-
The leaf-disc method was initially created to obtain transgenic plants by infecting tobacco leaf explants with Agrobacterium tumefaciens, and the method was simple and effective[1]. However, considering that excellent cultivars or promising application varieties are expected as the initial material, mature and practical transgenic systems are still essential for obtaining more transgenic lines. Watermelon, one crop of the Cucurbitaceae family, plays an irreplaceable role as an economically important fruit crop worldwide. Watermelon has been studied as a model crop for fruit quality due to its unique diversity in color, shape and size, flavor, texture, aroma, and nutrient content[2]. Therefore, personalized watermelon breeding is particularly important.
Watermelon genetic transformation was first reported in 1994, meanwhile transgenic lines expressing GUS reporter gene were successfully obtained[3]. Thereafter, studies were carried out to improve the regeneration level and transformation rate of watermelon, including genotype, Agrobacterium strains, culture conditions, and chemical agents. Wild watermelon germplasms were more suitable for tissue culture than cultivated watermelon, and shoot regeneration rate were higher[4]. In addition, triploid was superior to diploid in watermelon adventitious bud induction[5]. Various Agrobacterium strains have been applied to watermelon transformation, while there was not explicit exposition regarding the optimum strain[6−10]. Adding acetosyringone (AS) in the Agrobacterium inoculum could improve the sensitivity of strains to watermelon explant wounds and enhance the ability of infection[6, 11]. The infection efficiency and survival rate of explants was the highest after dark cultivation at 23−28 °C for 2−3 d[9,12]. Naphthalene acetic acid (NAA) and 6-benzylaminopurine (6-BA) have been widely used for explants shooting and rooting[7−9, 13, 14], while timentin (TMT), an antibacterial antibiotic, has been applied to many crops, but there was little research on watermelon. In addition, the herbicide glufosinate-ammonium (Basta) was often used as a resistance screening agent in watermelon genetic transformation studies[8, 15].
When particular genes, such as disease resistance and insect resistance genes, are introduced by means of the transformation system, the transgenic plants keep their original traits, meanwhile, new valuable agronomic traits are added. In addition, approaches for efficient gene-targeting are significant for analyzing functional genes included in the plant genome and producing genetically engineered crops[16]. Previous studies suggested that the CRISPR/Cas9 system[17] was used as a highly efficient and specific tool for accurate genome editing[18]. Because its genome sequence is available[19], the CRISPR/Cas9 system should represent a good prospect for exploring gene functions, improving fruit quality and breeding in watermelon[20]. Transgenic lines with both Zucchini yellow mosaic virus (ZYMV) and Papaya ringspot virus type W (PRSV-W) resistance genes were obtained, and provided great potential for quality breeding of commercial watermelon varieties[7]. Similarly, transgenic lines resistant to Cucumber mosaic virus (CMV) also showed its importance in the watermelon industry[14]. By overexpressing ClTST2, a QTL-mapped gene, the molecular mechanism of sugar accumulation in watermelon fruit was systematically elucidated[8]. In addition, gynoecious watermelon lines obtained by editing ClWIP1, provided a theoretical and technical basis for improving watermelon yield and breeding elite lines[21].
Prior to this investigation, although watermelon transformation protocols have been described in several studies[13, 14, 21, 22], the efficiency remained as low as 1.67%[9], and research on the optimization of the watermelon transformation system has not yet been reported in detail. An efficient regeneration and transformation system has five characteristics: high efficiency, good reproducibility, simplicity, rapidity and wide applicability[23]. This study attempts to establish an efficient watermelon transformation system and provide a more reasonable way to introduce foreign genes into watermelon.
-
In order to improve the efficiency of seedling germination, gradient tests were carried out for the disinfection time and cultivation time in the dark. The seeds were disinfected with a mixture of 75% alcohol and 6% sodium hypochlorite. The time of disinfection with alcohol was 35 s. The time of disinfection with sodium hypochlorite was designed with three gradients: 8, 12 and 16 min. After sterilization, seeds were rinsed three times with sterile water. The cultivation time of sterilized seeds was carried out in three gradients of 2, 3 and 4 d. Combined with disinfection conditions and dark incubation time, nine different treatments were designed. The seeds were cultured in the seedling culture medium (Supplemental Table S1) at 28 °C under the same dark conditions. The results showed that average germination rates of the seeds in the nine treatment groups (Supplemental Table S2) was between 20% and 100%. There were no significant differences between the germination rates of seeds cultured for 3 d and 4 d when disinfection time was consistent. Combined with the germination states, the highest germination rate can be obtained by sterilizing with 6% sodium hypochlorite for 12 min, and then culturing for 3 d (Supplemental Fig. S1a). The reasons for poor germination rate may be due to the long disinfection time and bacterial lesions resulting from short disinfection time.
Effects of Agrobacterium tumefaciens strains on transformation efficiency
-
According to the results of previous studies, a widely used culture condition was performed in the following experiments[9]. The explants were inoculated in Agrobacterium tumefaciens inoculum at 28 °C, and the initial OD600 of Agrobacterium tumefaciens inoculum was 0.8. After 4 d dark co-cultivation, GFP transient fluorescence was observed. When Agrobacterium strain AGL1 was used for genetic transformation, only weak instantaneous fluorescence was observed (Fig. 1a1, b1). When strain GV3101 was used, medium fluorescence was detected at edges of the whole cotyledon, but stronger fluorescence was found at edges of the whole cotyledon by using strain EHA105 (Fig. 1a3, b3). The results showed that watermelon 'YL' was insensitive to AGL1 strain. Both GV3101 and EHA105 strains could be applicative for the genetic transformation system in watermelon.
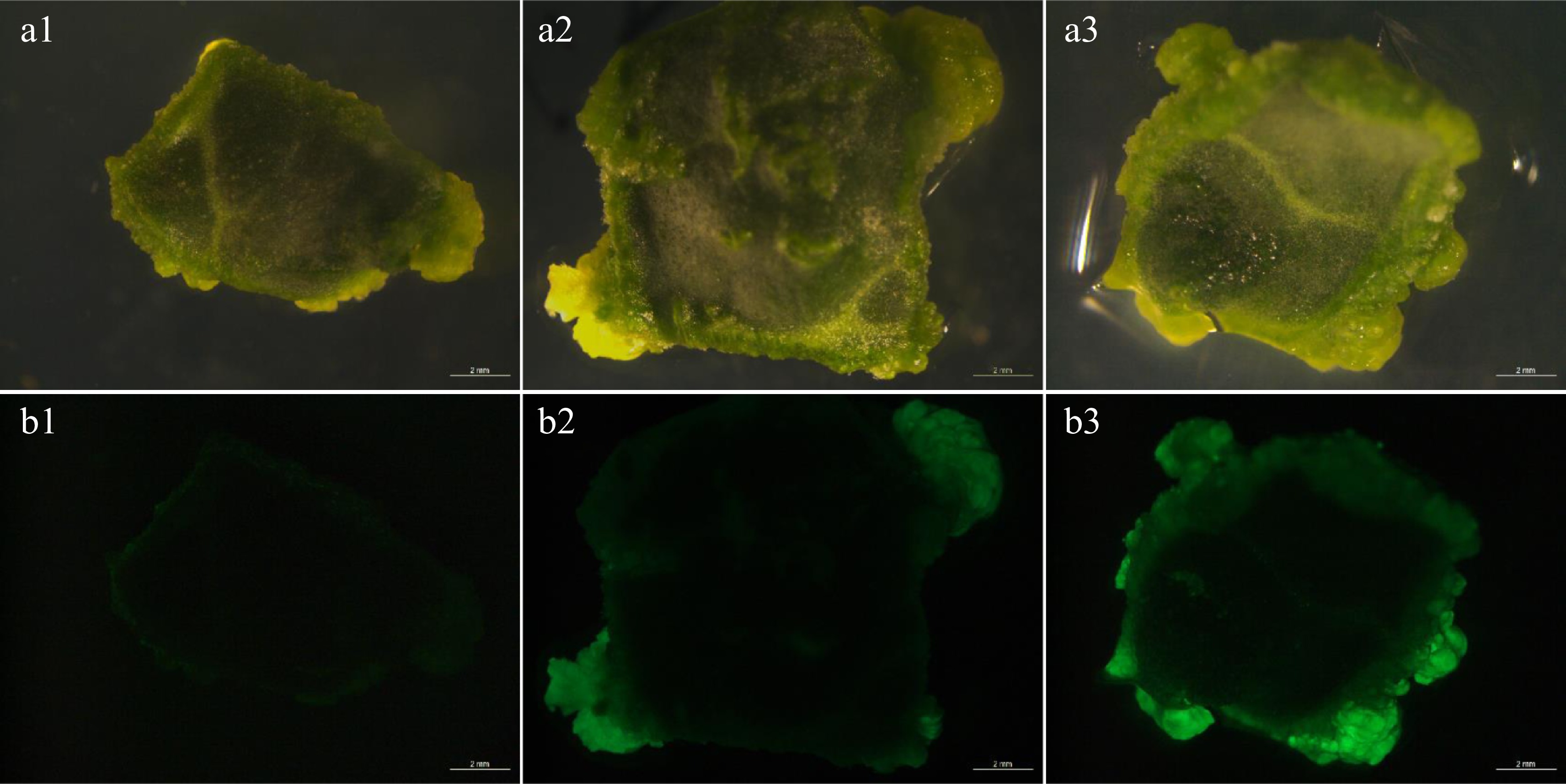
Figure 1.
Transient fluorescence of explants. (a1)−(a3) Bright view. (b1)−(b3) GFP view. (b1) The explants showed weak fluorescence after AGL1 infection. (b2) The explants showed moderate strong fluorescence after GV3101 infection. (b3) The explants showed strong fluorescence after EHA105 infection. Bar = 2 mm.
Effects of acetosyringone (AS) concentration in Agrobacterium inoculum on infection efficiency
-
To determine which concentration of AS in Agrobacterium inoculum could produce the highest genetic transformation efficiency in watermelon. The AS concentrations were separated into five gradients, which were 0, 100, 150, 200, 250 and 300 µM. In each gradient, 120 explants were inoculated. As shown in Table 1, the fluorescence efficiencies of the explants were 45.3%−85.0% due to increased concentrations of AS in the infection solution (Supplemental Table S3). It was also found that the fluorescence efficiency and differentiation efficiency of watermelon explants were significantly different. Fluorescence signals of calli from 100−200 µM AS treatments were acceptable (Fig. 2). However, the infection solution containing 250 or 300 µM AS had large negative effects on germination ability and survival rate of germinated callus, and finally led to a low transformation rate. Our results demonstrated that 200 µM AS in the infection solution was most suitable for genetic transformation of watermelon.
Table 1. Fluorescence efficiencies, brightness and differentiation states of watermelon explants under different AS concentrations.
AS treatment
(µM)Fluorescence efficiency (%) Number of fluorescent explants Fluorescence brightness Callus differentiation state 0 45.3 ± 0.05d 54 ++ The calli were green and compact. There was no contamination and little vitrification 100 68.8 ± 0.03b 82 +++ The calli were densely arranged with dark green, fluorescent speckles and little vitrification 150 75.0 ± 0.02b 90 ++++ The calli were densely arranged and appeared dark green with bright fluorescence and little vitrification 200 85.0 ± 0.05a 102 +++++ The calli were densely arranged and appeared dark green with bright fluorescence and little vitrification 250 69.3 ± 0.04b 83 ++++ Most calli were densely arranged, and a few turned pale and yellowed with water stain 300 54.7 ± 0.05c 65 ++++ The calli were closely arranged, partly vitrified with dark green and contaminated by bacteria Fluorescence efficiency = Number of fluorescence explants/Total explants number × 100%. Values are means (three independent experiments) ± standard errors (SE), and different letters indicate significant differences between treatments according to Duncan's multiple test (P < 0.05). 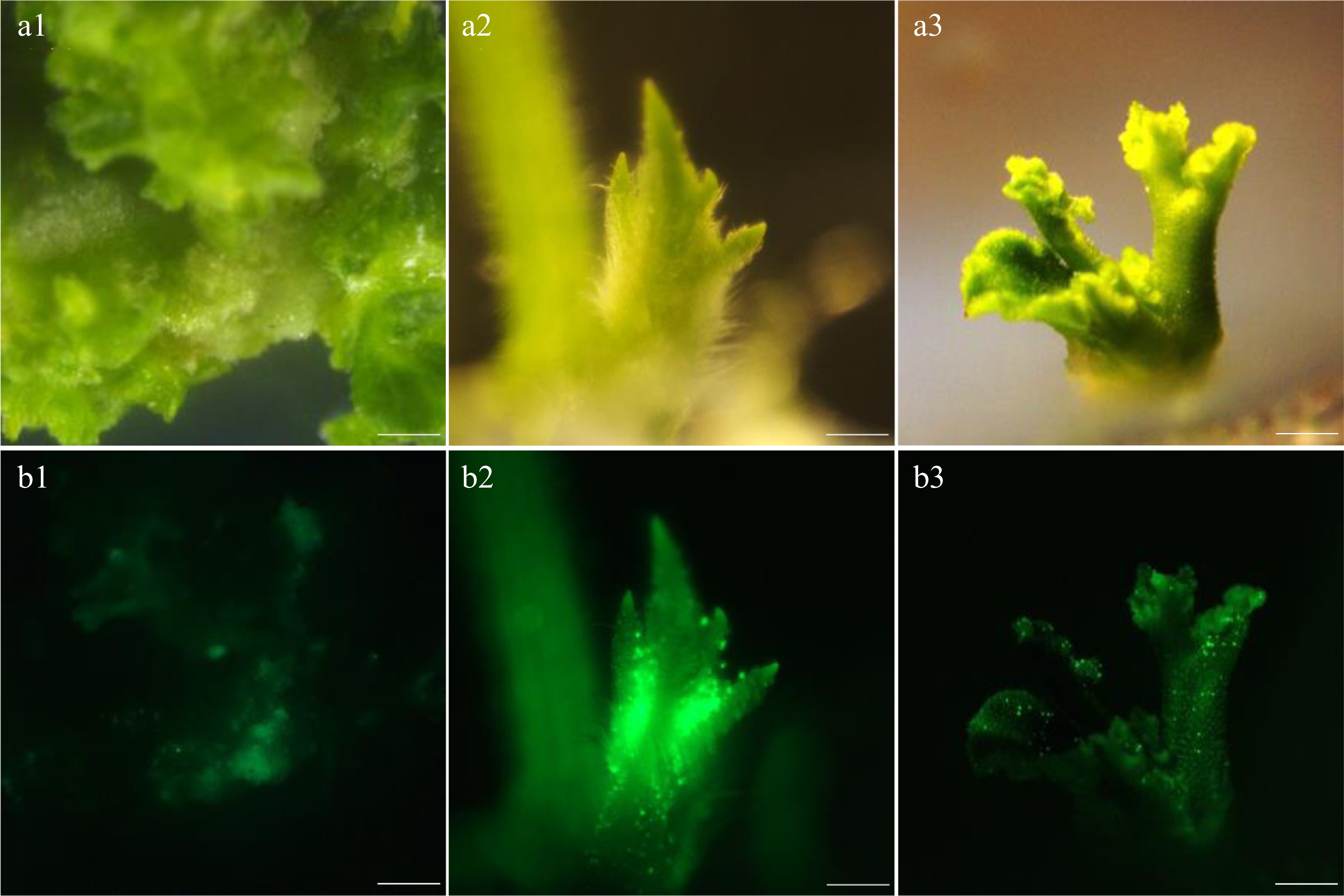
Figure 2.
Fluorescence intensity of the callus under different concentrations of AS. (a1)−(a3) Bright view. (b1)−(b3) GFP view. (a1), (b1) AS concentration at 100 µM. (a2), (b2) AS concentration at 200 µM. (a3), (b3) AS concentration at 300 µM. Bar=1 mm.
Effects of Agrobacterium tumefacien concentrations in the inoculum and co-culture time on transformation efficiency
-
The co-cultivation stage is the key to improving infection efficiency in genetic transformation. During the middle and later periods of co-cultivation, brighter GFP fluorescence meant higher transformation success rate. Due to the browning explants auto-fluorescence, we need to ensure that explants were kept alive and free of pollution as much as possible even though bright fluorescence was found.
To improve the infection efficiency, four concentrations of A. tumefaciens (EHA105) infection solution were set as 0.6, 0.2, 0.02 and 0.005 at OD600[7, 24]. The co-culture time was set at three gradients of 2, 3 and 4 d. Therefore, 12 associations were listed in total, named A-L respectively (Table 2). Explants, which were cut into eight pieces longitudinally, were immersed in the inoculum for 10 min. The results showed that GFP fluorescence of explants was the strongest when OD600 of A. tumefaciens infection solution was 0.6 and 0.02, but the number of fluorescent explants was more at OD600 = 0.02. The fluorescence efficiency of explants was relatively high when cultured in co-cultivation medium for 3 and 4 d (Fig. 1b). However, when the explants were co-cultured for 4 d, there was pollution around the explants, and the edges showed serious browning. The growth states of each combination are shown in Table 3.
Table 2. Transient fluorescence efficiencies and brightness of watermelon explants under different concentrations of Agrobacterium tumefaciens infection solution and co-culture times.
Combination Agrobacterium concentration (OD600) Coculture time
(days)Fluorescence efficiency
(%)Fluorescence brightness A 0.6 2 42.5 ± 1.5b +++ B 0.2 2 54.4 ± 1.4a ++ C 0.02 2 38.5 ± 1.8b + D 0.005 2 33.3 ± 0.3c + E 0.6 3 38.1 ± 1.1c + F 0.2 3 43.0 ± 2.0bc ++ G 0.02 3 79.1 ± 0.9a +++ H 0.005 3 47.1 ± 0.5b + I 0.6 4 35.5 ± 1.4c + J 0.2 4 44.5 ± 2.6b + K 0.02 4 79.1 ± 0.6a +++ L 0.005 4 51.5 ± 2.5b ++ Fluorescence efficiency = Number of fluorescence explants/Total explants number × 100%. Values are means (three independent experiments) ± standard errors (SE), and different letters indicate significant differences between treatments according to Duncan's multiple test (P < 0.05). Table 3. Growth states of watermelon explants under different Agrobacterium tumefaciens infection solution and co-culture times.
Combination Growth state A A small part died, and the margin of the surviving explants were yellow with obvious water-stained flora B The edges showed traces of a watery microflora C The explants were well developed D The explants were well developed E Some died with serious spillage, and fluorescence
explants were browningF A small portion were dead and the margins browned G The explants dilated well H The explants dilated well I Most died with serious spillage phenomenon, and
virtually all the edges brownedJ Partially dead with serious spillage and serious edge browning K Partially edged with yellow and with spillage phenomenon L The explants were well developed Effects of different concentrations of hormones in co-cultivation medium on callus regeneration
-
The direct differentiation method was used to optimize genetic transformation of watermelon, while the calli redifferentiation method was an optimization system developed on the basis of previous experience in this research[25]. According to the results of seedling germination, experiments of cotyledon dedifferentiation were conducted. The strong calli were more easily induced into adventitious shoots by 6-BA, which was beneficial to improve the efficiency of genetic transformation efficiency. The temperatures of cotyledon differentiation ranged from 26−28 °C. Concentration treatments of hormones were divided into 12 groups (Supplemental Table S4). We further selected 3 and 4 d (named as YL-3 and YL-4 in Supplemental Table S4) to culture explants in co-cultivation medium (Supplemental Table S5). The results indicated that the combination of 1.5 mg/L 6-BA without indole acetic acid (IAA) had the highest callus differentiation rate after 3 d of co-cultivation.
Effects of different concentrations of TMT and Basta on callus regeneration
-
Ticarcillin, one of the active components in TMT, was used as an antibacterial agent. Based on the differentiation and growth state of callus, TMT with a concentration of 50−350 mg/L could inhibit the growth of bacteria at an early stage. However, concentrations of TMT at 350 mg/L caused more severe aetiolation in the later stages of development, especially in the rooting stage, which could affect the growth of the roots (Supplemental Table S6). The addition of TMT with a concentration at 200 mg/L had no fungal contamination. Based on the standard of high proliferation rate and normal growth state, 200 mg/L TMT was considered as the best concentration, which was added to the recovery medium (Supplemental Table S7). In addition, 200 mg/L TMT could effectively inhibit the growth of Agrobacterium tumefaciens and no contamination was detected after regenerated explants were transferred to a TMT-free culture medium. TMT with a concentration higher than 350 mg/L resulted in yellowing phenomenon and seedling death, which indicates that excessive TMT has some toxic effect. We named the recovery stage after the recovery process, as shown in the Supplemental Fig. S1c.
The gene editing vector used in this experiment contained a marker gene that could be resistant to Basta, and resistance screening was used to screen the positive transgenic shoot (Supplemental Table S8). At concentrations of 0.4 and 1.4 mg/L Basta, the survival rate and fluorescence efficiency of callus were higher than those of other concentration gradients, although there was no obvious difference between them (Table 4). However, when the concentration of Basta was 2.4 mg/L, the callus vitrified seriously, and the redifferentiation of calls were inhibited. At 1.4 mg/L, the well-grown callus showed high fluorescence efficiency, and the brown callus was selected as the best concentration. We named the selection stage after the selection process (Supplemental Fig. S1d). The results revealed that the concentration of Basta higher than 1.4 mg/L caused the shorter survival time of explants.
Table 4. The survival rates and growth states of callus under different concentrations of Basta.
Basta concentration
(mg/L)Callus survival rate
(%)Fluorescence rate of
survival callus (%)Callus state 0.4 100.0 ± 0.0a 69.1 ± 4.1c The tissues appeared dark green and grew well 1.4 73.6 ± 0.4b 78.7 ± 0.3b Partial tissues appeared brown and most grew well 2.4 73.0 ± 3.0b 80.5 ± 1.5b Partial tissues were transparent and light green with serious vitrification 3.4 40.8 ± 1.2c 92.0 ± 2.9a Partial tissues died with serious yellowing phenomenon and obvious spillage 4.4 0.7 ± 0.3d 0.7 ± 0.3d Most tissues died, and the spillage was obvious Callus survival rate = Survival callus number/Total callus number × 100%; Fluorescence rate of survival callus = Survival callus number with fluorescence/Total survival callus number. Values are means (three independent experiments) ± standard errors (SE), and different letters indicate significant differences between treatments according to Duncan's multiple test (P < 0.05). Effects of hormone concentrations on adventitious shoot elongation
-
Adventitious shoot differentiation was easier in well-developed and dedifferentiated calli. In order to improve the regeneration rates of buds, three concentrations of 6-BA (0.05, 0.1, 0.15 mg/L) and two concentrations of naphthlcetic acid (NAA) (0.05, 0.1 mg/L) were applied to analyze the formation of adventitious elongated shoots (Table 5). When the concentration of 6-BA is 0.1 mg/L and NAA is 0.1 mg/L, the frequencies of shoot elongation was the highest (Fig. 3a). When 6-BA and NAA were higher than 0.15 mg/L and 0.1 mg/L respectively, the callus directly differentiated into aerial roots, and explants were prone to yellowing (Fig. 3b). When 6-BA and NAA were 0.1 mg/L and 0.05 mg/L respectively, shoot elongation was inhibited (Fig. 3c). The results indicated that 0.1 mg/L 6-BA and 0.1 mg/L NAA were the best concentrations to improve the elongation efficiency of adventitious shoots and keep their good growth states (Supplemental Fig. S1e). Shoot elongation medium is shown in Supplemental Table S9.
Table 5. Adventitious shoot elongation efficiencies and shoot growth states under different concentrations of hormones.
6-BA (mg/L) NAA (mg/L) Shoot regeneration
rate (%)Adventitious shoot growth state 0.05 0.05 10.3 ± 1.8c The adventitious shoots differentiated less and had serious vitrification 0.1 0.05 22.0 ± 1.0b Less differentiation, and vitrification was not serious 0.15 0.05 32.4 ± 2.6a Less differentiation, some shoots unable to continue to elongate and showed yellowing 0.05 0.1 29.2 ± 0.7b Less differentiation, calli partly yellowed and vitrified 0.1 0.1 64.5 ± 5.5a With more differentiation, calli partly vitrified without yellowing phenomenon 0.15 0.1 45.6 ± 3.8b More differentiation, shoots partly yellowed with a small amount producing air roots Shoot regeneration rate = Number of regerminated shoots/Total explants number × 100%. Values are means (three independent experiments) ± standard errors (SE), and different letters indicate significant differences between treatments according to Duncan's multiple test (P < 0.05). 
Figure 3.
Redifferentiation state of explants under different hormone concentrations. (a) The callus differentiated well under 0.1 mg/L concentration of 6-BA and 0.1 mg/L concentration of NAA. (b) The callus redifferentiated to form aerial roots under 0.15 mg/L concentration of 6-BA and 0.1 mg/L concentration of NAA. (c) The callus appeared to yellow and could not differentiate under 0.15 mg/L concentration of 6-BA and 0.05 mg/L concentration of NAA. Bar = 12 mm.
Effects of budding methods on positive plant rooting
-
In this study, it was found that adventitious buds could generate roots 7−14 d after being transferred to the rooting medium. (Supplemental Table S10). Based on these results, all optimized aspects were used in our following research to obtain an effective transformation frequency. A positive plant was obtained, which is shown in Supplemental Fig. S1f.
Verification of optimization efficiency of watermelon genetic transformation system
-
The above experimental results showed that the watermelon transformation system had been optimized to some extent. To verify the efficiency, we selected ClREC8 (Cla97C07G132920), ClACS1 (Cla97C01G017090) and ClACS7 (Cla97C03G066110) as target genes to perform gene knockout assay in watermelon. PCR amplification was performed using the primers Cas9-F and Cas9-R, and the results revealed the transgenic plants contained exogenous T-DNA inserts (Fig. 4). Furthermore, targeted gene sequences in the transgenic plants were sequenced, and the results in three target genes were partly shown (Fig. 5, Supplemental Fig. S2). As a result, in approximately 300 inoculated explants, we obtained a total of 45 T0 transgenic lines, among which 42 plants were successfully edited. The percentage of 93.3% meant that the optimized transformation system could also be used for watermelon gene editing.
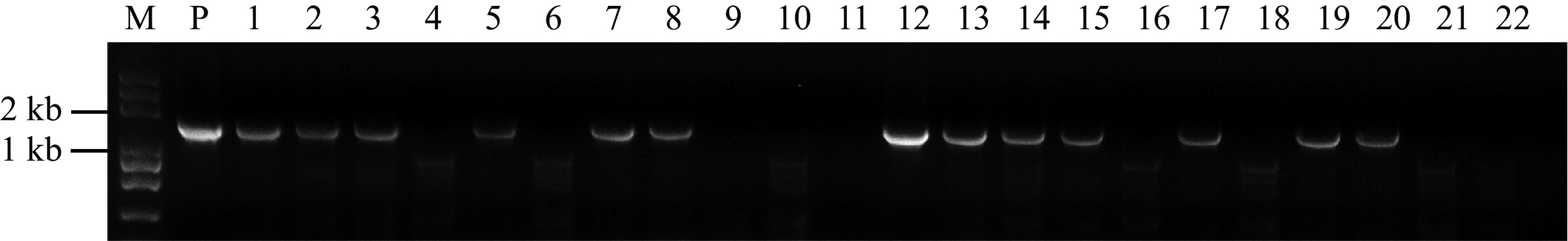
Figure 4.
PCR analysis of the transgenic plants. Genomic DNA isolated from putative transgenic plants were subjected to PCR amplification with Cas9 primers. Lane M, Trans2K Plus DNA Marker; Lane P, positive control (plasmid); Lanes 1, 2, 3, 5, 7, 8, 12, 13, 14, 15, 17, 19 and 20, putative transgenic watermelons; Lane 4, 6, 9, 10, 11, 16, 18, 21 and 22, Non-transgenic watermelon.
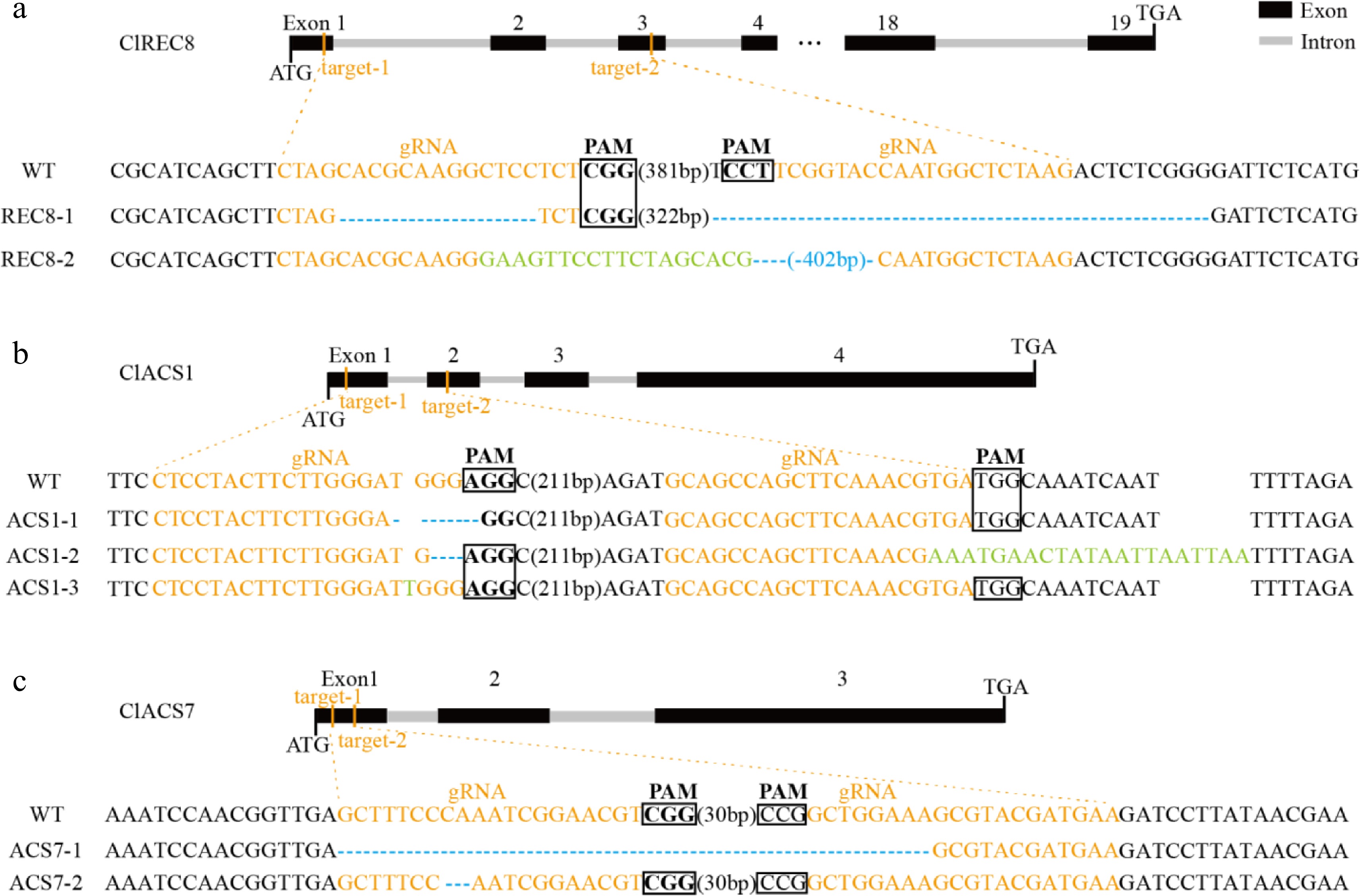
Figure 5.
Targeted mutagenesis of (a) ClREC8, (b) ClACS1, and (c) ClACS7 in the transgenic lines. The schematic diagrams illustrate sgRNA targeting the exons. The target sequences are shown in orange with protospacer adjacent motifs (PAM) sequence highlighted with black rectangles. Nucleotide deletions are shown with blue dashes, and inserted nucleotides are shown in green.
-
It is widely known that transgenesis and gene-editing technologies were made full use of for promoting plant research processes [25−27]. Considering the significance of the genetic stability of both transformed exogenous genes and regenerated plants, various methods have been tried for transformation, including particle acceleration[28], electroporation and polyethylene glycol permeabilization of protoplasts[29], and DNA transfer mediated by Agrobacterium. However, previous studies indicated that only few positive lines have been obtained due to difficulties in watermelon transformation[17, 30]. Extremely low transformation efficiency in watermelon transformants was largely due to the high degree of escape and chimeric shoot[14].
In this study, we optimized the watermelon genetic transformation system to improve the genetic transformation efficiency. The growth process of young seedlings grew promisingly compared with old seedlings[31], however, both tender aged and over aged seedlings are not suitable for better yield[32]. The watermelon seedling age, which was of great significance for callus regeneration, was the key factor as to whether induction was successful and highly effective or not. The explants state determined the impregnation efficiency of the subsequent Agrobacterium-mediated method. Poor explants state usually causes problems such as bacterial growth, vitrification or redifferentiating inability in the cultivation process. The experimental results showed that explants with good viability could be obtained after a disinfection time of 12 min and cultivation time of 3 d.
The findings of this study were consistent with other reports of watermelon transformation with the Agrobacterium-mediated method[3]. AS, a phenolic compound which was a known vir gene inducer especially in monocotyledon plants, plays a crucial role in the transferring process[33]. However, a high quantity of AS was toxic to the plants, which could be mainly due to the presence of alcohol as a solvent[34]. Germplasm differences showed different sensitivity to AS. In many other crops, such as wheat, cucumber and sesbania, the addition of AS to Agrobacterium inoculum and co-cultivation medium has been shown to improve conversion efficiency[35−37]. Previous studies indicated that the highest concentration of AS used for transformation of various plants was between 100−200 µM[38−40]. We selected an AS concentration at 200 µM to improve infection efficiencies of strain and fluorescence efficiency.
Due to germplasm differences possessing diverse genotypes, efficiencies of watermelon genetic transformation were also different[41]. It was also essential to select the appropriate Agrobacterium strain for different watermelon germplasms to improve their ability in infecting explants. In previous studies, types of Agrobacterium strains used in the genetic transformation of cucurbitaceae crops were different, including AGL1, EHA105 and GV3101[6, 42]. Our study indicated that watermelon germplasm 'YL' was sensitive to Agrobacterium strain EHA105, which was the basis for improving the transformation efficiency.
Agrobacterium concentration and coculture time were considered as the key to improving the genetic transformation efficiency. Too low a c oncentration could not infect explants smoothly, whereas too high a concentration led to bacterial overgrowth, causing explants death. Too short coculture time could not ensure T-DNA transferred into the explant cells, and integrated into the genome. Instead, bacterial overgrowth resulted in explants not being able to continue to grow, and transformation efficiency was reduced. In terms of Agrobacterium population density, the fluorescence efficiency of watermelon explants was higher at an OD600 of 0.02. The total time was fixed at 10 min, which was the same time reported previously for immersion in Agrobacterium inoculum[43]. The optimum working concentration of Agrobacterium used in this study was relatively low, and the fluorescence was brighter when co-cultured for 3 d. This may be related to the sensitivity of explants from different germplasms. However, if infected by lower concentrations, the efficiency cannot be improved. In contrast, co-cultivation for too long would cause bacteria colonies proliferation and explants death.
Efficiencies of watermelon transformation were affected by many other factors, and certain parameters were slightly different in a variety of watermelon germplasms[44]. Taking watermelon germplasm ‘YL’ as the original material, this study further explored hormone concentrations for watermelon callus development. The explants differentiation rate was the highest with 1.5 mg/L 6-BA and no IAA. In addition, adventitious shoots could be induced to differentiate and even elongate by using the hormone combination of 0.1 mg/L 6-BA and 0.1 mg/L NAA, and growth states were good. The results indicated that ‘YL’ might not be sensitive to IAA.
Antibiotics could effectively inhibit a portion of Agrobacterium to ensure good growth of explants. TMT, an antibiotic agent that is a combination of ticarcillin and clavulanic acid, can positively replace other antibiotics like carbenicillin and cefotaxime in tissue culture[45, 46]. Because it is stable in solid agar medium and can keep effective for longer than 70 d, TMT could be an alternative antibiotic for suppressing Agrobacterium growth in transformation effectively and improving regeneration potential compared with other antibiotics such as carbenicillin[47, 48]. Low concentration of TMT could not arrest bacteria proliferation so that explants cannot continue to grow. In contrast, high concentrations of TMT would not only inhibit the bacterial growth but also inhibit the growth of explants, resulting in a decline in transformation efficiency. Our results demonstrated that the most appropriate concentration of TMT was 200 mg/L, which could facilitate shoot regeneration.
A codon-modified phosphinothricin acetyltransferase gene, which confers resistance to the herbicide glufosinate-ammonium, was used as the selectable marker[49]. Glufosinate-ammonium is able to block the activity of an enzyme used in the biosynthesis of amino acid glutamine, which has been reported in different species[50−52]. The study demonstrated that Basta had a pivotal effect on the screening of positive callus and shoot at 1.4 mg/L. The screening method could greatly improve the detection efficiency, and reduce the workload.
The experiments provided preparation for the subsequent obtaining of knocked lines and reduced the problems such as poor differentiation ability of callus, yellowing phenomenon, and vitrification in watermelon. Meanwhile, fluorescence efficiencies of the watermelon callus, which was conducive to the fluorescent shoots, were improved. The pH and humidity of each medium should be strictly controlled in the experiment, and the callus growth states should be observed regularly and repeatedly.
Recently, a study revealed that a mutation in the miR396 microRNA region included in ClGRF4 gene led to efficiency up to 67.27% in watermelon, no matter which genotype was applied for transformation[53]. Nevertheless, our results suggested that watermelon transformation efficiency (from explant to transgenic plant) was increased to 12.5% using the optimized system in this study. Multigene knockouts as well as gene replacements have not yet been widely studied in watermelon and other plants[54]. Our research made it possible to optimize the watermelon multi-genetic transformation system more efficiently in the future.
-
The watermelon germplasm 'YL', cultivated at 36o−39o N and 107o−111o E from a warm semi-humid climate, has strong drought resistance and moderate resistance to Fusarium wilt, and was used as the original material in this experiment. The materials were planted in a greenhouse at the College of Horticulture, Northwest A&F University (Shaanxi, China).
A. tumefaciens strains and binary vector construction
-
Three A. tumefaciens strains including AGL1[55], GV3101[6] and EHA105[56] were tested in this study. The binary CRIPSR/Cas9 vectors pBSE402 (carrying a Bar and a GFP genes, both of which were driven by 35S promoters) was modified from pBSE401 provided by Dr. Qijun Chen from China Agricultural University (Beijing, China), and the vectors were constructed as described[7,16]. Positive regenerated plants were detected by polymerase chain reaction. The PCR primers Cas9-F (5’-GCAGCTCTCCAAGGACACAT-3’) and Cas9-R: (5’-CGTGAGTTCTTCTGGCCCTT-3’) were designed using Primer Premier 5 software. The PCR was conducted using Taq PCR Mix (GenStar, China), and performed in an optical 96-well plate with 2720 Thermal Cycler (Applied Biosystems, USA). The positive results were further sub-cloned with pUC18 plasmids, sequenced, and analyzed according to Kaur et al.[57].
Vitro culture and regeneration system
-
Plants were regenerated from watermelon cotyledons[58, 59]. Adventitious shoots formed on the proximal cut edges of the cotyledonary explants[60]. Cotyledons of sterile watermelon seedlings aged 2−5 d were cut into small pieces of 0.5 cm, then inoculated on MS medium with 6-BA to dedifferentiate into calli, on which a lot of adventitious shoots appeared after 2 weeks. Adventitious shoots were then transferred to the rooting medium for 2−3 weeks. The culture conditions were 28 °C, 16 h day, and 3,000 lx light intensity.
Optimization of the genetic transformation system
-
The experiment was carried out from several aspects to optimize the transformation system, including different seedling ages, Agrobacterium strains, AS and Agrobacterium concentrations of the inoculum, concentrations of antibiotic TMT and Basta selection pressure, and concentrations of added hormones.
Data availability
-
The data that support the results are included in this manuscript and its supporting information files. Other relevant materials are available from the corresponding author upon reasonable request.
This work was supported by Young Talent fund of University Association for Science and Technology in Shaanxi, China (20210202, to JS), and the Science and Technology Innovation Team of Shaanxi (2021TD-32, to ZL).
-
The authors declare that they have no conflict of interest.
- Supplementary Table S1 Seeding medium ingredients.
- Supplementary Table S2 Germination rates and states of watermelon seeds under different disinfection and cultivation times.
- Supplementary Table S3 Infection solution ingredients (pH=5.4).
- Supplementary Table S4 Differentiation efficiencies of watermelon cotyledons under different concentrations of hormones.
- Supplementary Table S5 Co-cultivation medium ingredients (PH=5.8).
- Supplementary Table S6 Callus growth states at different concentrations of TMT.
- Supplementary Table S7 Recovery medium ingredients (pH=5.8).
- Supplemental Table S8 Selection medium ingredients (PH=5.8).
- Supplementary Table S9 Shoot elongation medium ingredients (PH=5.8).
- Supplementary Table S10 Rooting medium ingredients (PH=5.8).
- Supplementary Fig. S1 Optimized stages of watermelon transformation system. (A) Seeding stage. (B) Co-cultivation stage. (C) Recovery stage. (D) Selection stage. (E) Shoot elongation stage. (F) Rooting stage. Bars=12mm.
- Supplementary Fig. S2 Sequencing results of the PCR products in partial knocked lines of ClREC8, ClACS1 and ClACS7.
- Copyright: © 2022 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Cao L, Wei W, Shen J, Xu Z, Li Z. 2022. Study on the optimization of transformation systems in watermelon. Vegetable Research 2:12 doi: 10.48130/VR-2022-0012
Study on the optimization of transformation systems in watermelon
- Received: 20 December 2021
- Accepted: 22 August 2022
- Published online: 21 September 2022
Abstract: To date, the genetic transformation system of watermelon has remained inefficient. In this study, the genetic transformation system of watermelon mediated by Agrobacterium tumefaciens was optimized, including different seedling ages, strains of Agrobacterium tumefaciens, concentrations of acetosyringone infected solution, co-culture time, selection pressure of antibiotics timentin and glufosinate, and concentrations of hormones 6-benzylaminopurine, indoleacetic acid and naphthylacetic acid in the corresponding culture medium. Our results suggested that cotyledons would be used as explants, disinfected with 6% sodium hypochlorite for 12 min, cultured for 3 d, and then infected with Agrobacterium inoculum (Agrobacterium EHA105) containing 200 µM acetosyringone and 0.02 of final OD600. The explants differentiated into adventitious shoots in the medium with 1.5 mg/L 6-benzylaminopurine and 200 µM timentin. Positive adventitious shoots were obtained through further screening by 1.4 mg/L herbicide glufosinate-ammonium, and were induced by 0.1 mg/L naphthalene acetic acid into independent plants. Our system improves the genetic transformation efficiency of watermelon and provides a technical basis for continuous acquisition of watermelon transgenic plants.
-
Key words:
- watermelon /
- genetic transformation /
- hormone /
- optimization /
- transgenic













