-
In higher eukaryotic genomes, approximately 90% of the genetic information can pervasively transfer to RNAs[1]. More than 75% of the transcripts do not have protein-coding potential and are classified as non-coding RNAs (ncRNAs)[2,3]. Long non-coding RNAs are a group of ncRNAs with a transcript length of more than 200 nt[4]. Compared with that of mRNAs, their transcript level is generally low and has strong tissue or condition expression specificity[4]. In addition, the sequence conservation of lncRNAs is very low across plant species, which may result from rapid sequence evolution[5]. Most lncRNAs have also been found to be transcribed by RNA pol II, while the others are produced by pol III, IV, and V[6,7]. Based on their location relative to adjacent protein-coding genes in the genome, the lncRNAs are classified into five types: sense lncRNA, antisense lncRNA, bidirectional lncRNA, intronic lncRNA (incRNA), and large intergenic lncRNA (lincRNA)[8]. Each lncRNA is produced by a specific mechanism and can act in cis or trans to regulate gene expression through diverse modes at chromatin, transcription, post-transcription, translation, and post-translation levels[9,10]. With the wide applications of high throughput RNA-sequencing technology, thousands of lncRNAs have been identified in diverse plant species. They act not only as regulators of basic cellular mechanisms but also participate in the regulation of developmental processes as well as biotic and abiotic stress responses[5,11−14]. In recent years, the research on the function of lncRNAs in vegetable crops has gradually increased. This paper reviews the characteristics of lncRNAs and their biological functions in vegetables.
-
LncRNAs refer to ncRNAs longer than 200 nt, sometimes in a range of tens of kilo-nucleotides. By comprehensive comparative analysis of lncRNAs among 37 species, we found that the length of lncRNAs fluctuates greatly among different species, ranging from 550.83 nt of mean length in Brassica rapa to 12,053.52 nt in Manihot esculenta[15]. Most plant lncRNAs identified so far are polyadenylated and 5'-capped. However, there are some non-polyadenylated lncRNAs[4,16]. In comparison with those polyadenylated lncRNAs, the length of non-polyadenylated lncRNAs is shorter, the transcript abundance is lower, and the specificity in response to stresses is stronger[17]. Like most proteins, the structure of some lncRNAs is simple, while others appear to have a complex but poorly understood secondary/or tertiary structure, which is generally believed to be necessary for their function. There are two classes of functional elements in lncRNA: One is necessary for physical interactions with partner nucleic acids or proteins, and the other governs the secondary and/or tertiary structure, which further directs interaction partners' binding sites[18].
Expression features
-
The transcript abundance of lncRNAs is generally low, only 1/30 to 1/60 of the average mRNA expression level[8]. However, there are also exceptions: In a previous study, we found that some lncRNAs had very high expression abundance after comprehensive analysis of the lncRNAs in 37 species[15]. Furthermore, there are significant differences in lncRNA expression patterns across species[15]. Most lncRNAs reside in the nucleus, while they can also export to the cytosol or other organelles, such as mitochondria, which was demonstrated by ribosome profiling and RNA FISH[19]. In the nucleus, lncRNA may perform its function in either cis or trans mode; it has been suggested that lncRNAs with low transcript abundance may work in cis, while those transcribed at a higher level are likely to act in trans[20].
The expression of lncRNAs was highly specific in different tissues and developmental stages. For example, in cabbage, lncRNA BoNR8 was specifically expressed in the epidermal tissue of the elongation region of germinating seeds[21]. In tomato (Solanum lycopersicum), 4,079, 4,135, and 4,311 lncRNAs that were expressed in tomato fruits at the mature green, breaker, and breaker plus 7 days, respectively, were identified by integrating 134 datasets. Only 20 lncRNAs were expressed in all three developmental stages[22]. It was proposed that the apparent specificity was partly attributed to the generally low expression level of lncRNAs as well as limitations in detection by standard mRNA-sequencing protocols[23]. Most lncRNA sequences are weakly conserved. This shows that only a small part of lncRNA in Chinese cabbage has high homology with lncRNA in other Brassica crops[24]. Based on the analysis of lncRNA from five monocot and five dicot species, it was found that lncRNA had higher sequence conservativeness at the intra-species and sub-species levels but lower inter-species conservativeness[25].
-
With the rapid development of next-generation sequencing (NGS) technology, RNA-Seq has become the first choice for studying the whole transcriptome due to its advantages of high throughput, high accuracy, high sensitivity, and low cost, which has also greatly facilitated the development of lncRNA identification and prediction[26]. However, the construction and sequencing of general transcriptome libraries cannot separate the sense strand and the antisense strand, therefore, a strand-specific RNA-seq (ssRNA-seq) technique was developed to facilitate the identification of transcript orientations[27]. Although NGS techniques are effective, they still suffer from several drawbacks. One major disadvantage is short read lengths, and it is difficult to ensure the accuracy of reconstructed transcripts during assembly[28]. Single-molecule real-time sequencing technology (SMRT) is a third-generation sequencing method that can overcome these limitations and generate long reads without further assembly[29,30]. The third-generation sequencing technology (isoform sequencing, ISO-seq) based on the SMRT sequencing platform has recently been applied to analyze the full-length transcriptome and lncRNA prediction of various species[31−34]. In addition, in order to solve the problem of high error rate of SMRT, the 'SMRT + NGS' sequencing joint analysis method, which uses high-quality, high-coverage NGS to correct SMRT data, has been more and more widely used[35−37]. ChIP-seq technology, which combines chromatin immunoprecipitation (ChIP) and NGS, provides massive data for the identification of transcription factor binding sites, and it can also be used to identify lncRNA targets of specific transcription factors[38].
-
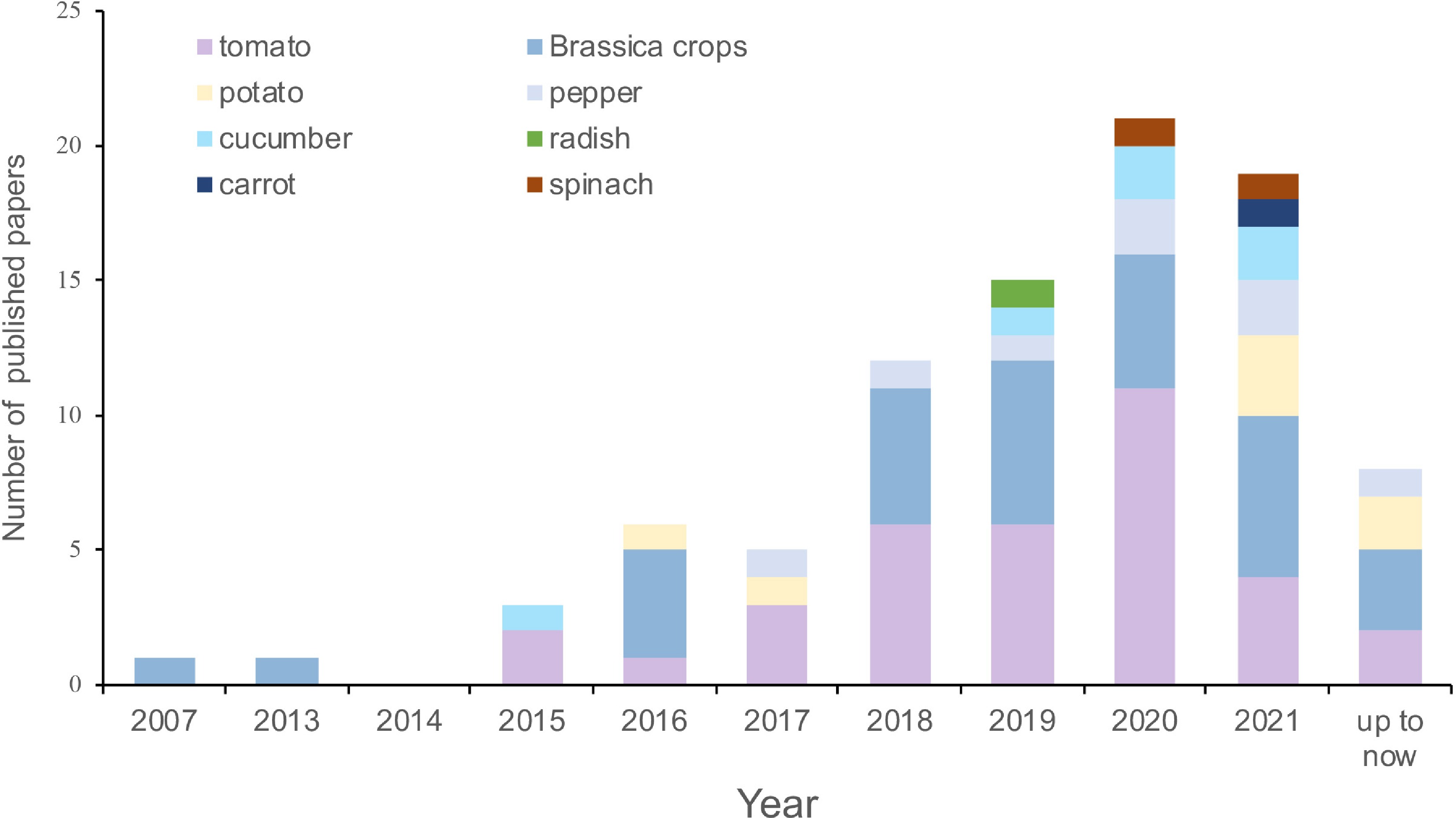
As an important new regulatory factor, in recent years, the function of lncRNA in vegetable crops has received attention. Here, we summarize the studies involving lncRNA research for some important vegetables, including tomato, Brassica crops, cucumber (Cucumis sativus L.), pepper (Capsicum annuum L.), carrot (Daucus carota L.), radish (Raphanus sativus L.), potato (Solanum tuberosum L.), and spinach (Spinacia oleracea L.), which were also the most studied among the various vegetable species. It was found that the first report about lncRNAs on these vegetables was the discovery of BcMF11 in 2007, which was predicted as an ncRNA associated with pollen development of Chinese cabbage[39]. Then in 2013, the function of BcMF11 was further explored[40]. Based on our statistics, there are fewer than 100 relevant studies in the literature to date (Supplemental Table S1). From 2017 to 2020, the number of published papers increased gradually, then decreased slightly in 2021 (Fig. 1, Supplemental Table S1). Among the studied species, studies on tomato were the most common (35 papers), followed by Brassica crops (32 papers), with relatively few reports on the other six vegetable crops: 8, 7, 6, 2, 2, and 1 for pepper, potato, cucumber, spinach, carrot, and radish, respectively (Fig. 1, Supplemental Table S1).
-
With the advances in genomic and bioinformatic techniques, lncRNAs in vegetable crops were suggested to be involved in various biological processes, and in our study, these processes were mainly categorized into three groups, including growth and development, abiotic stress, as well as biotic stress (Table 1). In different species, lncRNAs were found to be related to various developmental events, such as fruit ripening, vernalization, anther or pollen development, anthocyanin biosynthesis, and sex differentiation[37,41−44]. Moreover, lncRNAs were implicated in a variety of abiotic stress responses, such as drought, heat, chilling, and salt stresses[45−48]. In addition, lncRNAs may play an important role in plant immunity[49−52] (Table 1). Even though a large number of lncRNAs were identified by high-throughput sequencing and suggested to be associated with different physiological processes, only a small portion of lncRNAs have been assessed by functional analysis using molecular biology approaches (Fig. 2, Table 2). The regulation modes of plant lncRNAs in different biological processes are complex and variable[14]. Among them, the interaction between lncRNAs and miRNAs was the most reported relationship in plants. First, lncRNAs can function as an endogenous target mimic (eTM) to sequester miRNAs via base pairing to complementary sites, therefore blocking the interaction of miRNAs and their potential targets[53]. These kinds of lncRNAs are also known as competitive endogenous RNAs (ceRNAs)[53,54]. Second, TMs with extensive complementarity to the 5' and 3' ends of endogenous miRNAs were recently found to trigger miRNA destruction in animals, a process known as target-directed miRNA degradation (TDMD)[55−57]. Similarly, by expressing a short tandem target mimic (STTM) in plants, specific endogenous miRNAs can be disrupted. This technology was developed to investigate the function of specific miRNAs[58]. Furthermore, the F-box protein HAWAIIAN SKIRT (HWS) was found to be involved in the degradation pathway and may play a role in the clearance of RNA-induced silencing complexes (RISCs)[59]. Third, some lncRNAs were discovered as precursors of miRNAs, which positively regulate the maturation of miRNAs[54]. Lastly, some lncRNAs can bind and be cleaved by the sequence of complementary miRNAs, that are further processed into phased small-interfering RNAs (phasiRNAs) and guide RNA silencing[54].
Table 1. List of long non-coding RNAs (lncRNAs) identified in major vegetable crops.
Roles Species Pathways Approaches LncRNAs
numberDE-LncRNAs number Ref. Growth and development Solanum lycopersicum Fruit ripening RNA-seq − 378 [41] Solanum lycopersicum Fruit ripening ssRNA-seq 3,679 677 [61] Solanum lycopersicum Fruit expansion and
ripeningssRNA-seq 17,674 tissue- and stage-dependent [60] Solanum lycopersicum RIN target lncRNAs; fruit ripening ChIP-seq & RNA-seq 187 − [38] Solanum lycopersicum Fruit ripening integrate 134 data sets 79,322 tissue- and stage- specificity [22] Capsicum chinense Jacq. Fruit ripening RNA-seq 20,563 1,1826 [64] Capsicum annuum Fruit ripening RNA-seq 11,999 366 [65] Capsicum annuum Fruit development ssRNA-seq 2,505 1,066 [66] Brassica rapa Vernalization RNA-seq 1,961 254 [42] Brassica rapa var. pekinensis Vernalization RNA-seq 2,088 549 [68] Brassica
campestris ssp. pekinensisVernalization ssRNA-seq 1,858 151 [69] Brassica rapa Pollen development RNA-seq 12,051 14 [43] Brassica rapa ssp. pekinensis Anther development SMRT 407 − [34] Brassica campestris ssp. pekinensis Anther development RNA-seq 2,384 1,344 [72] Brassica rapa ssp. pekinensis Cytoplasmic male sterility RNA-seq 3,312 529 [74] Brassica
campestrisMale sterile RNA-seq 13,879 361 [73] Capsicum annuum Cytoplasmic male sterilitye RNA-seq 10,655 1,137 [75] Solanum lycopersicum Sperm cell lineage
developmentssRNA-seq 31,931 cell/tissue-type specificity [76] Capsicum annuum Anthocyanin biosynthesis ssRNA-seq − 172 [44] Solanum tuberosum Anthocyanin Biosynthesis ssRNA-seq 4,376 1,421 [80] Solanum tuberosum Anthocyanin Biosynthesis RNA-seq 1,072 6 [81] Daucus carota Anthocyanin biosynthesis RNA-seq 7,288 639 [82] Solanum lycopersicum Trichome formation ssRNA-seq 1,303 196 [83] Solanum tuberosum Potato tuber sprouting RNA-seq 3,175 723 [87] Spinacia oleracea Flowering RNA-seq 1,141 111 [89] Spinacia oleracea Sex differentiation PacBio Iso-seq & RNA-seq 500 42 [37] Growth and development Brassica napus Oil
biosynthesisssRNA-seq & RNA-seq datasets 8,905 13 [90] Brassica oleracea
var. capitataCuticular wax
biosynthesisRNA-seq 4,459 148 [91] Abiotic stress Solanum lycopersicum Drought response RNA-seq 521 244 [45] Solanum lycopersicum drought-response RNA-seq 67,770 3,053 [103] Brassica napus Drought response RNA-seq - 477/706 [104] Solanum tuberosum Drought response NGS & SMRT 3,445 − [105] Brassica rapa Heat response ssRNA-seq 4,594 1,686 [46] Brassica juncea Heat and drought response RNA-seq 7,613 1,614 [107] Brassica rapa Heat response RNA-seq 18,253 1,229 [15] Brassica rapa ssp. pekinensis Heat response RNA-seq 278 65 [106] Brassica rapa ssp. chinensis
(NHCC)Cold and heat response RNA-seq 10,001 2,236 [108] Cucumis sativus Heat response RNA-seq 2,085 108 [109] Raphanus sativus Heat response ssRNA-seq − 169 [110] Solanum lycopersicum Chilling injury RNA-seq 1,411 239 [47] Capsicum annuum Chilling injury RNA-seq 9,848 380 [111] Solanum lycopersicum Fruit cracking RNA-seq 2,508 − [112] Solanum pennellii and M82 Salt response ssRNA-seq 1,044 154/137 [48] Cucumis sativus Waterlogging response RNA-seq 3,738 922/514/1,476/
1,270[115] Cucumis sativus Phosphate-deficiency
responsessRNA-seq 14,277 22 [121] Brassica
napusCadmium toxic response ssRNA-seq 5,038 301 [126] Biotic stress
Solanum tuberosum Phytophthora infestans resistance RNA-seq 2,857 133 [49] Solanum lycopersicum Phytophthora
infestans resistanceRNA-seq 28,256 688 [130] Solanum lycopersicum L3708 Phytophthora
infestans resistanceRNA-seq 9,011 196 [131] Solanum lycopersicum TYLCV resistance ssRNA-seq 1,565 529 [50] Solanum lycopersicum TYLCV resistance ssRNA-seq 2,056 345 [138] Biotic stress
Solanum lycopersicum Bacillus subtilis SL18r-induced tomato resistance against Botrytis cinerea RNA-seq − 55/34/15 [51] Solanum lycopersicum Pseudomonas putida Sneb821- induced tomato resistance against Meloidogyne incognita RNA-seq 3,371 78 [140] Solanum lycopersicum Pst resistance RNA-seq 2,609 Different in each comparison [52] Solanum lycopersicum PSTVd resistance RNA-seq 6,726 44 [141] Brassica campestris ssp.chinensis Makino Plasmodiophora brassicae resistance RNA-seq 1,492 114 [143] Brassica napus Plasmodiophora brassicae resistance ssRNA-seq 4,558 530 [144] Brassica rapa ssp. pekinensis Downy mildew resistance RNA-seq 3,711 − [145] Brassica napus Sclerotinia sclerotiorum resistance RNA-seq 3,181 931 [142] Brassica rapa Fusarium oxysporum
resistanceqPCR [146] Capsicum
annuumPhytophthora
capsica resistanceRNA-seq 2,388 607 [147] Solanum tuberosum Pectobacterium carotovorum resistance ssRNA-seq 1,113 559 [148] Solanum tuberosum Potato Virus Y resistance and heat stress RNA-seq 4,007 421 [149] Cucumis sativus Powdery mildew resistance ssRNA-seq 12,903 119 [150] Others Solanum lycopersicum Ethylene signaling RNA-seq 397 12 [151] Solanum
pimpinellifolium LA1589,
S. lycopersicum Heinz1706Lycopersicon specificity ssRNA-seq 413/709 92/161 [152] Brassica napus, B. oleracea and
B. rapaSpecies divergence RNA-seq 1,885/1,910/
1,299186 /157/161 [153] Capsicum chinense Heterosis effect ssRNA-seq 2,525 1,932/ 593 [154] Cucumis hytivus Allopolyploidization RNA-seq 2,206 1,328 [155] Brassica rapa, B. carinata, and
B. hexaploidPolyploidization RNA-seq 2,725/1,672/
2,810725 [156] 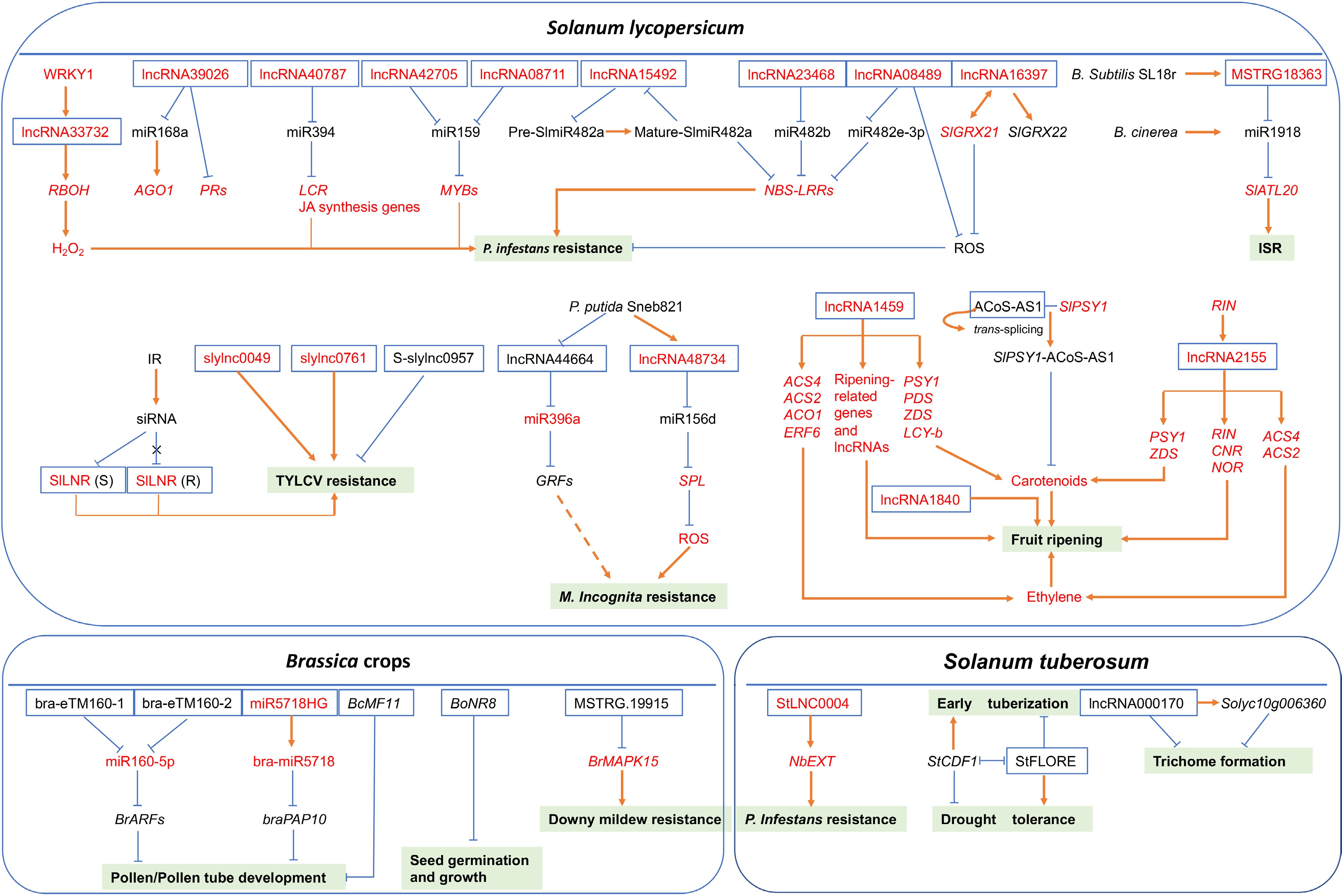
Figure 2.
The predictive model of regulatory mechanisms of lncRNAs with known functions under various developmental events or stress conditions in different vegetable crops. Full-line arrows represent positive regulatory interactions, blunt-ended bars represent negative regulation and dotted-line arrow indicates that the regulatory mechanism is unclear. White boxes with blue lines represent lncRNAs, light green boxes represent different developmental events or stresses. The letters in red denote positive regulators, while the letters in black denote negative regulators.
Table 2. Summary of functionally validated lncRNAs in major vegetables.
Species LncRNA name Biological functions Interaction targets References Solanum lycopersicum lncRNA000170 Trichome formation Solyc10g006360 [83] lncRNA1459, lncRNA1840 Fruit ripening − [61, 62] lncRNA2155 Fruit ripening RIN [38] ACoS-AS1 Trans-splicing; carotenoids biosynthesis SlPSY1 [63] lncRNA33732 Resistance to Phytophthora infestans RBOH [132] lncRNA16397 Resistance to Phytophthora infestans SlGRX21, SlGRX22 [130] lncRNA15492 Resistance to Phytophthora infestans Sl-miR482a [133] lncRNA08489 Resistance to Phytophthora infestans miR482e-3p [134] lncRNA23468 Resistance to Phytophthora infestans miR-482b [135] lncRNA39026 Resistance to Phytophthora infestans miR-168a [136] lncRNA40787 Resistance to Phytophthora infestans miR394 [137] lncRNA42705, lncRNA08711 Resistance to Phytophthora infestans miR159 [131] slylnc0049, slylnc0761 Resistance to TYLCV − [50] S-slylnc0957 Resistance to TYLCV − [138] SlLNR1 Resistance to TYLCV − [139] MSTRG18363 Bacillus subtilis SL18r-induced tomato resistance against Botrytis cinerea miR1918 [51] lncRNA44664, Pseudomonas putida Sneb821- induced tomato resistance against Meloidogyne incognita miR396 [140] lncRNA48734 Pseudomonas putida Sneb821- induced tomato resistance against Meloidogyne incognita miR156 [140] Brassica oleracea BoNR8 Seed germination; root and silique growth − [21] Brassica rapa bra-eTM160-1, bra-eTM160-2 Pollen development miR160-5p [43] Brassica rapa ssp. pekinensis MSTRG.19915 Resistance to downy mildew BrMAPK15 [145] Brassica campestris bra-miR5718HG Pollen tube growth miR5718 [73] BcMF11 Pollen development; male fertility − [39, 40] Solanum tuberosum StFLORE Tuber development; drought response StCDF1 [88] StLNC0004 Resistance to Phytophthora infestans NbEXT [49] Participation in the growth and development of vegetables
Fruit development and ripening
-
Based on previous studies, many lncRNAs were found to be involved in fruit development and the ripening process of vegetable crops. Tomato is a model plant to study flesh fruit development and ripening, and emerging evidence has shown that lncRNAs play crucial roles in this process[22,38,41,60,61],. It was found that lncRNAs may function as ceRNAs of miRNA, interfering with the expression of genes associated with ethylene and carotenoid pathways or directing the methylation of some critical genes involved in fruit ripening[41]. Silencing of either lncRNA1459 or lncRNA1840 resulted in a repressed tomato fruit ripening process[61]. The knockout mutant of lncRNA1459 was obtained by using the clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (Cas9) system, which, in addition to severely delayed fruit ripening, significantly reduced ethylene biosynthesis and lycopene accumulation compared with wild-type, and meanwhile the expression of fruit-ripening-related genes and lncRNAs was also impaired[62]. RIPENING INHIBITOR (RIN) is one of the known core regulators of fruit ripening, in vivo and in vitro experiments have shown that lncRNA2155 could be targeted by RIN, lncRNA2155 knockout mutant exhibited delayed fruit ripening and the expression of ripening-related transcription factors, ethylene and carotenoids biosynthetic genes were also declined[38]. The ripening process of the tomato fruit is generally accompanied by the accumulation of carotenoids, and phytoene synthase (PSY) is the rate-limiting enzyme of carotenoid biosynthesis. Evidence showed that the trans-splicing between lncRNA ACoS-AS1 and its cognate sense transcript SlPSY1 may be responsible for the loss of function of SlPSY1, which further resulted in the yellow color of fruit in Solanum Lycopersicum var. cerasiforme accession PI 114490[63]. ACoS-AS1 was found to be an essential regulator of the trans-splicing event by generating ACoS-AS1 mutate, which gave rise to red fruit color in PI 114490[63]. Pepper is also an important vegetable worldwide and a model plant for studying the ripening process of non-climacteric flesh fruits. Yang et al. systematically identified 20,563 lncRNAs during three fruit development stages in C. chinense Jacq[64]. Among these, 11,826 were differentially expressed with 5,918 upregulated and 5,908 downregulated[64]. To investigate the regulatory roles of non-coding RNAs in bell pepper fruit ripening, Zuo et al. conducted RNA-seq to explore the expression pattern of lncRNAs in the bell pepper fruit ripening process, and 366 lncRNAs were discovered to exhibit distinct expression patterns in mature green and red ripe fruit[65]. LncRNAs were also involved in hot pepper fruit development, which was verified by comparative analysis of the lncRNA transcript abundance in successive fruit development stages[66].
Vernalization
-
Plants have evolved mechanisms to sense their environment and alter their growth and development for adaptation accordingly. Most varieties of Brassica vegetables must undergo low-temperature vernalization to realize the transition from vegetative growth to reproductive growth[67]. This process is crucial for floral organ formation as well as flowering time regulation. By conducting comparative transcriptome analysis, some lncRNAs were found to be differentially expressed before and after vernalization in Brassica crops[42,68,69]. Furthermore, some lncRNAs were identified as key lncRNAs involved in vernalization through bioinformatic analysis. For instance, in B. rapa, the antisense transcript of BrFLC and BrMAF, which act as repressors of flowering, may play a role in the transcriptional response to vernalization[68]. In Chinese cabbage, the vernalization-related lncRNAs, cirRNAs, miRNAs, and mRNAs were screened for ceRNA network construction, and several lncRNAs were identified as valuable candidates in the vernalization pathway based on this network[69].
Pollen development and male sterility
-
Male plant sterility, broadly defined as the inability to produce dehiscent anthers, functional pollen, and viable male gametes, opens up new avenues for the utilization of heterosis. In 1763, male sterility was first observed by German botanist Joseph Gottlieb Kolreuter, and more than 610 plant species have been reported to be sterile[70,71]. At present, many Brassica crops have abundant male sterility variant materials, which have been widely utilized in production, but the molecular mechanism of male sterility is still elusive. Pollen abortion is a phenotypic feature of male sterility; therefore, a more in-depth exploration of the molecular regulation mechanism of pollen or anther development is an effective method to understand male sterility. LncRNAs were identified as participants in the process of pollen/anther development and male sterility in Brassica crops[34,43,72−74]. For example, an RNA-seq experiment was performed to investigate the dynamic gene expression changes during successive pollen development stages of B. rapa. It is worth noting that 14 lncRNAs were revealed to be strongly co-expressed with 10 function-known coding genes which were related to pollen development. In particular, further exploration of these lncRNAs demonstrated that two lncRNAs, braeTM160-1 and bra-eTM160-2, were negatively involved in pollen formation and male fertility by acting as eTMs of miR160-5p, which further released the transcript of ARF genes[43]. Another study performed whole transcriptome sequencing to enclose the regulatory network of pollen development in different B. campestris sterile lines, of which bra-miR5718HG was demonstrated to reduce the expression of miR5718 and upregulate purple acid phosphorylase 10 (braPAP10), thus inhibiting the growth of pollen tubes and influencing seed set[73]. BcMF11 is a lncRNA that was strongly expressed in the floral organs, and it was confirmed to play an essential role in pollen development by conducting antisense RNA strategy-mediated downregulation of BcMF11 transcript, which leads to abnormal pollen development[39,40]. In addition to Brassica crops, lncRNAs are regarded as a critical regulator in the floral bud development process in pepper through performing RNA-seq and bioinformatic analysis of the transcript abundance in the cytoplasmic male sterility (CMS) line and maintainer line, which laid the foundation for further study of the molecular mechanisms underlying CMS[75]. Moreover, by conducting strand-specific RNA sequencing (ssRNA-seq), lncRNAs were found to be involved in sperm cell lineage development in tomato[76].
Anthocyanin biosynthesis
-
Anthocyanins are important pigments that are beneficial to health and have major contributions to the quality of fruit[77,78]. At present, the biosynthetic pathway of anthocyanins is well understood, and key regulatory genes have been identified in many species[79]. However, the role of lncRNAs in anthocyanin biosynthesis remains unclear. It is known that anthocyanins are accumulated under light exposure, and in pepper, 172 differentially expressed lncRNAs were identified on the light-exposed and shaded surface of pepper fruit[44]. In potato, Tang et al. found 1,421 differentially expressed lncRNAs between purple- and yellow-fleshed potato tubers. Furthermore, through constructing a lncRNA–mRNA interaction network, lncRNAs such as XLOC_060098 and XLOC_017372 were identified as positive regulators in anthocyanin biosynthesis by target anthocyanin-associated genes[80]. LncRNAs were also implicated in the anthocyanin biosynthesis of potato leaves[81]. Gene annotation suggested that lncRNAs could regulate the expression of PAL, F3H, and CHS, which are critical genes in the anthocyanin biosynthesis pathway and thus modulate the color of potato leaves[81]. Carrot is also an important vegetable that has been cultivated for thousands of years. Carrots were originally purple, and modern yellow varieties were domesticated from mutants lacking anthocyanins. By comparative analysis of the expression profile of lncRNAs in two carrot genotypes with a strong difference in anthocyanin accumulation in roots, Chialva et al. identified 639 lncRNAs with distinct expression patterns between these two genotypes, of which the natural antisense transcript of DcMYB7 was suggested to play an important role in anthocyanin pigmentation[82].
Others
-
LncRNAs are also involved in other developmental events. In young tomato stems, 196 lncRNAs were discovered to be differentially expressed between woolly mutant LA3560 (Wo) and its non-woolly segregants (WT). Among them, lncRNA000170 and its cognate sense transcript, Solyc10g006360, exhibited a common expression trend, and overexpression of either of them could inhibit type I trichome formation[83]. Sprouting is the key factor leading to a quality deterioration of potato tubers and other huge storage losses[84]. Many studies have attempted to reveal the molecular mechanisms underlying potato sprouting[85,86]. Among them, the expression of 723 lncRNAs was distinct in potato tubers from dormancy to sprouting, and these lncRNAs may function by affecting the cellular components and cellular metabolic processes of potato apical buds[87]. Furthermore, a lncRNA named StFLORE together with its counterpart StCDF1 was found to be involved in tuber development and drought response by creating StFLORE knockout mutants and overexpression lines[88]. In spinach, several well-known flowering-related genes such as ELF, COL1, FLT, and FPF1 and also some important flowering transcription factor genes such as MYB, WRKY, GATA, and MADS-box were potential targets for lncRNAs[89]. Based on PacBio Iso-seq and Illumina RNA-seq data, Li et al. discovered 42 differentially expressed lncRNAs in male and female spinach flowers, suggesting the role of lncRNAs in sex determination[37]. In cabbage, Wu et al. identified a lncRNA homologous to Arabidopsis AtR8, BoNR8. Studies have shown that BoNR8 could respond to abiotic stress and negatively regulate seed germination and root and silique growth[21]. B. napus is a conventional oil crop with high economic value. Some lncRNAs were found to be important regulators in oil biosynthesis after comparative analysis of lncRNAs at multiple seed development stages and co-expression analysis[90]. Moreover, lncRNAs were implicated in cuticular wax biosynthesis in cabbage[91].
Participation in abiotic stress responses
-
Plants are constantly affected by adverse environmental factors. To survive under various abiotic stresses, plants have to rapidly activate defense mechanisms and adapt to stressful environments[92−94]. Among them, lncRNAs have been reported to be involved in multiple abiotic stress responses[5,95−99].
Drought is an important stress factor that affects the normal growth and development of plants. The research on the effects of drought stress on the growth and development of vegetables and crops has always been one of the hotspots in the field of stress research[100−102]. In a previous study, a total of 244 lncRNAs were identified and characterized in drought-exposed tomato leaves[45]. Some of them may act as eTMs of miRNAs or through lncRNA–mRNA interactions to respond to drought stress[45]. According to strand-specific RNA-seq, 67,770 lncRNAs were discovered at different anther development stages of tomato, of which 3,053 were drought-responsive[103]. In drought-tolerant B. napus Q2 and drought-sensitive B. napus Qinyou8, 477 and 706 lncRNAs were differentially expressed between the two genotypes under drought stress and rehydration treatment, respectively[104]. Furthermore, a co-expression network of lncRNAs and mRNAs was constructed for functional prediction of these lncRNAs[104]. In potatoes, the role of lncRNAs under drought stress was also explored. A total of 3,445 lncRNAs were identified in different periods of drought stress, and function enrichment analysis indicated that they may be involved in drought response by modulating the 'ubiquitin-mediated proteolysis' pathway[105].
In the 21st century, the frequent occurrence of extreme high-temperature events will bring a great threat to agricultural production. Growing evidence showed that lncRNAs may play an essential role in heat resistance in Brassica crops[15,46,106−108]. In Chinese cabbage, lncRNAs could interact with mRNAs and miRNAs to form a network that affected plant hormone pathways and responded to heat stress[46]. In B. juncea, lncRNAs can also respond to heat and drought stress by functioning as putative targets of miRNAs, or through interaction with abiotic-stress-related transcription factors[107]. Furthermore, in our previous study, 1,229 differentially expressed lncRNAs were identified as being heat-responsive in Chinese cabbage; they can confer thermotolerance by affecting the 'protein processing in the endoplasmic reticulum' and 'plant hormone signaling' pathways, as well as the expression patterns of HSPs and ABA receptor PYL genes[15]. The role of lncRNAs in cucumber and radish under heat stress has also been explored. In cucumber, a total of 2,085 lncRNAs were found to be differentially expressed when exposed to heat stress, and some of them may have executive functions by acting as ceRNAs to compete for miRNA binding sites with mRNAs[109]. Radish is a semi-hardy vegetable, and high temperature is one of the greatest threats to its growth and development. Through performing ssRNA-seq, 169 lncRNAs were predicted to be heat-responsive and one lncRNA–miRNA–mRNA combination was constructed that provided valuable clues for further studies to elucidate their precise functions[110].
Low-temperature storage is a common storage method for fruits and vegetables after harvest, but for cold-sensitive vegetables such as tomatoes and peppers, improper storage will often cause serious chilling damage. The regulatory relationship between lncRNA and fruit chilling stress has also been investigated in previous studies[47,111]. Combined with RNA-seq and bioinformatic analysis, 239 lncRNAs involved in chilling injury were identified in tomato, some of which may function by targeting chilling-injury-related genes[47]. In particular, a complex regulatory network composed of miRNAs, lncRNAs, and their regulatory targets was established to fully understand the molecular mechanism of lncRNAs in chilling stress response[47]. Likewise, 380 chilling-responsive lncRNA were identified in bell pepper, and their potential targets and relationship with miRNAs, circRNAs, and mRNAs were also assessed to uncover the influenced pathways and processes[111].
LncRNAs also play important roles in other types of abiotic stresses. In tomatoes, fruit cracking occurs easily under abiotic stresses. Plants have evolved defense mechanisms and regulatory networks to combat this damage. Xue et al. investigated the expression profiles of mRNAs and lncRNAs at different stages of saturated irrigation-treated tomato fruits, and some lncRNAs (XLOC 16662, XLOC_033910, etc.) were identified as participants in regulating tomato fruit cracking via a lncRNA–mRNA (hormone–redox–cell wall) network[112]. By examining the differences in the expression of lncRNAs before and after salt treatment in wild and cultivated tomato materials, Li et al. screened some salt-induced LncRNAs, which may affect tomato salt tolerance by regulating the expression of hormone-pathway-related genes[48]. Cucumber is characterized by a shallow root system. Limited availability of oxygen often occurs during the cucumber cultivation period in unfavorable environmental conditions, one of which is excess water in the soil, which causes leaf wilting, chlorosis, and necrosis and decreased growth rates and yields due to the lack of available oxygen required to support aerobic respiration[113,114]. Through conducting high-throughput RNA-seq, 71 lncRNAs were predicted as members participating in acquiring hypoxia tolerance under long-term waterlogging stress in cucumber, and some of them may function by interacting with miRNAs[115]. In plants, phosphorus is a macronutrient essential for plant growth and yield and plays an important role in nucleic acid, phospholipid composition, energy transfer, and signal transduction[116]. Available forms of phosphorus (phosphate, Pi) are generally low in soil, and many plant species have evolved complex adaptive responses to maintain Pi homeostasis[117−120]. LncRNAs were implicated in maintaining phosphate homeostasis in cucumber. Grafting studies combined with RNA-seq identified 22 lncRNAs that could serve as systemic signals during the early Pi deficiency response and can move a long distance from the source region into sink tissues[121]. Cadmium (Cd), a toxic heavy metal, is one of the main inorganic pollutants in the environment[122,123]. It can be freely absorbed and accumulated by plants, resulting in the disruption of nutrient homeostasis, the recurrence of toxicity symptoms, and interference with many physiological processes[124,125]. LncRNAs were also involved in mediating cadmium toxic response and detoxication in B. napus. Of the 5,038 lncRNAs identified, 301 were cadmium-responsive[126].
Participation in biotic stress responses
-
Vegetables often suffer from various biotic stresses during their growth and development, such as infection by fungi, bacteria, viruses, and nematodes[127]. Late blight is one of the most devastating diseases affecting Solanaceae crops and can cause a massive reduction in or even the extinction of potato and tomato production[128,129]. Phytophthora infestans is the causal agent of late blight; therefore, it is of great significance to study the resistance mechanism of tomato and potato to P. infestans. Based on the published RNA-sequencing data, Cao et al. discovered 133 lncRNAs involved in the resistance of P. infestans in potatoes and their regulatory mechanisms by constructing an interaction network[49]. It was remarkable that after transient transformation of StLNC0004 into tobacco, the expression of extensin (NbEXT) was activated, accompanied by the enhancement of resistance to P. infestans[49]. In tomato, the role of lncRNAs in P. infestans resistance has been widely explored[130,131]. Tomato lncRNA33732 activated by WRKY1 is positively involved in tomato resistance to P. infestans by inducing the expression of RESPIRATORY BURST OXIDASE (RBOH) and increasing H2O2 accumulation during early infecting stages[132]. lncRNA16397 could induce the expression of SlGRXs, resulting in a reduction in the accumulation of ROS and damage to the cell membrane, which in turn enhances tomato resistance[130]. Sl-lncRNA15492 acts against P. infestans infection via inhibiting the expression of mature Sl-miR482a, which could target Sl-NBS-LRR resistance genes[133]. Additionally, lncRNAs could function as ceRNA to modulate the expression of resistance-related genes by decoying miRNAs in the tomato-P. infestans interaction. Among them, lncRNA23468 and lncRNA08489 could decoy miR482b and miR482e-3p, respectively, to affect the expression of NBS-LRR genes[134,135]. lncRNA39026 can positively regulate Argonaute proteins 1(AGO1) by decoying miR168a and improve the transcript level of PR genes[136]. lncRNA40787 can suppress the expression of miR394, thereby improving the transcript abundance of LCR and changing the expression of JA-related genes[137]. Furthermore, some lncRNAs could modulate the expression of resistance-related transcription factors by decoying miRNAs, thus enhancing tomato resistance[131].
Apart from the role of lncRNAs in P. infestans resistance, they were also implicated in yellow leaf curl virus (TYLCV) infection responses[50,138]. Wang et al. identified 529 lncRNAs that could respond to TYLCV infection in the resistant tomato breeding line CLN2777a, and several lncRNAs could serve as miRNA target mimics involved in disease resistance. Two of the lncRNAs, slylnc0049 and slylnc0761, that exhibited a substantial increase after TYLCV inoculation, were functionally characterized by virus-induced gene silencing (VIGS), and it was found that silenced tomato plants accumulated more virus than controls[50]. Furthermore, the role of lncRNAs in virus resistance in TYLCV-susceptible tomato line JS-CT-9210 was explored, and silencing of S-slylnc0957 resulted in improved resistance of tomato to TYLCV infection[102]. In addition, in tomato, the host lncRNA SlLNR1 in susceptible but not in resistant cultivars could interact with viral siRNA which was derived from intergenic region (IR) of TYLCV genome, thereby affecting virus accumulation and disease development during TYLCV infection[139].
LncRNAs can also mediate Bacillus subtilis SL18r-induced tomato resistance to Botrytis cinerea, in which MSTRG18363 may modulate the expression of SlATL20 by decoying miR1918, thereby triggering the process of induced systemic resistance (ISR) against pathogens[51]. Yang et al. identified 78 lncRNAs that were implicated in Pseudomonas putida Sneb821-induced tomato resistance to Meloidogyne incognita, of which lncRNA44664 and lncRNA48734 could decoy miR396 and miR156, respectively, to competitively inhibit the expression of their target genes, thereby conferring resistance to M. incognita infection[140]. In addition, according to a comprehensive assessment of lncRNA expression profiles, lncRNAs were found to be involved in the immune response against Pseudomonas syringae pv. tomato (Pst) and potato spindle tuber viroid (PSTVd) in tomato[52,141].
As with all crops, Brassica species are constantly threatened by biotic stresses during production, resulting in huge economic losses. The role of lncRNAs in Brassica crops in mediating responses to Plasmodiophora brassicae, Hyaloperonospora brassica, Sclerotinia sclerotiorum, and Fusarium oxysporum was explored[142−146]. For instance, in Chinese cabbage, by comparing the lncRNA expression profiles before and after P. brassicae infection, 114 differentially expressed lncRNAs were identified, and 16 of them were predicted to interact with 15 defense-responsive genes based on the expression correlation between lncRNAs and mRNAs[143]. The role of lncRNAs in P. brassicae response was also explored in B. napus, of which 530 lncRNAs were found to exhibit distinct expression patterns in clubroot-susceptible and clubroot-resistant lines[144]. Downy mildew is an important oomycete disease threatening the production of Brassica vegetables worldwide. It was found that lncRNAs may participate in the disease defense response by regulating the expression of resistance-related genes[145]. Furthermore, silencing lncRNA MSTRG.19915 induced the expression of BrMAPK15 and improved resistance to downy mildew[145]. Additionally, 931 lncRNAs were involved in S. sclerotiorum infection response in B. napus[142]. Following F. oxysporum f. sp. conglutinans (Foc) inoculation, the expression of natural antisense lncRNAs was positively correlated with their cognate sense genes in B. rapa[146].
Comprehensive analysis of the expression of lncRNAs in Phytophthora capsici-resistant grafted peppers and susceptible samples revealed a total of 607 differentially expressed lncRNAs[147]. These lncRNAs participate in disease resistance responses in part through a lincRNA–miRNA–mRNA interaction network that regulates the expression of disease defense-related genes[147]. LncRNAs were also involved in resistance to Pectobacterium carotovorum and potato virus Y (PVY) in potato[148,149]. Kwenda et al. identified 559 lncRNAs that are P. carotovorum-responsive, and 17 lncRNAs were highly correlated with 12 defense-related genes through co-expression analysis[148]. A systematic RNA-seq analysis explored a comprehensive landscape of 4,007 lncRNAs in tomato infected by PVY at normal and elevated temperature status, of which 12 lncRNAs participated in stress response regulation by recruiting complex mechanisms based on eTM[149]. Cucumber downy mildew (DM) is the most serious epidemic disease in the production of cucumbers in solar greenhouses. After the onset of the disease, most of the leaves of the cucumbers can be withered, and the cucumber fields will turn yellow. To reveal the resistance mechanism of this disease, Nie et al. have performed ssRNA-seq and miRNA-seq to explore the roles of lncRNAs, mRNAs, and miRNAs in DM resistance[150]. According to the expression profiles in resistant and susceptible cucumber lines, a total of 119 lncRNAs were identified to be associated with DM resistance since their expression changed after inoculation with DM. Furthermore, a lncRNA–miRNA–mRNA interaction network was set up to reveal the action mode of lncRNAs in DM response[150].
Participation in other biological processes
-
Apart from participating in growth and development, in response to abiotic and biotic stresses, lncRNAs are also involved in many other biological processes in vegetable crops, such as ethylene response and species divergence[151−153]. Heterosis is a universal phenomenon in biology. The hybrid generation obtained by crossing different strains, varieties, and even different species often exhibits stronger growth rates and metabolic functions than its parents. Allopolyploid is a manifestation of hybrid vigor, which is obtained by doubling the chromosomes of hybrids produced by crossing different species. LncRNAs were found to be implicated in heterosis and displayed distinct expression patterns in allopolyploids and their parents[154,155]. In pepper, 1,932 lncRNAs were identified to be associated with heterosis, and a co-expression network was constructed to illustrate the functional modes of lncRNAs[154]. In Cucumis, the allotetraploid Cucumis hytivus was produced by chromosome doubling after crossing cultivated cucumber C. sativus with wild-type C. hystrix. Through systemic analysis of the transcriptome, 1,328 lncRNAs were found to be activated following hybridization. Some of their cis-regulatory targets were involved in the regulation of biological chloroplasts, and the others may be associated with epigenetic regulation of leaf verticillium and enhanced photosynthesis[155]. The function of lncRNAs in allopolyploidization was also explored in Brassica genus. Wang et al. discovered 725 differentially expressed lncRNAs between Brassica hexaploid and its parents, and the lncRNAs in the hexaploidy exhibited a significant paternal expression bias. The lncRNA–mRNA interaction network was constructed to visually display the relationship between lncRNAs and their potential target genes. Furthermore, the lncRNAs may perform their roles partially by functioning as ceRNAs or miRNA precursors[156].
-
The wide application of high-throughput RNA sequencing has provided revolutionary ways to discover novel lncRNAs[157,158]. In this review, we introduced the structures and expression features and highlighted the biological functions of lncRNAs in major vegetable crops. This work shows that lncRNAs could participate in a wide range of biological processes, including many development events, such as vernalizaion, fruit ripening, pollen or anther development, anthocyanin biosynthesis, flowering, and sex differentiation. LncRNAs were also confirmed to be involved in a serious of abiotic and biotic stress responses, such as drought, heat, cold, salt, chilling, P. infestans, TYLCV, Pst, and PSTVd (Table 1). However, in comparison with the research on humans and animals, the research involving plants is still in its infancy[159,160]. Although genome sequencing data have been reported for dozens of plants, annotations in most plant species lack information on lncRNAs, and studies of lncRNA functions are limited to only a few model angiosperms[161]. Among the species we explored in vegetable crops, the lncRNA research was mainly concentrated on tomato and Brassica crops, while few studies existed for other vegetable species. Furthermore, the function annotated lncRNAs are limited and only confined to a few cases (Fig. 2 & Table 2). Therefore, it is imperative to expand the research of lncRNA into other vegetables, and more efforts should be made towards a systematic analysis of the regulatory roles of non-coding RNAs in biological processes. Furthermore, the application of traditional reverse genetics based on highly efficient and stable plant genetic transformation systems, such as over-expression and RNAi as well as CRISPR/Cas9 gene-editing technology, would enrich our understanding of the precise function of lncRNAs in plants[162,163].
LncRNA regulates the function of its target genes in a cis or trans manner through various mechanisms of interaction with DNA, RNA, or proteins[164−166]. lncRNAs often work in highly intricate networks to regulate plant growth and development, as well as stress responses[167−169]. Several tools have been developed to predict the function modes of lncRNA, for example, the lncRNATargets platform was conducted to predict the interaction of lncRNAs and mRNAs, SpongeScan for lncRNAs and miRNAs, TF2LncRNA for lncRNAs, and transcription factors and RegRNA for identification of functional sites of lncRNAs[170−173]. Among them, the interaction between lncRNAs and miRNAs has received a lot of attention. In this paper, we outline many examples of the lncRNA functions as ceRNAs involved in fruit ripening, pollen development, resistance to P. infestans infection, salt stress tolerance, etc.[41,48,72,131]. Furthermore, the crosstalk network between lncRNAs and miRNAs was constructed by bioinformatics researchers for different vegetables under various experimental conditions, which expanded our knowledge of the function modes of lncRNAs. It is generally believed that the specific spatial structures of lncRNAs affect their interactions with other molecular elements, and the functional motif is necessary for physical interaction with various partners[174,175]. Therefore, it is of high importance to further explore the sequence motifs and secondary/tertiary structures, which is essential for fully elucidating the mechanisms of lncRNA regulation and developing new methods to predict lncRNA targets. These studies will provide a new perspective on the involvement of lncRNAs in the complex gene regulatory networks of plant growth and development and stress responses.
This work was supported by the National Natural Science Foundation of China (32172583), Natural Science Foundation of Hebei (C2021209005, C2021209019), and the China Postdoctoral Science Foundation (2020M673188, 2021T140097).
-
The authors declare that they have no conflict of interest.
-
# These authors contributed equally: Nan Li, Yujie Wang
- Supplemental Table S1 Published papers on lncRNAs in major vegetable crops.
- Copyright: © 2022 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Li N, Wang Y, Zheng R, Song X. 2022. Research progress on biological functions of lncRNAs in major vegetable crops. Vegetable Research 2:14 doi: 10.48130/VR-2022-0014
Research progress on biological functions of lncRNAs in major vegetable crops
- Received: 24 June 2022
- Accepted: 27 September 2022
- Published online: 27 October 2022
Abstract: With the advances in genomics and bioinformatics, particularly the extensive application of high-throughput sequencing technology, a large number of non-coding RNAs (ncRNAs) have been discovered, of which long ncRNAs (lncRNAs) refer to a class of transcripts that are more than 200 nucleotides in length. Accumulating evidence demonstrates that lncRNAs play significant roles in a wide range of biological processes, including regulating plant growth and development as well as modulating biotic and abiotic stress responses. Although the study of lncRNAs has been a hotspot of biological research in recent years, the functional characteristics of plant lncRNAs are still in their initial phase and face great challenges. Here, we summarize the characteristics and screening methods of lncRNAs and highlight their biological functions in major vegetable crops, including tomato, Brassica genus crops, cucumber, pepper, carrot, radish, potato, and spinach, which are implicated in the interaction of lncRNAs and miRNAs. This review enhances the understanding of lncRNAs' roles and can guide crop improvement programs in the future.
-
Key words:
- Long non-coding RNAs /
- Vegetable crops /
- Biological functions