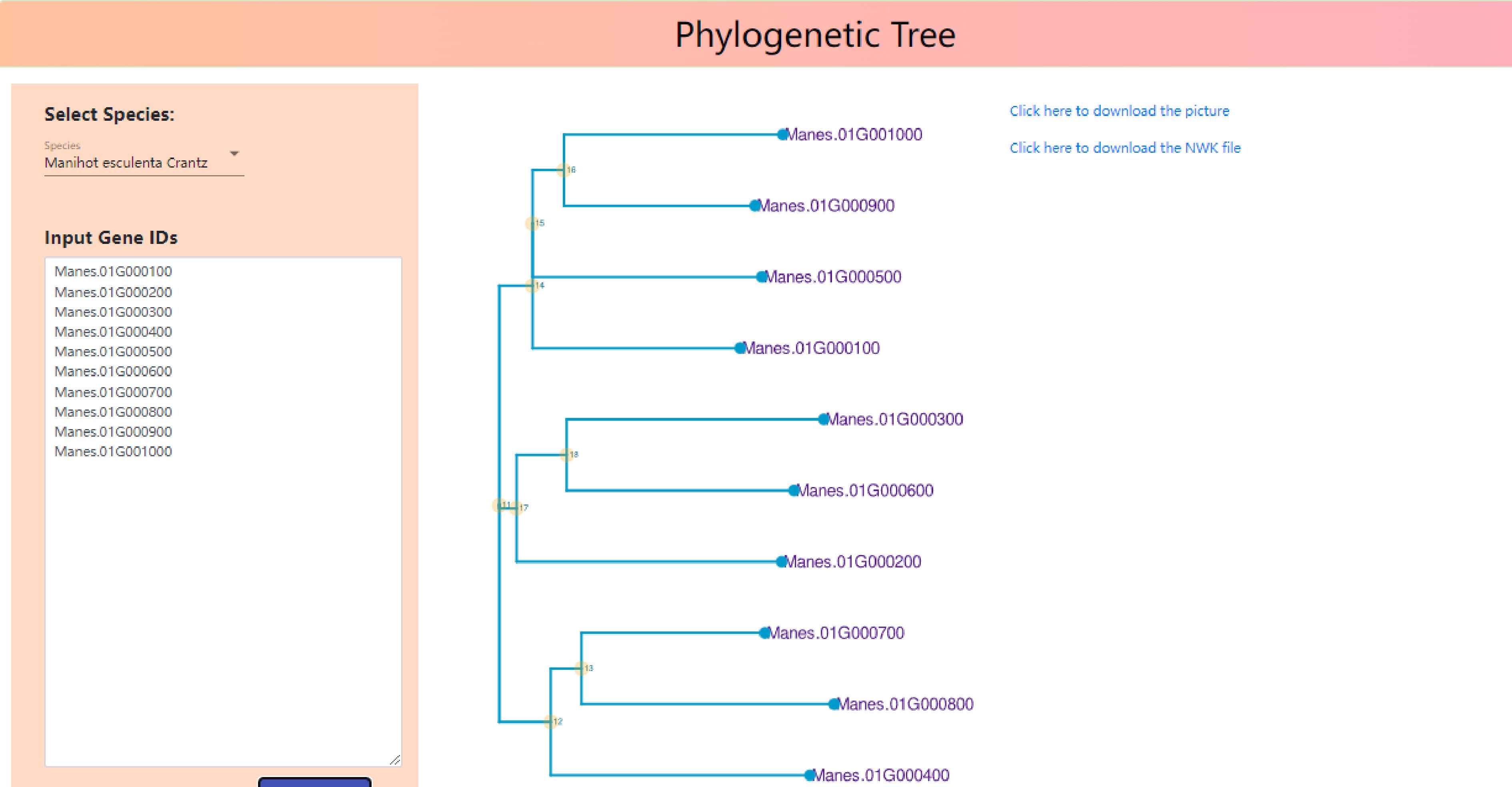
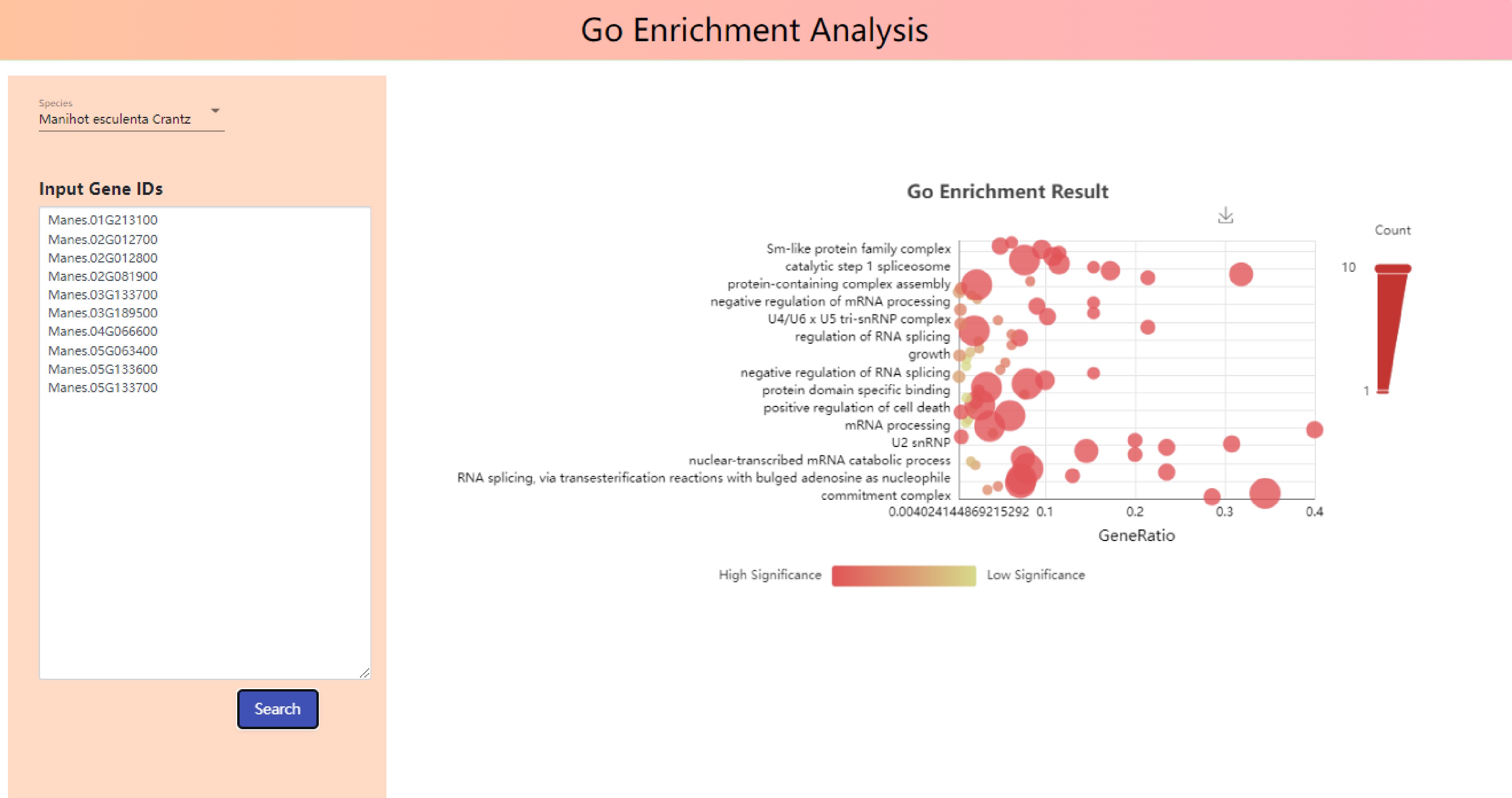
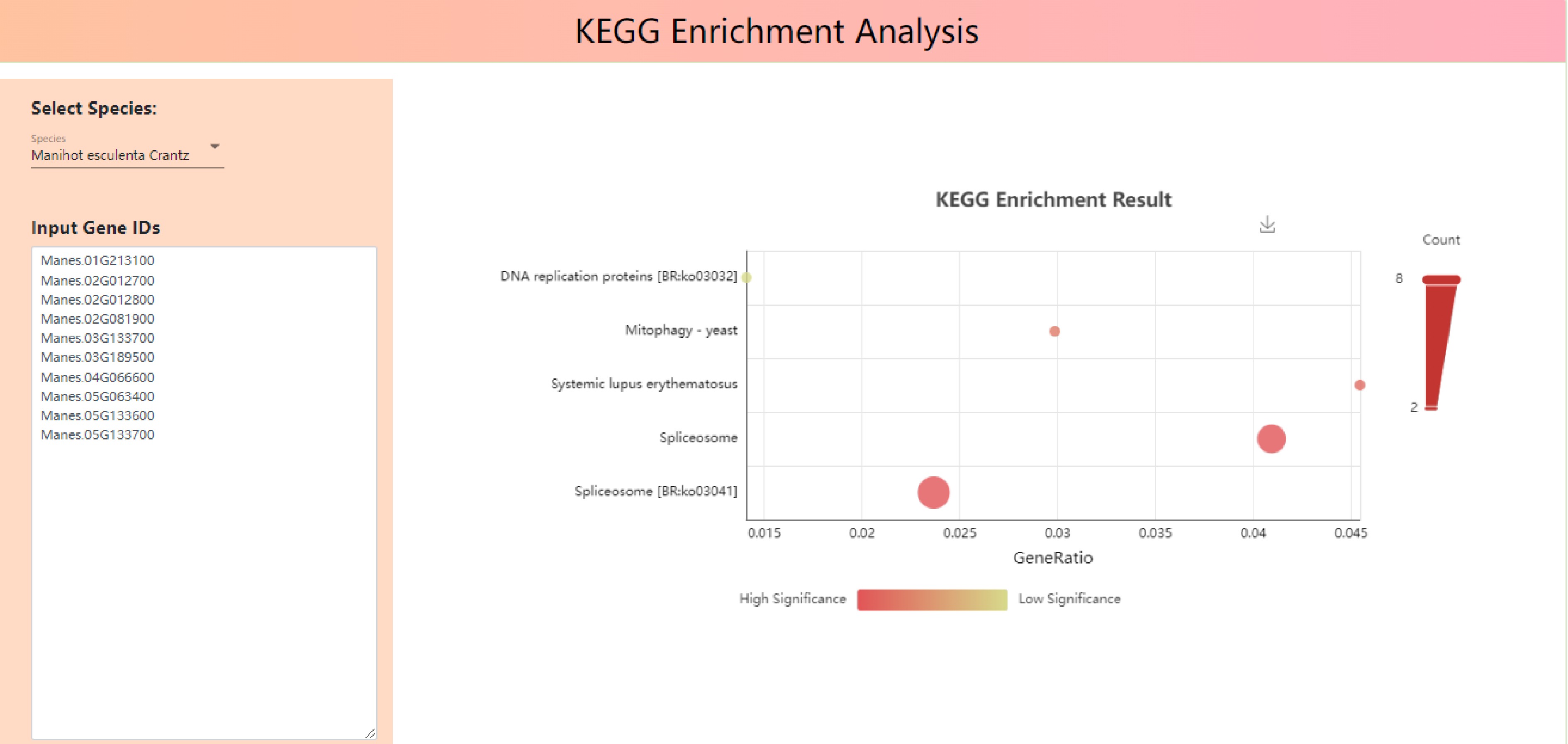
The tools module includes phylogenetic tree drawing, GO function enrichment analysis, Kegg enrichment analysis, and Manhattan map. The tools are all drawn in R language on the server background. R packages such as ggplot2, ggtree, treeio were used to complete data processing and image rendering in the background, providing users with the most reliable recognition results. Users click the corresponding function in 'Tools', select the species, enter the ID of the gene to be mapped in the blank below the species, and click the 'Search' button to obtain the functional relationship diagram of related genes and the phylogenetic relationship between species genes.
As shown in Fig. 7, Input Gene IDs into input gene IDs, 'Manes.01G000100', 'Manes.01G000200', 'Manes.01G000300', 'Manes.01G000400', 'Manes.01G000500', 'Manes.01G000600', 'Manes.01G000700', 'Manes.01G000800', 'Manes.01G000900', 'Manes.01G0001000', phylogenetic tree, GO functional enrichment bubble map and Kegg metabolic pathway enrichment bubble map were drawn (Fig. 7).
It can be seen that in the phylogenetic tree, Manes.01G0001000 and Manes.01G000900, Manes.01G000300 and Manes.01G000600, Manes.01G000700 and Manes.01G000800 have close homology (Fig. 8).
The top ten enrichment functions in the GO functional enrichment bubble chart are commitment complex, spliceosomal complex assembly, nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay, nuclear-transcribed mRNA catabolic process, exonucleolytic, RNA splicing, via transesterification reactions, RNA splicing, via transesterification reactions with bulged adenosine as nucleophile, regulation of alternative mRNA splicing, via spliceosome, spliceosomal snRNP assembly, mRNA splicing, via spliceosome and P-body.
There are three pathways enriched in the bubble diagram of Kegg metabolic pathway, namely Spliceosome [BR:ko03041], Spliceosome, Systemic lupus erythematosus and Mitophagy - yeast (Fig. 9).
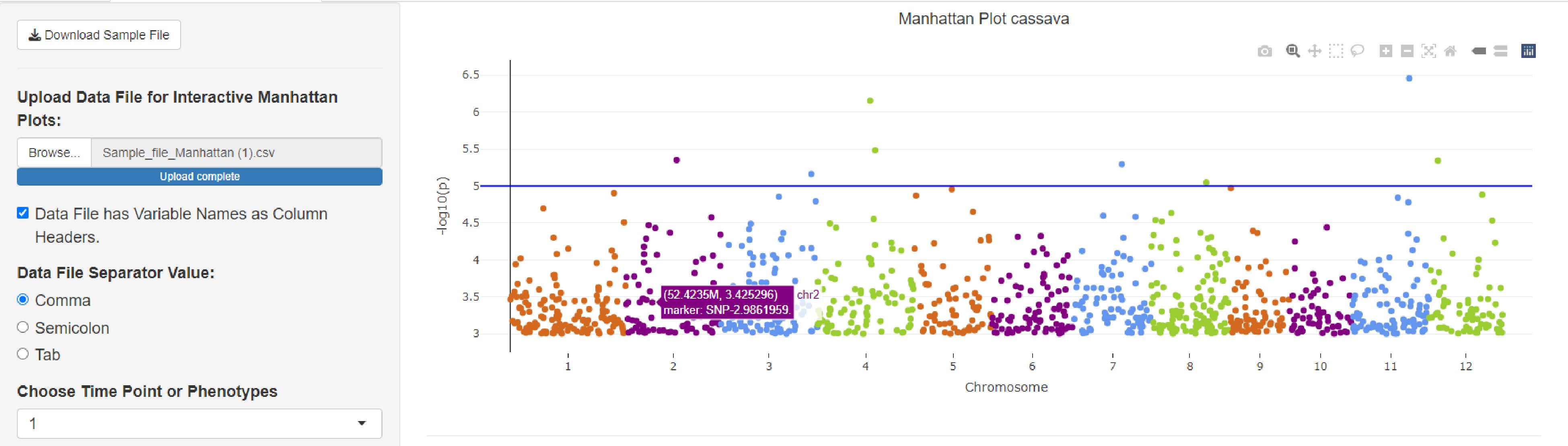
The Manhattan map is drawn using ShinyAIM[18] software, which can download the character data provided by ourselves or upload the file to generate an interactive Manhattan map. Users can click 'GWAS' in 'Tools' to enter the Manhattan map drawing interface, download the Manhattan map summary table of GWAS analysis results of tropical fruit traits provided in the database, or upload their own data in the specified format to complete the Manhattan map drawing. Click 'Browse' to upload the data to be analyzed. When the upload is completed, 'the upload complete' will be displayed. Users can check whether the table header is included or not.
After uploading, you can select the traits for analysis and set an appropriate -log 10 p-value threshold for filtering based on your requirements. The generated Manhattan map can be interactive. The mouse on each point will display the SNP information of that point and you can freely drag the selection area, zoom in and out to move and save the picture. As shown in the figure, the -log 10 p-value threshold was set to 5, and a total of 8 significantly related SNP sites were screened out, among which there were two significant sites on chromosome 4 (Fig. 10).
Data availability statement
-
The data used in this study are available from the corresponding author upon request
-
Currently, in the absence of a comprehensive breeding program, it takes a long time to breed varieties and new varieties with marketable traits. In the early stages of breeding, much time, space and resources are invested in selection and genetic progress after the initial crossing with the parent genotype. Many important agronomic traits of tropical crops are quantitative traits, which are controlled by a large number of microgenes. TropCRD promotes the breeding of tropical crops by constructing the genetic map of tropical crops and seeking QTL loci related to traits. On the other hand, genetic variation has been widely used in human diseases, identification of genetic loci related to important agronomic or economic traits in animals and plants, cloning of important functional genes, marker-assisted selection breeding, etc. TropCRD can be used in crop genetic breeding through the retrieval of genetic variation, including single nucleotide polymorphism (SNP) and small fragment insertion/deletion (InDel). TropCRD is important for the accurate identification of germplasm resources and the discovery of excellent alleles[19].
The Tropical Crop Resource Database is the first database built based on tropical crops. Different from other databases, it takes crops growing in tropical regions as research objects and covers the genomic data of crops in tropical regions around the world. It provides valuable resources for researchers to obtain and analyze genome sequence data, variation data and functional annotations of tropical crops. There are bioinformatics analysis tools, which provide molecular basis for the study of tropical crops. Secondly, the tropical crop resource database is the first database that combines Hyper-seq sequencing technology and builds tropical crop BLAST database based on Hyper-seq data.
In addition, the database still has a lot of room for improvement and expansion. The first is the integration of different types of data. At present, only genome sequence data, variation data and functional annotations are included in the database. However, other types of data, such as transcriptome data, epigenetic data, and metabolome data, can also provide valuable insights into the molecular mechanisms behind these important fruit traits. So integrating these different types of data into a tropical fruit DNA database could greatly improve their usefulness. Second is the standardization of data formats and annotations. There is a lack of standardization of the formats and annotations currently used in different tropical crop databases. This can make it difficult for researchers to compare data across different databases and can lead to inconsistencies in data analysis. Therefore, the establishment of standardized format and annotation of data can improve the interoperability of data and enhance the repeatability of research results. Finally, expanding the database to include more species could greatly improve their usefulness. The current database focuses on a few key species, such as Carica papaya L., Musa nana Lour., Ananas comosus and Mangifera indica L.. However, there are many other tropical crops that have not yet been fully characterized at the genomic level. Thus, expanding the database to include more species could provide researchers with new insights into the genetic basis of these important traits in fruits.
With the continuous development of sequencing technology, data in the field of tropical crops is also increasing. Therefore, multi-omics data are systematically integrated and analyzed by using a database as the carrier, and different data formats are visualized by using bioinformation tools, which is convenient for botanists and breeders to mine the genes of related agronomic traits and promote the development and utilization of excellent variety traits and speed up breed selection. It not only provides an important platform for the future genetic breeding of tropical crops, but also provides an important reference for other crops to integrate and use multi-omics data to promote the development of the breeding industry. In the future, the tropical crop database will be updated with additional data on tropical species, as well as more multidimensional omics data and further data analysis tools to provide important support for tropical crop breeding research.
This research was supported by Sanya Yazhou Bay Science and Technology City (SCKJ-JYRC-2022-65), Science and Technology special fund of Hainan Province (ZDYF2022XDNY149) and National Key R&D Program of China (2019YFD1000500).
-
The authors declare that they have no conflict of interest.
-
Received 29 May 2023; Accepted 5 June 2023; Published online 30 June 2023
-
The research materials are tropical crop varieties with high economic value and wide sources.
Use multiple tools and software to build a website framework.
It has multiple functions such as sequence alignment, molecular labeling and data visualization.
Provide a more comprehensive and accurate data base for relevant scientific research, and promote the in-depth development of tropical crop research.
-
# These authors contributed equally: Jianjia Xiao, Hai Liu, Yangyang Tian
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press on behalf of Hainan University. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Xiao J, Liu H, Tian Y, An P, Liu B, et al. 2023. TropCRD (Tropical Crop Resources Database): the multi-tropical crop variation information system. Tropical Plants 2:9 doi: 10.48130/TP-2023-0009
TropCRD (Tropical Crop Resources Database): the multi-tropical crop variation information system
- Received: 29 May 2023
- Accepted: 05 June 2023
- Published online: 30 June 2023
Abstract: TropCRD (www.tropical-resources.org.cn) is organized on a crop basis with current Manihot esculenta Crantz, Hevea brasiliensis, Saccharum officinarum L., Ananas comosus, Mangifera indica L. and other tropical crop species. TropCRD is currently divided into four modules: genome module, variation module, molecular marker module and tools module. These data and tools are available for further research by geneticists and tropical crop breeders, including the application of marker-assisted selection techniques and genomic selective breeding to tropical crop breeding. It provides important support for tropical crop breeding research, and also provides valuable resources for obtaining and analyzing the genome sequence data, genetic variation and functional annotation of tropical crops.
-
Key words:
- Tropical crops /
- Database /
- Crop breeding