-
Cucumber is an agronomically important vegetable crop cultivated worldwide. The cucumber fruit belongs to the pepo fruit that can be eaten fresh or processed into pickles, in which the fruit length is a predominant trait that directly affects crop yield and product appearance quality[1]. Fruit length is also an important index to distinguish cucumbers from different market types. There were significant differences in fruit length (5−60 cm) among different cucumber germplasm, with multiple genetic factors participating[2]. In a recent study, we elucidated that the natural variation in CRABS CLAW (CsCRC) contributes to fruit length divergence in cucumber[3].
CsCRC gene is a positive regulator conferring the allelic variations in the cucumber germplasms with different fruit length. CRC protein belongs to the YABBY family, which contains a conserved C2C2 zinc-finger and a DNA-binding YABBY domain[4−8]. In Arabidopsis, CRC signals were detected in the floral tissues, the nectary, the abaxial region of the carpel, the receptacles, sepals, and petals[9]. In contrast, we found CsCRC expression displayed a more concentrated signal in the cucumber carpel, developing ovary and nectary[3]. CRC has an active role in carpel development, nectary formation, floral meristem determinacy, and sex determinance[4,10,11]. Arabidopsis crc null mutants showed wider and shorter siliques with unfused carpels, reduced ovules, and lack of nectary[9,12]. Overexpression of CRC resulted in sepals developing some carpel characters and gynoecium transforming into cylindrical structures comprised of stigma and style in Arabidopsis[4]. To explore whether CsCRC has a redundant role with CRC in Arabidopsis, we first investigated the ectopic expression of CsCRC in wild type Landsberg erecta (Ler) and found an intense staining of proCsCRC:GUS in the developing stigma (Fig. 1a). We then transformed the CsCRC driven by the 35S promoter into the crc-1 mutant and selected three transgenic lines for phenotype observation (Fig. 1b). Overexpression of CsCRC rescued the unfused-stigma phenotype in crc-1 (Fig. 1b, c), but was unable to rescue the short siliques phenotype, suggesting that CsCRC has a partially conserved role in gynoecium development similar to CRC gene in Arabidopsis, while displaying a unique function in fruit length regulation in cucumber.
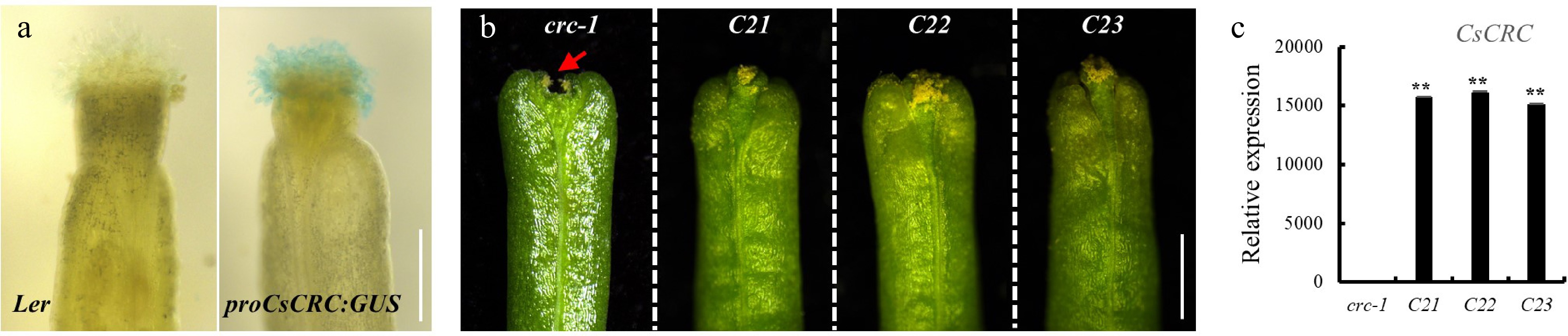
Figure 1.
Ectopic expression of CsCRC in Arabidopsis. (a) proCsCRC:GUS reporter activity in wild-type Arabidopsis Ler showing intense staining in stigma. (b) Ectopic expression of CsCRC rescued the unfused-stigma phenotype in Arabidopsis crc-1 mutant. (c) RT-qPCR analysis of CsCRC overexpression in crc-1 mutant. The arrow indicates the stigma defect in crc-1. **: significance at P < 0.01. Three biological replicates and three technique replicates were performed for each RT-qPCR analysis. Scale bar = 1 mm.
The gynoecia defects in the crc mutant can also be partially restored by auxin activity[13]. CRC participates in auxin homeostasis by inhibiting TORNADO (TRN) transcription, which modulates auxin maxima in Arabidopsis[14]. In cucumber, CsCRC acts as a positive regulator in fruit length elongation through activating the downstream auxin responsive protein CsARP1 and promotes cell expansion. To investigate the auxin role in cucumber fruit length regulation, we performed exogenous IAA and polar auxin-transport inhibitor N-1-naphthylphthalamic acid (NPA) treatments in cucumber inbred line R1461. IAA treatment resulted in elongated fruit length and enlarged cell size, whereas NPA treatment gave rise to the opposite effects, suggesting that auxin promotes fruit elongation via cell expansion in cucumber (Fig. 2a−c). Interestingly, the expression of CsARP1 and CsCRC were significantly induced by IAA and repressed by NPA (Fig. 2d). These data suggests that auxin promotes fruit elongation through elevated cell expansion and enhanced CsCRC and CsARP1 expression in cucumber.
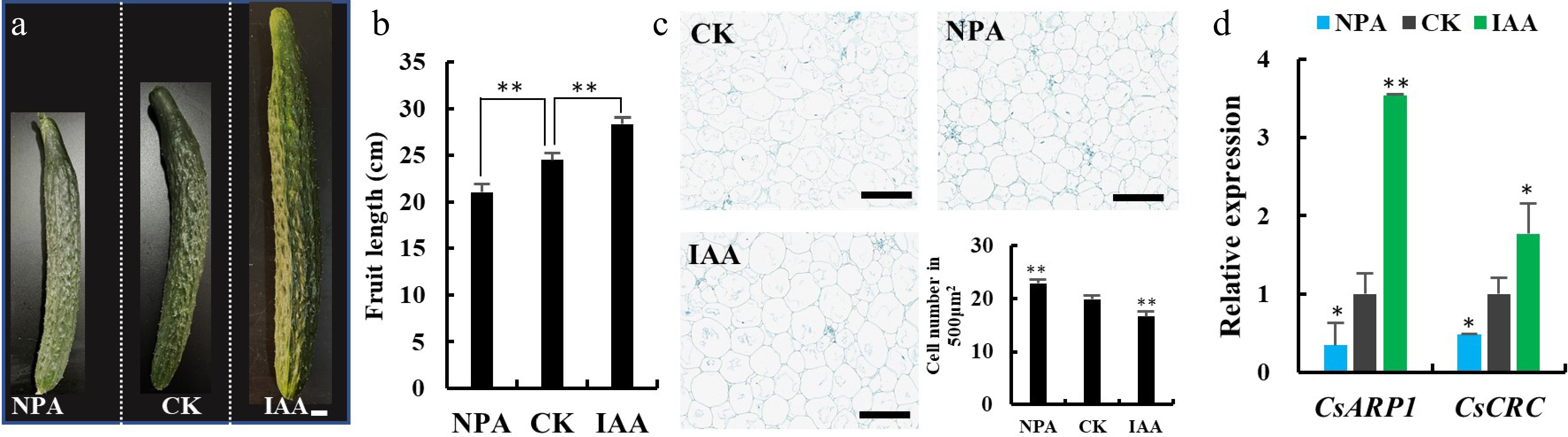
Figure 2.
Auxin promotes fruit elongation and activates the expression of CsCRC and CsARP1. (a) Cucumber fruits at commercial stage upon IAA and NPA treatments. (b) Quantification of fruit length at 10 DAA after treatment. (c) Longitudinal sections in the mesocarp fruits at 10 DAA. Cell numbers in the mesocarp of 10DAA fruit were counted. (d) The relative expression of CsARP1 and CsCRC in cucumber ovaries upon treatment. Scale bars are (a) 1 cm and (c) 200 μm. *: significance at P < 0.05; **: significance at P < 0.01. Three biological replicates and three technical replicates were performed for each RT-qPCR analysis.
Auxin signaling pathways has an interplay mechanism with light signal that antagonistically regulates plant growth[15]. In a previous study, we revealed that the rare CsCRCA allele only exists in a few semi-wild Xishuangbanna cucumber with short or round fruits. The Xishuangbanna area is close to the tropics and has more light intensity than northern China. The phytochromes, which are stimulated by light, promote the degradation of the PHYTOCHROME INTERACTING FACTOR (PIF) proteins and enhance the accumulation of auxin transcriptional repressors[16]. We analyzed the 2038-bp promoter of CsCRC, and found one G-box and two E-boxes, which are the specific binding sites for PIFs (Fig. 3a). We then conducted Y1H assay and found that CsPIF1 can directly bind to the promoter of CsCRC (Fig. 3b). To investigate whether CsPIF1 regulates CsCRC expression in vivo, transient expression assays were performed in tobacco leaves, in which the 2038 bp upstream promoter region of CsCRC fused into the luciferase (LUC) gene was used as the reporter, and the coding sequence of CsPIF1 driven by the 35S promoter was used as the effector (Fig. 3c). The LUC activity was significantly enhanced upon co-transformation of 35S:CsPIF1 with proCsCRC:LUC (Fig. 3d). This data supports the fact that CsPIF1 directly promotes the expression of CsCRC. Therefore, we presumed that the strong light intensity in Xishuangbanna reinforces CsPIF1 degradation, thus inhibiting CsCRC expression and results in reduced fruit length.
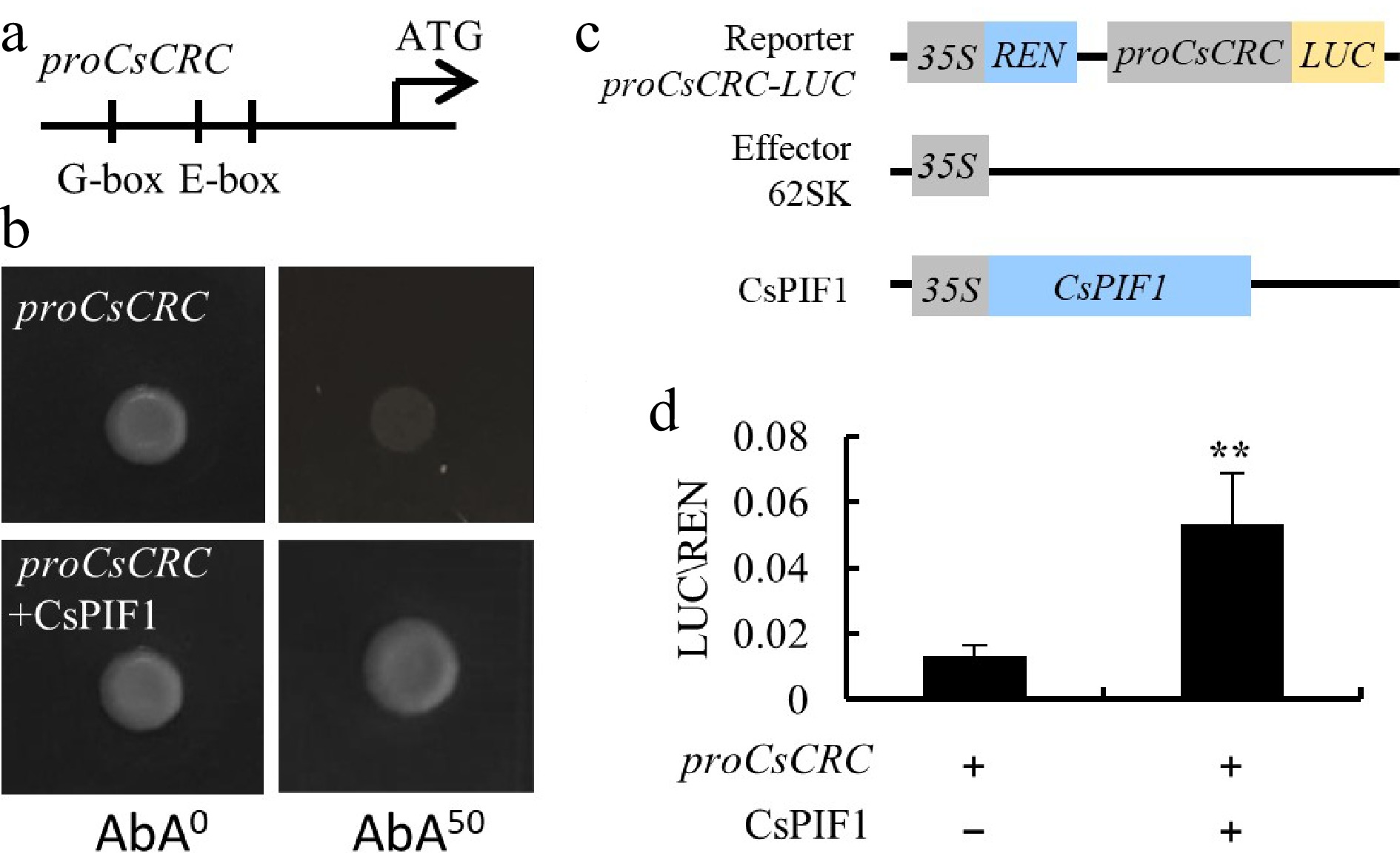
Figure 3.
CsPIF1 is the upstream regulator of CsCRC. (a), (b) Y1H assay showed that CsPIF1 bound to the G-box at the promoter of CsCRC. (c) Schematic diagram of the reporter and effector constructs used for LUC/REN assays. (d) LUC activity measurement after transient expression of proCsCRC:LUC with 35S:CsPIF in tobacco leaves. Error bars represent standard errors. '**: significance at P < 0.01.
Fruit development is fine-tuned by multiple genes involved in several aspects of the reproductive growth phase. YABBY gene family contains six members: CRC and INNER NO OUTER (INO), regulate reproductive growth, while other vegetative YABBY genes, FILAMENTOUS FLOWER (FIL), YAB2, YAB3, and YAB5, control leaf-derived organ development[17]. In Arabidopsis, CRC can enhance SPT function in controlling carpel development[18]. Another reproductive YABBY gene INO is expressed in floral organs and has an important role in seed formation[5,19]. To explore their potential role in the CsCRC-mediated cucumber fruit length regulation pathway, we performed protein interaction assays. Yeast two-hybrid analyses showed that CsCRC directly interacted with CsSPT1 and CsINO in vitro (Fig. 4a). The interactions were confirmed by performing BiFC in tobacco leaves (Fig. 4b). Therefore, CsSPT1 and CsINO may interplay with CsCRC to participate in cucumber fruit development. Further elucidation of the CsCRC-mediated gene network will shed a light on the molecular mechanisms affecting fruit development in cucumber.
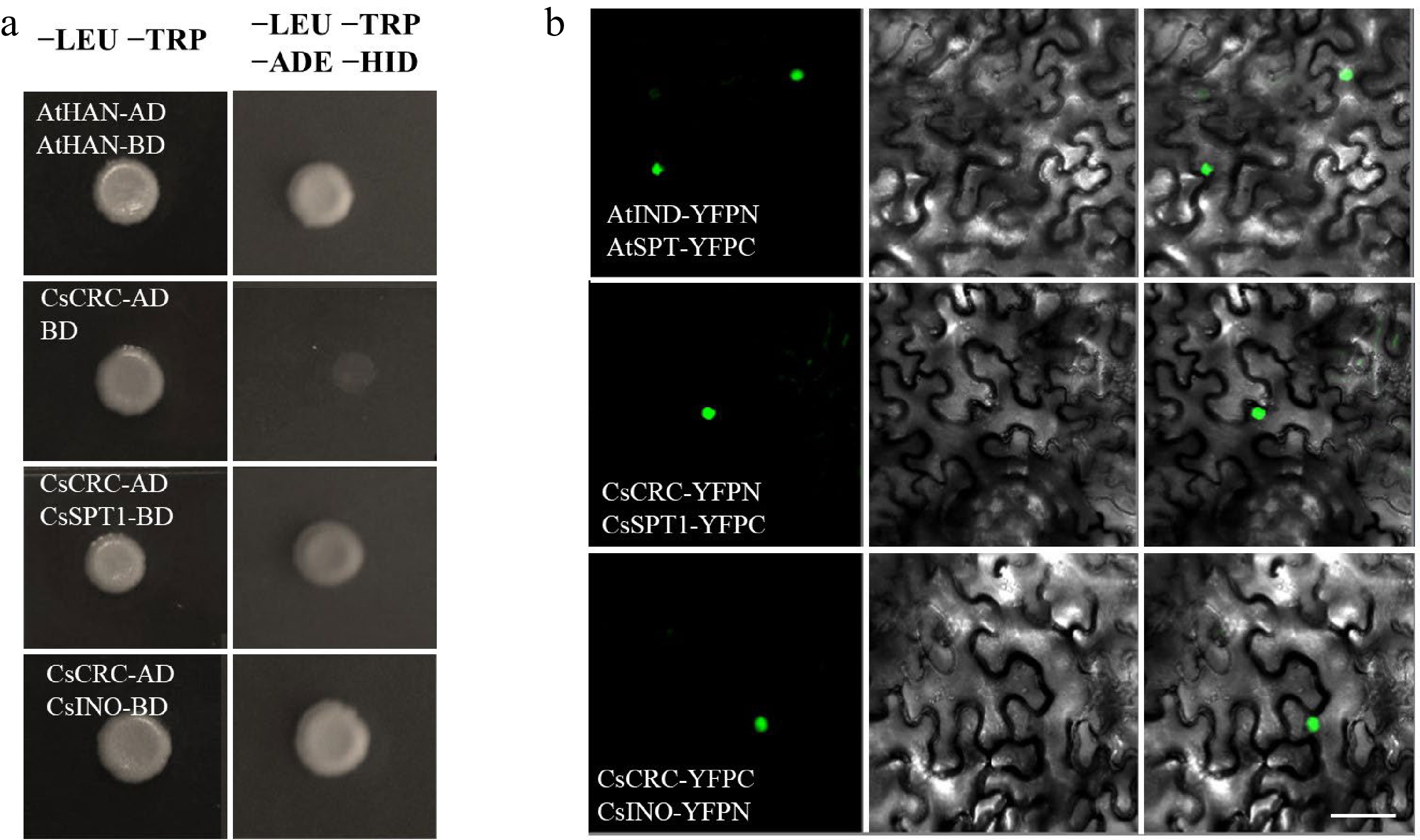
Figure 4.
Protein interactions detected with Y2H and BiFC assays. (a) Y2H assay showed the interaction of CsCRC with CsSPT1 and CsINO. The interaction of AtHAN-BD and AtHAN-AD was used as the positive control. (b) BiFC assay. The combination of AtIND-YFPC and AtSPT-YFPN was used as a positive control. Dark-field, bright-field and merged channels are shown sequentially from left to right. Scale bar = 50 μm.
-
The Landsberg erecta (Ler) Arabidopsis and the crc-1 mutant were obtained from Zhang's lab. The Arabidopsis were cultivated in a growth chamber under 16/8 h light/dark at 22 °C. Cucumber inbred line R1461 were grown in the glasshouse of Inner Mongolia University in Hohhot under standard environment control. The GUS reporter gene recombinant construct and CsCRC overexpression vector were transferred into Agrobacterium and transformed into Ler and crc-1 mutant.
RT-qPCR
-
The young ovaries of R1461 were used for RNA extraction. The synthesized cDNA was used as the template for qPCR. The SYBR Premix Taq Mix (Takara) was used for RT-qPCR assay and an Applied Biosystems 7500 real-time PCR system was used for statistical analysis. Arabidopsis ACTIN2 and cucumber UBI were used as reference genes, respectively. For each RT-qPCR analysis, three biological replicates and three technical replicates were performed.
Yeast One-Hybrid assay
-
CsPIF1 coding sequence was cloned into the pGADT7 vector (Clontech, CA, USA). The putative PIFs binding sites were found at the promoter of CsCRC and then cloned with the pAbAi vector (Clontech, CA, USA). The resultant constructs were introduced into the yeast Y1H Gold strain and selected by different AbA (Aureobasidin A) concentration on the SD/-LEU (Synthetic Dropout Medium/-Leucine) medium (Clontech, CA, USA).
LUC/REN assay
-
The 2038 bp upstream sequence of CsCRC was inserted into pGreenII 0800 vector and used as reporter. The coding sequence of CsPIF1 was cloned and constructed with pGreenII 62-SK vector. The constructs were introduced into Agrobacterium tumefaciens strain GV3101 together with pSUPER and p19 plasmid[20]. The bacterial fluid of effector: reporter (9:1) construct cell mixture was injected into young leaves of Nicotiana benthamiana. The LUC/REN ratio was measured by luciferin (Promega, USA). A strain of the proCsCRC and empty effector was used as a negative control.
Yeast Two-Hybrid assay
-
Coding sequence of CsCRC, CsSPT1, and CsINO genes were inserted into pGADT7 and pGBKT7 vectors. All constructs were transformed into yeast strain AH109 for preparation. Yeast two-hybrid assays were performed according to the description of Matchmaker TM GAL4 Two-Hybrid System 3 & Libraries (Clontech, CA, USA). Combination of each gene with blank vector pGADT7 and pGBKT7 were used as negative control. The primers used for Y2H vector construction are listed in Supplemental Table S1.
Bimolecular Fluorescence Complementation (BiFC) assay
-
The coding sequence of CsCRC, CsSPT1, and CsINO which are lacking a stop codon were inserted into the fluorescence vectors pSPYCE-35S and pSPYNE-35S. Those constructs were transformed into Agrobacterium GV3101. Each combination of two proteins was injected into the young leaves of tobacco. The infected tobacco leaves were sampled and observed through Zeiss LSM 510 Meta confocal laser microscopy under 488 nm excitation wavelength. AtIND-YFPC and AtSPT-YFPN interaction was used as a positive control[21].
This study was supported by the National Natural Science Foundation of China (32202513, 32025033, and 31930097) and Natural Science Foundation of Inner Mongolia Autonomous Region (2021BS03002).
-
Xiaolan Zhang and Gen Che are the Editorial Board members of journal Vegetable Research. They were blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of these Editorial Board members and their research groups.
- Supplemental Table S1 List of primers used in this study.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Che G, Song W, Zhang X. 2023. Gene network associates with CsCRC regulating fruit elongation in cucumber. Vegetable Research 3:7 doi: 10.48130/VR-2023-0007
Gene network associates with CsCRC regulating fruit elongation in cucumber
- Received: 09 December 2022
- Accepted: 04 January 2023
- Published online: 02 March 2023
Abstract: In cucumber (Cucumis sativus L.), fruit length is a crucial agronomic trait that affects appearance quality and crop yield. Different cucumber germplasms exhibit a large variation in fruit length, and the underlying candidate genes and regulatory mechanisms need to be further characterized. Recently we showed that an essential gene in modulating the cucumber fruit length, the YABBY transcription factor CRABS CLAW (CsCRC), activates its downstream target gene auxin responsive protein CsARP1 to promote cell expansion and fruit elongation. Here, we show that auxin activity and three fruit development-related genes are involved in the CsCRC-regulated gene network in cucumber. We found that a PHYTOCHROME INTERACTING FACTOR (CsPIF1) protein elevates CsCRC transcription by directly binding to its promoter. The protein interaction assays showed that a reproductive YABBY transcription factor INNER NO OUTER (CsINO) and a bHLH protein SPATULA (CsSPT1) have direct interaction with CsCRC, suggesting their redundant role in cucumber fruit development.
-
Key words:
- CsCRC /
- gene network /
- cucumber /
- fruit development













