-
Trichomes, specialized epidermal appendages, are widely distributed and can be found on approximately 30% of vascular plant species[1]. They grow on plant epidermal tissues as specialized structures and are diverse in form and size, consisting of one cell or multiple cells, like branched or unbranched. While some trichomes hold glands others do not. As the outermost structures, trichomes are thought to be the result of ecological adaptation of plants during long-term evolution, helping them cope with biotic and abiotic stresses. On the one hand, the presence of multicellular trichomes can reduce UV damage, water loss, and increase tolerance to freezing[2]. On the other hand, trichomes protect against predators (including fungi, bacteria, viruses, and insects) by synthesizing or secreting toxic secondary metabolites[3].
Plant glandular trichomes are capable of synthesizing or secreting a variety of important compounds, such as amino acids, polysaccharides, terpenoids, alkaloids, and polyphenols[4,5]. These compounds not only confer plants with specific smells, but can also be used as natural pesticides, medicines, perfumes and more. For example, wild tomatoes (Solanum habrochaites) are resistant to pests through physical structures or chemical compounds like sesquiterpenes[6]. The anti-malarial drug, artemisinin, could be synthesized, stored and secreted by the annual wormwood glandular trichomes[7]. The psychoactive and therapeutic anesthetic, cannabinoids, is extracted from the glandular trichomes of Cannabis sativa[8]. Glandular trichomes are hence described as a 'chemical factory'. Terpenoids that are repellent or toxic to pests are abundant and diversely. They can also attract predators or parasitic natural enemies[9,10]. A large number of terpenoid metabolites are conserved throughout the plant session and essential for growth and development[4,11].
Efficient isolation and purification of high-quality trichomes is necessary to identify more regulatory genes controlling trichome development. Several methods have been explored for the isolation of Arabidopsis trichomes , including separation by dissecting forceps[12], isolation with glass microcapillary tubes[13], collection by laser microdissection[14], and freezing paintbrush collection[15]. Recently, some researchers proposed an efficient and high-quality method with two steps. Firstly, plant seedlings are gently agitated for a long time in the presence of the cationic chelator EDTA, which mitigates the interaction of the epidermal trichomes with the epidermis. This is followed by centrifugation using a discontinuous sucrose gradient to obtain purified trichomes. This method will be applicable to the isolation of glandular and non-glandular trichomes in tomato and tobacco[16].
Recently a lot of excellent research articles and reviews related to the genetic control of trichome formation and metabolites biosynthesis have been published extensively[17−22]. However, to complement these excellent literatures, this review focuses on the roles and regulatory mechanisms of multicellular trichomes in common vegetable crops mainly focusing on tomato and cucumber, as well as the regulatory mechanisms of specialized metabolites produced by glandular trichome. Lastly, this review outlines the challenges and directions for further comprehensively understanding specialized metabolites.
-
Multicellular trichome structures exhibit diverse in different morphology in different plants, or even in different parts of the same plant. Trichomes can be categorized into glandular and non-glandular trichomes, depending on the presence or absence of the glandular head. Glandular trichomes typically consist of a head that secretes specific metabolites, a stem that supports the head, and a base that connects the stem to the epidermal cells. Conversely, the model plant Arabidopsis thaliana has only single-celled non-glandular trichomes. Glandular trichomes can be further classified into two types: peltate and capitate, based on their morphology and structure[23]. Both capitate and peltate types have a basal cell, but capitate trichomes typically have longer stalks and smaller heads compared to peltate trichomes. Non-glandular trichomes generally differ only in size while capitate trichomes mainly secrete non-volatile or less volatile substances that exude onto the trichome surface. In tobacco or wild tomatoes, the thick metabolites produced from glandular trichome heads drip down the trichome stems, becoming a trap for insects, which get stuck to the leaves and eventually die[24]. In contrast, peltate trichomes in mint (Mentha sp.) and sage (Salvia officinalis) have a sub-epidermal storage compartment located above the glandular cells. When insects touch these trichomes, the cuticle ruptures and the metabolites in the storage compartment are released into the apical space[25]. With the availability of complete genomic information and advanced genetic transformation techniques, tomatoes have emerged as a model plant for studying the molecular mechanisms underlying multicellular trichome formation and metabolite synthesis. Next, we present an overview of the classification of multicellular trichomes in common vegetable crops.
In tomato, the multicellular trichomes play a considerable role in resisting various stresses, which have been extensively studied by many researchers. There are seven distinct types of trichomes, categorized into glandular and non-glandular trichomes. Type I, IV, VI, and VII are glandular, whereas type II, III, and V are non-glandular[26] (Fig. 1). Further, types I and IV are capitate trichomes and types VI and VII are peltate trichomes. In wild tomato relatives, an intercellular storage cavity allows metabolites to accumulate and release when glandular cells break off. However, in cultivated types, type VI glandular trichomes have a four-leaf clover shape, consisting of four glandular cells and a smaller inner lumen with reduced storage capacity for compounds[27]. Interestingly, type IV glandular trichomes, which are shorter in length, are found exclusively in wild varieties such as Solanum habrochaites and Solanum pennellii. Notably, trichomes of type I, VI, and VII are common in both wild varieties and cultivars[28]. Overall, the various types of trichomes in tomatoes serve distinct functions, and studying their morphology and composition can reveal important insights into the plant's defense mechanisms.
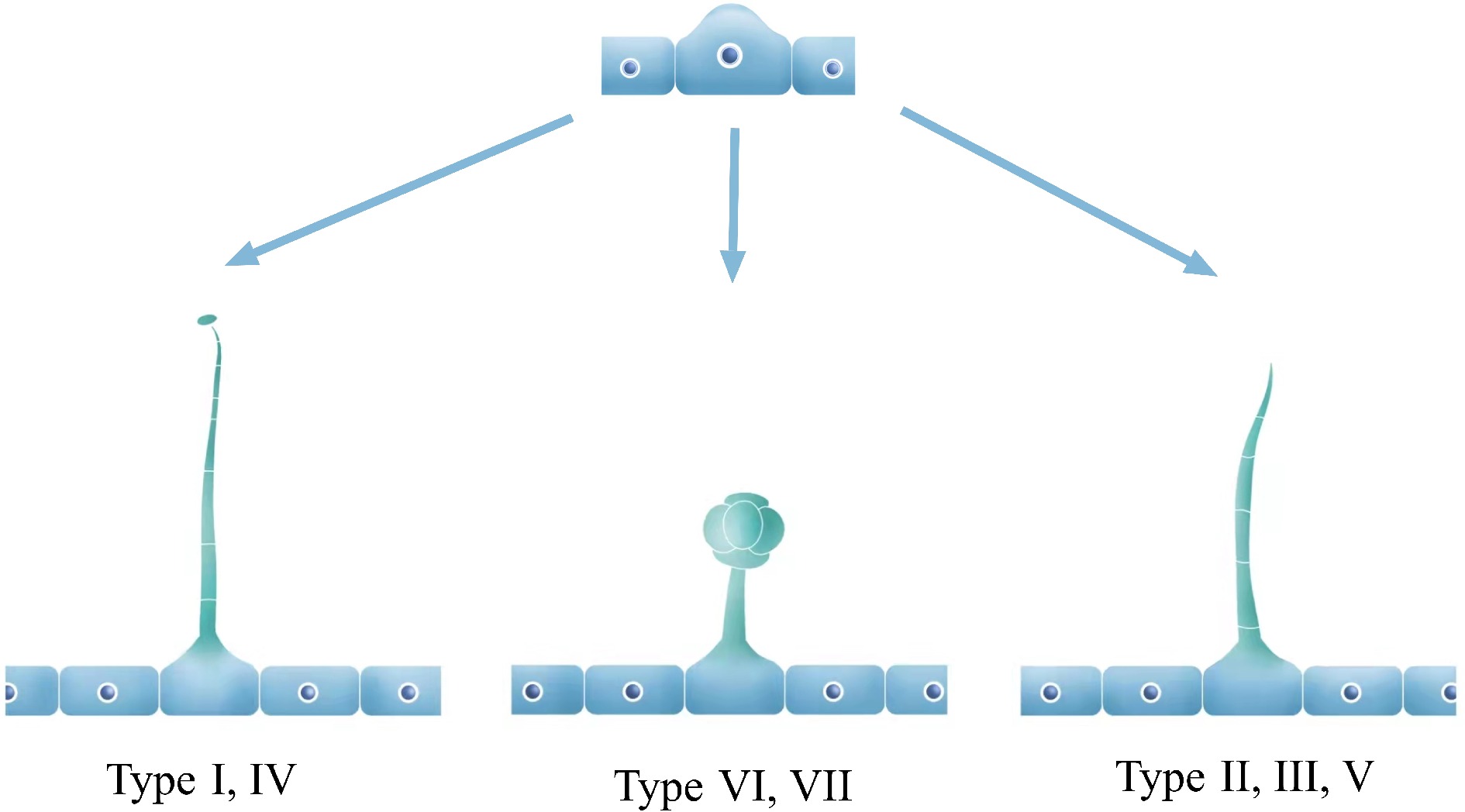
Figure 1.
Model diagram of tomato epidermal trichomes. In tomato, epidermal trichomes that protrude outward from the epidermal cells usually develop into three common types: long glandular trichomes (Type I, IV), short glandular trichomes (Type VI, VII), and long non-glandular trichomes (Type II, III, V).
In cucumber, trichomes originating from epidermal cells which are highly specialized structures with distinct morphologies. Xue et al. classified trichomes into eight different types (I-VIII, Fig. 2) based on the microscopic morphology of fruit trichome in 200 cucumber varieties[29]. Of these, only types I and VI are glandular, with type I trichomes being peltate structures consisting of a short monochromatic stalk of 3−4 cells and a head of 4−5 cells. These glandular trichomes are mostly distributed on the fruit surfaces and are associated with the production of mineral elements[30,31]. Type I trichomes are present on the fruit of all cucumber varieties except for csgl1 and csgl3 glabrous mutants[29]. Type VI trichomes, on the other hand, are capitate and have longer stalk cells than type I, with 4 or 5 cellular glands in the head. Type II non-glandular trichomes are also known as 'fruit spines', an important quality trait[32].
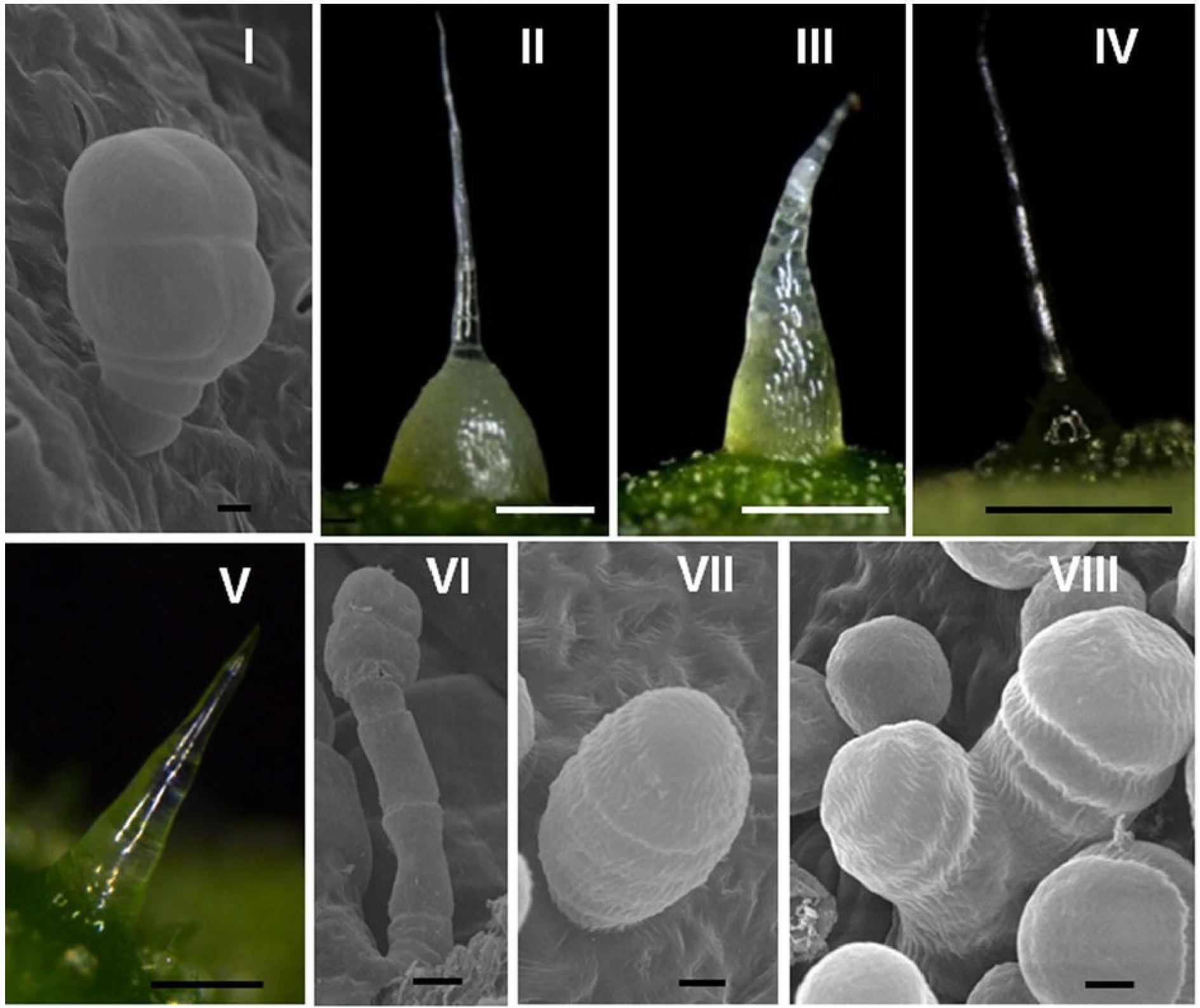
Figure 2.
Epidermal trichomes of eight types of cucumber. Bar = 500 μm (II–IV); 100 μm (V); 20 μm (VI); or 5 μm (I, VII, and VIII). This image is quoted from Xue et al.[29]
Recently, researchers found that four types (types I, IV, VI, and VII) are glandular and five types (types II, III, V, VIII, and IX) are non-glandular in pepper. Type I trichomes have a long multicellular stalk with a glandular cell at the tip, occurring only on the stems and leaves of Capsicum chinensis. Among the pepper trichome types, type VI and type VII exhibit similar length and structure, but with varying glandular cell number and morphology at later stages of development. Types VIII and IX are only observed at the top margin of the petals, with shorter non-glandular trichomes and larger and rounder stem cells than other types[33]. Overall, understanding the morphology and composition of trichomes in cucumber and pepper varieties can provide insights into their specialized functions and the underlying mechanisms of fruit quality traits.
-
In recent years, there have been several excellent studies attempting to describe the different developmental stages of trichomes in different crops[21,31,34]. Han et al. divided the developmental processes into three stages: fate determination or initiation, branching, elongation and maturation[17]. Feng et al. broadly divided trichome development into four stages: identity determination, initiation, morphogenesis, and maturation[35]. For multicellular trichomes, it can be classified into five stages which are initiation, start of division, formation of tip with simultaneous gland head conversion, continued elongation with completion of gland head development, and completion of basal development with metabolic activity[21]. Regardless of the developmental stages, all of them express similar developmental mechanisms. The initiation and morphogenesis of glandular trichomes are currently well researched. Therefore, here we focus on an overview of the regulatory mechanisms of glandular trichome initiation and morphogenesis.
Regulatory mechanisms of multicellular trichome initiation in tomato
-
The initiation step of the multicellular trichomes is significant as it determines the subsequent developmental process. Genes that regulate epidermal trichome development have been cloned and functionally validated in a variety of plants. A multicellular organism produces many cell types during its development, and cells in different locations, sensing diverse signals, respond through intracellular signalling pathways, ultimately produce different cell fates. The subsequent differentiation process of trichome cells appears to be even more complex. Identifying the regulatory mechanisms of multicellular trichome development is essential for understanding metabolite synthesis. Genes, phytohormones, and environmental factors all play a vital role in regulating glandular trichome initiation.
The genes that regulate multicellular trichome initiation are transcription factors, cell cycle proteins and a range of regulatory complexes. In tomato, there are four main types of transcription factors that regulate glandular trichome initiation, such as Homeodomain-Leucine Zipper (HD-ZIP), Zinc Finger Proteins (ZFPs), basic helix-loop-helix (bHLH), and v-myb avian myeloblastosis viral oncogene homolog (MYB). Studies have demonstrated that the formation of type I trichomes is regulated by a dimer composed of HD-ZIP transcription factor Woolly (Wo) and the C2H2 ZFP transcription factor Hair (H). B-type cycB2, plays important roles in the transition of G2-to-M. Among which, SlCycB2 affecting the development of almost all non-glandular trichomes (type III and type V), along with the glandular ones of type I and type VI. The interaction of Wo with SlCycB2 may initiate multicellular trichome development, and the Wo-H-SlCycB2 complex may regulate the initiation of type I glandular trichome, but this has not yet been confirmed[36−38].
A recent study found that SlZFP8-like (SlZFP8L) interacts directly with H and positively affects the density and length of tomato trichomes by regulating SlZFP6, which is the target gene of H. Moreover, SlZFP8L can also interact with Wo to regulate trichome initiation[20]. We have also identified a novel HD-ZIP IV transcription factor, Lanata (Ln), that triggers the hairy phenotype through a missense mutation. Ln has been demonstrated to interact with Wo and H, and furthermore, SlCycB2 represses the transcriptional activation of SlCycB3 via Ln and vice versa[19]. The bHLH transcription factor MYC1 regulates the formation of type VI glandular trichome in tomato. MYC1 affects the development of these trichomes and positively regulates the synthesis of monoterpenes in stems and leaves trichomes, while simultaneously promoting the synthesis of sesquiterpenes in leaves but opposite in stems[39]. Silencing the gene SlMX1, encoding a SlMIXTA like MYB transcription factor, resulted in multiple trichomes on leaves[40], while overexpressing Mixta-like reduced the density of type VI glandular trichomes but did not affect that of type I and IV trichomes[41]. However, a renent study by Ying et al. found that overexpressing SlMIXTA-like in tomato fruit enhanced trichome formation[42]. This suggests that epidermal trichome may show different results in different plant regions. In addition to genes, long non-coding RNAs also regulate trichome formation. Among them, lncRNA000170 inhibits the formation of type I trichome on the lower stems of the adult transgenic plants after overexpression[43].
In addition, studies have shown that the development of multicellular trichomes is triggered by phytohormones and among them, Jasmonic acid (JA) plays an important role in tomato trichome initiation. Jasmonate ZIM-domain (JAZ) proteins are vital in the JA signalling pathway, functioning as inhibitor that suppresses glandular trichome development. In JAZ2 overexpressing plants, the transcript levels of Wo and SlCycB2 were significantly reduced in stem trichomes[44]. The woolly (wo) is a loss-of-function allele of the HD-ZIP IV transcription factor, and the wo-MYC1 regulatory module can be repressed by JAZ2 via a competitive binding mechanism[45]. Moreover, SlJAZ2 inhibits the activity of H and H-like (HL) through physical interactions. H and HL directly inhibit the expression of THM1, which encoding a negative regulator of trichome formation[46]. SlJAZ4 is a negative regulator, while the HD-ZIP gene SlHD8 is a downstream regulator of JA signaling that promotes trichome elongation. The module SlHD8-SlJAZ4 can mediate JA-induced trichome elongation in tomato[47]. Furthermore, overexpression of the bHLH transcription factor gene bHLH95 regulates the formation of trichomes through two Gibberellic acid (GA) biosynthesis genes, GA20ox2 and KS5[48]. In addition, the auxin response factor SlARF4 directly targets two R2R3 MYB genes, SlTHM1 and SlMYB52. Among these, SlTHM1 is specifically expressed in type II and VI trichomes and negatively regulates their formation in tomato leaves, while SlMYB52 is specifically expressed in type V trichomes and negatively regulates the formation of type V trichomes in tomato leaves. Both SlTHM1 and SlMYB52 work by targeting SlCycB2. Besides, increasing trichome density confers resistance to spider mites in tomato[49]. Subsequently, it was also found that downregulation of SlMYB75 increased the formation of type II, V and VI trichomes. Further investigation revealed that SlARF4 directly targets and represses the expression of SlMYB75, and SlMYB75 protein interacts with and activates the expression of SlMYB52 and SlTHM1. More importantly, SlMYB75 can directly target and increase the activity SlCycB2[50]. In addition, phytohormone-related genes such as SlIAA15, SlARF3,SlARF4 and JAI-1 are involved in the formation of glandular trichomes in tomato[49,51,52].
Until now, transcription factors that regulate various types of trichomes have been studied in depth in tomato. Among them, the transcription factors regulating tomato type I and VI trichomes are the most abundant. Additionally, phytohormones such as auxins, JA and GA, play a significant role in regulating the development of tomato trichomes (Fig. 3).
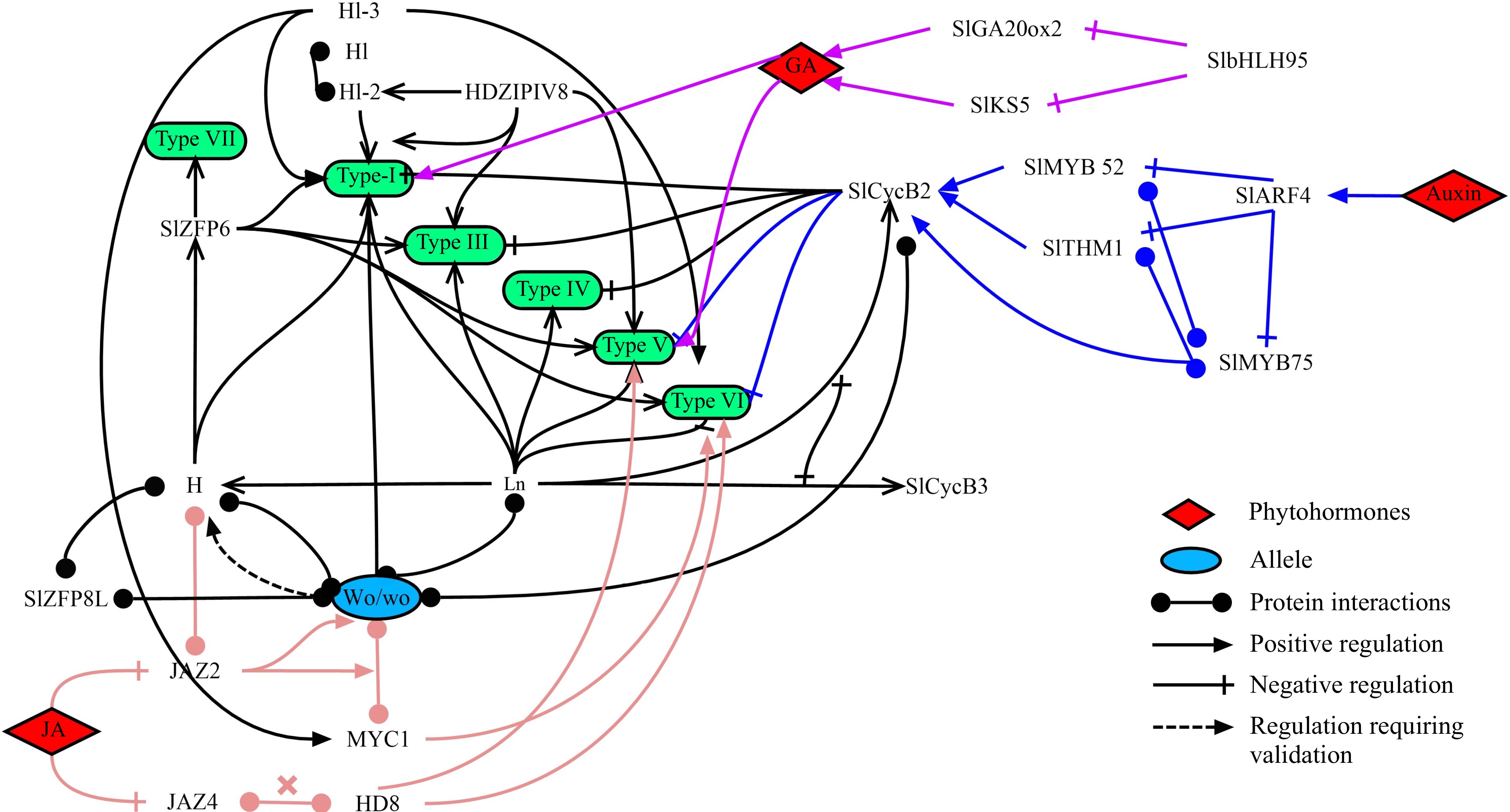
Figure 3.
A model for regulating different types of trichome development in tomato. Different types of tomato trichomes are presented in the green box. The colored lines correspond to different phytohormones-related regulatory pathways. The transcription factors Wo, H, Ln and CycB2 play an essential role in trichome development.
Regulatory mechanisms of multicellular trichome initiation in cucumber
-
In cucumber, four main categories of genes regulating multicellular trichome initiation are MYB, HD-ZIP, ZFPs, and WD-repeat (WDR) proteins. The HD-ZIP IV transcription factor genes, trichome-less (Tril) and its allele GLABROUS3 (CsGL3), play an important role in the fate determination and initiation of multicellular trichomes, exhibiting completely glabrous phenotypes on cucumber leaves, stems, flowers, and fruits when mutated[53,54]. Mutant phenotypic analysis suggests that the HD-ZIP transcription factors TINY BRANCHED HAIR (CsTBH), Micro-trichome (CsMICT) and GLABROUS1(CsGL1) are three alleles localized to Csa3M748220 and it may be involved in trichome morphogenesis but not in trichome fate determination and initiation[35,53,55]. The five genes are all mutant genes identified in cucumber and Tril/CsGL3 has an epistatic effect on TBH/CsGL1/MICT[56,57]. Silencing of TRANSPARENT TESTA GLABRA1(CsTTG1), encoding the WDR protein inhibits fruit spines formation[58], while molecular and genetic analyses suggest CsTTG1 has similar roles with CsMICT and CsGL1, the key trichome formation factors during trichome initiation[32]. The transcription factor TBH can bind to the promoter of the cucumber 1-aminocyclopropane-1-carboxylate synthase (CsACS) gene and regulate its expression. This modality regulates trichome development in cucumber type I and type II via the ethylene (ETH) pathway[59]. MYB transcription factors CsMYB6 and CsTRY both negatively regulate the initiation of cucumber trichomes. The gene MYB6 is located upstream of CsTRY and MYB6 negatively regulates CsTRY expression. Meanwhile, the CsMYB6-CsTRY complex negatively regulates the formation of trichomes in cucumber fruits[60,61]. The CsMYB6 was identified as one of the differentially expressed genes (DEGs) between the tiny branched hair (tbh) mutant and the WT. The gene CsGA20ox1 is presumed to encode a GA 20-oxidase that degrades active GA, and CsGA20ox1 expression was upregulated in the csgl1 mutant. Researchers speculate that CsGL1 may indirectly regulate the expression of CsMYB6 and CsGA20ox1, however, further study is needed to confirm this theory[56,62].
Recent research has revealed that the expression of CsMYB6 is strongly downregulated in the csgl1 mutant[61], speculating that CsGL1 may positively regulate MYB6 expression. The WDR protein gene CsTTG1 is highly expressed in cucumber ovary, and silencing the gene inhibits trichome germination, indicating its ability to negatively regulate trichome initiation in cucumber. At the same time, CsTTG1 also interacts directly with Mict to regulate the initiation of trichomes[32]. The tuberculate (Tu) fruit gene encoding C2H2 ZFP, could not expressed in the glabrous and tuberless mutant line gl, which contained the Tu gene. This indicates that GL1 has an epistatic effect on Tu. Studies showing that Tu may promote cytokinin (CTK) biosynthesis in fruit warts[63]. Recently, researchers discovered that the bHLH transcription factors HECATE2 (CsHEC2) can work together with GL3 and Tu to promote the formation of cucumber fruit warts including trichomes and nodules, by promoting CsCHL1 which is a CTK synthesis gene[64]. More recently, a new gene spine base size 1 (CsSBS1), encoding C2H2 ZFP, has been identified which forms a trimeric complex with CsTTG1 and CsGL1 at the protein level, thus regulating trichome formation via ETH signalling pathway. The gene CsGL1 was found to have an epistatic effect on CsSBS1 during this process. During the regulation of cucumber glandular trichome initiation, CsTBH can bind to and regulate the expression of the promoter of the cucumber 1-aminocyclopropane-1-carboxylate synthase (CsACS) gene, thus CsTBH can regulate type I trichomes in cucumber fruit via the ETH pathway[29,59,65]. Dong et al. recently used virus-induced gene silencing analysis and transcriptional data to show that CsbHLH95 is involved in glandular trichome formation[48]. Based on these studies, a model for the regulation of trichome development in cucumber is proposed. TTG1 and CsSBS1 can interact with TBH/Mict/CsGL1 to regulate trichome development, but the exact molecular mechanisms and any other interacting genes remain unknown. The gene CsGL3/Tril is located upstream of key trichome development genes, but whether it regulate other genes that may be involved in trichome development, such as TTG1 and CsSBS1, requires further study (Fig. 4). Although GA, CTK and ETH are primarily involved in trichome development, a deeper investigation is necessary to determine if other phytohormones are also involved in trichome formation.
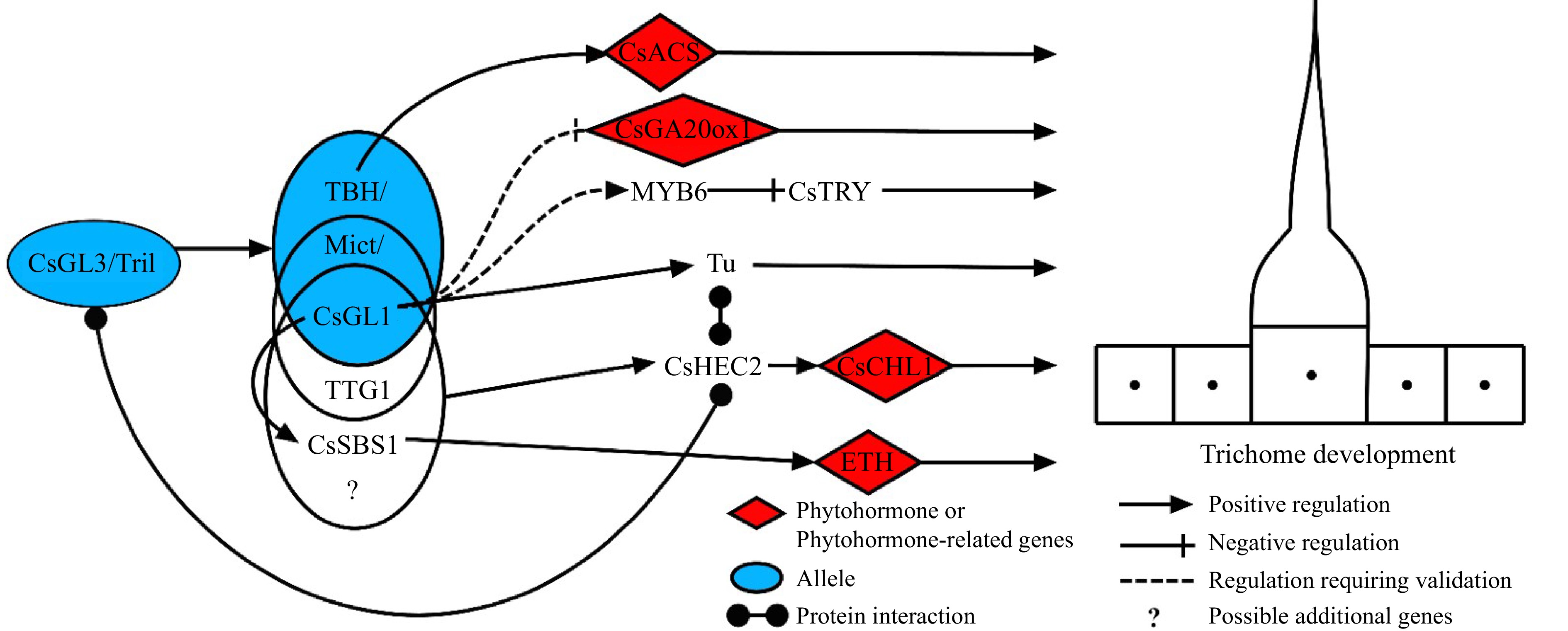
Figure 4.
A proposed cucumber trichome regulatory scheme. Tril/CsGL3, TBH/CsGL1/MICT are key transcription factors regulating trichomes in cucumber. In particular, the transcription factors in the blue circles represent different alleles of a gene, in which Mict can interact with TTG1 to regulate the initiation of trichomes[32]. CsSBS1 can form a trimeric complex with CsTTG1 and CsGL1, thereby regulating trichome formation, and the gene CsGL1 has an epistatic effect on CsSBS1[65]. In addition, these transcription factors affect trichome development by regulating the expression of downstream genes or modulating phytohormone signaling pathways.
Regulatory mechanisms of multicellular trichome initiation in pepper
-
Genes regulating trichome development in peppers are not well-documented. The pepper trichome locus 1 (Ptl1), which is associated with Capsicum annuum L. CM334 trichome formation, was localized to an 80-kb bacterial artificial chromosome (BAC) clone on chromosome 10 in 2010, but no candidate genes were selected[66]. Two candidate genes controlling trichome formation in bell pepper, TRICHOME BIREFRINGENCE-LIKE 5 and GLABROUS INFLORESCENCE STEMS, were identified using the scantwo permutation and stepwiseqtl methods from R/qtl in 2018. Similarly, these genes are also located on chromosome 10[67]. Recently, researchers screened 11 DEGs associated with trichome development using Illumina- and PacBio SMRT-based RNA-Seq, providing a basis for future characterization of trichome formation in peppers[68]. More recently, the researchers cloned the key gene Hairiness, which encodes the C2H2 ZFPs and is located on chromosome 10. This gene controls the formation of multicellular non-glandular trichomes (types II, III and V) and is 45% homologous to the H that controls type I trichome formation in tomato[33,38]. However, the genes mentioned above are only a few of critical genes related to the regulation of multicellular trichome development. There are many other genes that play a role in this process, and we provide a comprehensive list of the reported genes based on their types of transcription factor (positive or negative regulation) (Table 1).
Table 1. Genes involved in the development of trichomes in tomato, cucumber, pepper and soybean. Table adapted from Feng et al. [35]
Types TFs Species Function Effect Metabolite production Hormone involved Reference bHLH SlMYC1 Tomato Type VI formation P Terpenoids [39] CsbHLH1 Cucumber Glandular trichome formation P [21] HECATE2 Cucumber Trichome density P CTK [64] bHLH95 Tomato Trichome initiation N GA [48] MYB SlMX1 Tomato Glandular trichome density P; N Terpenoids, carotenoids, and phenylpropanoids [40,69] SlMixta-like Tomato Trichome Initiation N [41] CsTRY Cucumber Trichome density N [32,61] CsMYB6 Cucumber Trichome initiation N [60,61] MYB52 Tomato Types V formation N AUX [49] SlTHM1 Tomato Types II, V and VI formation N AUX/JA [46,49] MYB75 Tomato Types II, V and VI formation N Sesquiterpene AUX [49,50] HD-ZIP Wolly Tomato Type I density P Terpenoids JA, AUX and GA [36,70] SlHZ45 Tomato Trichome density (especially Type I,IV,VI) P [71] HDZIV8 Tomato trichome morphology P [72] Ln Tomato Trichome density [19] CsGL3/Tril Cucumber Trichome initiation P [53,54,73] Tbh/Mict/CsGL1 Cucumber Trichome morphogenesis P; N Flavonoids [56,57,62,74,75] CsGL2 Cucumber Trichome density P [71,76] SlCD2 Tomato Trichome formation (especially Type VI) P [70] SlHD8 Tomato Trichomes elongation P [47] CsSBS1 Cucumber Trichome development P ETH [65] Mict-L130F Cucumber Trichome morphogenesis N [77] Hairiness Pepper Type II, III, V formation P [33] ZFPs CsTu Cucumber Trichome formation P [63,73] Hair/Hair-like (HL)/ SPARSE Hair (SH) Tomato Type I formation P JA [38,46,78] SlZFP8 Like Tomato Trichomes initiation and elongation P [20] ZFP6 Tomato Trichomes density and length P [20] WD-repeat protein CsTTG1 Cucumber Trichome density P [32] Cyclin SlCycB2 Tomato Trichome density P [36,37] SlCycB3 Tomato Trichome formation P [19,36,37] WAVE regulatory complex Hairless (Hl) Tomato Trichome morphology N Sesquiterpenes, flavonoids [79] Hairless-2 (Hl-2) Tomato Trichome morphology N [72] ARPC1/Hairless-3 (Hl-3) Tomato Trichome morphology; Trichome density (especially Type I, IV) N Terpenoids [80] CHI CHI1 Tomato Trichomes density N Flavonoids [5] AUX/IAA SlIAA15 Tomato Type I, VI density P auxin [51] ARF SlARF3 Tomato Type I, VI density P auxin [51] SlARF4 Tomato Type II, V, VI formation P [49] JA related JAI-1 Tomato Type I, VI formation N JA [52] JAZ2 Tomato Trichome density N JA [44,46] JAZ4 Tomato Trichome development N JA [47] P1 Soybean glabrous [81] Ps Soybean sparse pubescence N [81] Pd1 Soybean dense pubescence P GA, CTK [81] Note: P: Positive; N: Negative. Environmental factors regulating the initiation of glandular trichomes
-
In addition to genes and phytohormones, the development of multicellular trichome is also affected by the environment. Wu et al. found that the higher the altitude, the higher the proportion of plants with glandular trichome[82], which is consistent with the fact that wild tomatoes are derived from higher ground and more resistant to adverse conditions. Drought stress causes a decrease in the density and size of glandular trichome in Artemisia annua[83], while salt stress increases the density of total glandular trichome on both sides of leaves of thorny mustard (S. tenuifolia). Additionally, in Arabidopsis thaliana, the formation of epidermal trichomes is stimulated under UV-B conditions[84]. However, plant-environment interactions are sophisticated, and plants have developed various mechanisms to adapt to varied and complicated environments. Therefore, more research is necessary to determine the relationship between the plant multicellular trichome and environment.
-
Cellular morphology and structure are largely dependent on the cytoskeletal network. Kang et al. observed severe distortion of trichomes in the recessive hairless (hl) mutation and found that the tomato Hl gene encodes a subunit (SRA1) of the WAVE regulatory complex, which controls nucleation of actin filaments in eukaryotic cells[79]. Later, the recessive hairless-2 (hl-2) mutation was found to cause severe distortions of all trichome types in tomato. The Hl-2 gene encodes NAP1, a subunit of the SCAR/WAVE complex. The HD-ZIP IV transcription factor, HD-ZIPIV8, can regulate the expression of Hl-2 and participates in organizing actin filaments in tomato[72]. Recently, it was shown that trichomes swell and distort, and the expression of actin-related protein Component1 (ARPC1) is lower in hl-3 tomato mutants. The Hl-3 gene regulates trichome density and morphology by encoding ARPC1, one of the subunits in the ARP2/3 complex, and also, researchers demonstrated that ARPC1 is Hl-3[80]. Studies show that various mutations in the SCAR, WAVE and ARP2/3 complexes cause abnormal trichome morphology found through genetic screening at different developmental stages of tomato glandular trichome, which induces actin-bound and distorted trichomes. Disruption of microtubules led to isotropic expansion, while disruption of actin filaments inhibits axial extension of trichomes, showing that microtubules and actin filaments regulate different aspects of trichome development[85]. Two types of abnormal trichomes in soybean, Glycine max distorted trichome mutant 1-1 and 1-2 (Gmdtm1-1 and 1-2), are caused by the involvement of Glycine max NCK-associated protein 1 (GmNAP1) in actin filament assembly[86]. These studies confirm the role of the cytoskeletal network in controlling the morphology of plant multicellular trichomes.
Role of cuticle deposition during multicellular trichome morphogenesis
-
The cuticle and trichomes cover the outside of the plant epidermis, serving as a certain defence mechanism. In mutants with altered cuticle synthesis or deposition, trichomes exhibit different types of abnormalities, such as altered trichome density and morphology (including size, branching, shape, etc.). In tomato sticky peel mutants, researchers noted the existence of SlCD2, encoding an HD-ZIPIV transcription factor, at the PE locus, which is associated with cuticle biosynthesis and epidermal trichome density in tomato[70]. RNA interference with the R2R3-MYB transcription factor SlMX1 results in reduced cuticle accumulation, whereas overexpression leads to greater cuticle accumulation and a greater number of glandular and non-glandular trichomes[69]. Another R2R3-MYB transcription factor, SlMixta-like, is a positive regulator of cuticle synthesis[87]. Reduced trichome density in overexpression SlMixta like lines suggests that SlMixta like negatively regulates trichome initiation[41]. The gene Wo regulates the density of type I glandular trichomes in tomato, and SlMYB31, a transcription factor involved in wax biosynthesis, can interact directly to influence the expression of genes in the wax and cuticle biosynthesis pathways[36,88]. In cucumber, a few genes associated with cuticle metabolism were significantly down-regulated in Mict, the glabrous mutant[74]. Recent studies illustrate that overlapping gene networks in Arabidopsis, tomato, cucumber or Artemisia, suggest an interaction between trichome development and cuticle deposition[89]. These results show that some genes have dual functions in mediating trichome initiation and cuticle biosynthesis. In conclusion, cuticle deposition plays an essential role in the morphogenesis of multicellular trichomes.
Molecular mechanisms controlling multicellular trichome morphogenesis
-
Besides the cytoskeleton and cuticle, several other genes can also commonly influence the morphology of multicellular trichomes. The C2H2 transcription factors H and SPARSE Hair (SH) play redundant roles in regulating trichome formation, as evidenced by reduction in tomato type I trichome density and complete disappearance of long-stalked trichomes after h/sh double mutation[78]. A recent article revealed the complexity of the regulation of multicellular epidermal trichome development in tomato. Rencently, researchers discovered that high concentrations of Wo promoted the formation of digitate trichomes (DT, type I−V) and repressed the differentiation of peltate trichomes (PT, type VI and VII)[90]. Wo was able to become more stable when the negative regulators of Wo, multicellular trichome repressor 1 and 2 (MTR1 and MTR2), were simultaneously knocked down. The high concentration of Wo regulated DT formation by promoting the expression of downstream target genes, including SlWox3b encoding WUSCHEL-RELATED HOMEOBOX (WOX) family transcription factor and MX1, while SlWox3b and MX1 inhibited the action of Wo on downstream target genes, such as LEAFLESS (LFS), by protein interactions, thus inhibiting PT formation. Trichome cells were maintained at the starting stage and stopped further differentiation if SlWox3b, MX1 and LFS were knocked down simultaneously[90]. In cucumber, a single nucleotide polymorphism (SNP) of the HD-ZIP I transcription factor gene Mict-L130F caused abnormal trichome development[77]. The regulation of morphogenesis in cucumber multicellular trichomes is influenced by the action of Tril, Mict and Tu[56,57,62]. Cucumber tbh mutants have small and branched trichomes with increased density and abnormal cell shape. Trichome development may be regulated by some unique pathways, such as the meristematic tissue gene CUP-SHAPED COTYLEDON3 (CUC3) and the polarity regulator SHOOT MERISTEMLESS (STM)[62]. Some more genes which affect the initiation and morphogenesis of multicellular trichomes are listed in Table 1.
-
In recent years, rapid developments in genomics and metabolomics and the use of more systematic research methods have led to better understanding of the biosynthesis and evolution of epidermal trichome metabolites. It is widely known that plants are often capable of producing or accumulating large amounts of metabolites, and different plants produce or accumulate different metabolites that perform various functions. Depending on the origin of precursors used to synthesize secondary metabolites and the structural characteristics of these metabolites, the metabolites produced by plant trichomes usually include terpenoids, acyl sugars, polyketides, fatty acids derivatives and phenylpropanoids etc. Tomato type I and IV trichomes mainly synthesize acyl sugars[91], while type VI trichomes primarily synthesize terpenoids, flavonoids, and methyl ketones[5,92,93]. Acyl sugars function as a direct defence to protect plants against pathogenic fungi and specialist herbivores[94]. Volatile terpenes, also known as isoprenoids, are the largest and most diverse plant volatile metabolites, which repel or toxic to herbivores, or attract predators and parasitic natural enemies[9,10,95]. Recently, it has been found that flavonoid deficient mutants may lead to increased production of reactive oxygen species (ROS) and suppress terpenoid biosynthesis. This suggests that flavonoids produced by tomato trichomes have the property of reducing oxidative damage induced by short-wave solar radiation[96]. In cucumber, the trichomes (fruit spines) are an important agronomic trait[29]. There are relatively fewer articles related to the synthesis of secondary metabolites of cucumber and pepper trichomes. It is speculated that the trichomes of cucumber and pepper may mainly act as a physical barrier to protect plants from biotic or abiotic stresses. Pan et al. used metabolomic and transcriptomic analyses to find that Mict may be associated with downstream functional genes (including CsTT4, CsFLS1, CsCER26, and CSMYB36) to directly bind to regulate the biosynthesis of flavonoids, lipids and cuticles in cucumber glandular trichomes[74]. Next, we will focus on the synthesis and regulatory mechanisms of secondary metabolites, including terpenoids, acyl sugars, flavonoids and other metabolites.
Biosynthesis of trichome specialized metabolites
Terpenoids
-
The head cells of tomato trichomes are capable of synthesizing terpenoids through different metabolic pathways. Two conserved C5 isoprenoids are the basic units for the synthesis of terpenoids, which refer to isopentenyl pyrophosphate (IPP) and its isomer dimethyl propenyl pyrophosphate (DMAPP). Plants have two pathways for the synthesis of IPP and DMAPP, the mevalonate pathway (MVA) and the methylerythritol phosphate (MEP) pathway. The MVA pathway occurs in the peroxisome, cytoplasm and endoplasmic reticulum, and the MEP pathway occurs in the plastid. All biochemical steps of both pathways have been demonstrated, and relevant genes have been cloned[97]. In tomato trichome plastids, the precursors of terpenoids synthesized from IPP and DMAPP are further converted into hemiterpenes (C5), monoterpenes (C10), sesquiterpenes (C15), and diterpenes (C20)[95,98]. The diversity of terpenoids in plants is generated by terpene synthases (TPSs). Among them, TPS5, TPS9, TPS12, TPS20 are associated with the synthesis of monoterpenes and sesquiterpenes in tomato glandular trichomes[97,99−101]. In wild tomatoes, type VI glandular trichomes predominantly accumulate volatile monoterpenes and sesquiterpenes, whereas cultivated tomato mainly accumulates monoterpenes. This suggests that sesquiterpenes have been lost or down-regulated during the evolution of the crop[92,99,100,102].
Acyl sugars
-
Acyl sugars are biosynthesised in the glandular trichomes of many Solanaceae plants. The biosynthesis of acyl sugars in wild tomato (Solanum pennellii) can be divided into two stages. The first step is the synthesis of fatty acyl chains, and then esterification of the synthesised acyl molecules to glucose (Glu) or sucrose (Suc). It is generally accepted that the sugar molecules in the acyl glycosidic backbone are derived from Glu or Suc. The esterification of hydroxyl groups at different positions of the sugar by acyl groups of varying lengths and branches is a crucial step in the biosynthesis of acyl sugars. In wild tomato, the acyl sugars are mainly 2, 3, 4-tri-O-acylated Glu esters and some Suc esters with fatty acids ranging from C4 to C12[103,104]. The Branched chain keto acid dehydrogenase (BCKDH) and acyl glycosyltransferase (ASAT) genes are now known to be involved in the biosynthesis of acyl sugars[105,106]. Schilmiller et al. identified a BAHD-type acyltransferase (SlAT2) in cultivated tomato, which recognizes triacyl sucrose and acetyl coenzyme A (CoA) as substrates and catalyses the synthesis of tetraacyl sucrose. Tissue-specific analysis showed that SlAT2 is specifically expressed in the apical cells of type IV glandular trichomes in tomato, which corresponds to the site of synthesis of tetraacyl sucrose[104]. A convertase (acyl sucrose fructosidase 1; ASFF1) has also been reported to produce acyl glucose from acyl sucrose[107].
Flavonoids
-
Flavonoids are a group of polyketides which are widely distributed and possess numerous biological functions. These compounds are derived from phenylalanine and malonyl-CoA[108,109], and typically feature a diphenylpropane (C6–C3–C6) backbone composed of two aromatic rings via a three-carbon chain[109,110]. The core network of flavonoid biosynthesis is largely conserved across land plants[111], and the associated genes enzymes have been well characterized. Most flavonoids are classified into several types including flavones, flavonols, flavanones, flavanols, anthocyanidins, and isoflavones. They are synthesized in the cell membrane and then transported to the vesicles for storage or to other destinations, where they can function as biologically active molecules[112].
Other metabolites
-
Methylone, secreted by wild tomato glandular trichomes, has potent insecticidal properties and is effective in killing pests such as spider mite, greenhouse whitefly, tomato fruitworm, Colorado potato beetle, and tomato needleworms[113]. Methyl ketones include 2-heptanone, 2-undecanone, 2-tridecanone, 2-nonanone and 2-pentadecanone, are the major forms, with 2-undecanone and 2-tridecanone. Among them, 2-tridecanone being the most prevalent[114,115]. The biosynthesis of methyl ketones in wild tomato Solanum habrochaites subsp. glabratum includes two steps. First, 3-ketoacyl-acyl carrier proteins (ACPs) are hydrolyzed to 3-keto acids by the action of the thioesterase methyl ketone synthase 2 (ShMKS2), utilizing intermediates from fatty acid synthesis in chloroplasts. Then, decarboxylase methyl ketone synthase 1 (ShMKS1) converts the 3-ketoacid to methyl ketone[116]. Researchers have attempted to transfer this synthetic mechanism from wild tomatoes to cultivated tomatoes for efficient insect resistance. It was found that transferring MKS1 and MKS2 in heterologous plants can produce methylone in young leaves, however, severe growth defects were observed. Unfortunately, no more methyl ketones were produced as the plants matured[117].
Regulatory network of trichome specialized metabolites
-
Regulating the metabolites produced by multicellular trichome can be done through three modes. Mode I involves directly regulating the enzymes and substrates in the synthetic pathway. Mode II involves influencing genes associated with an enzyme or substrate in the synthetic pathway. Finally, Mode III involves affecting genes associated with trichome development. The following is a detailed introduction.
Mode I
-
Numerous enzymes and substrates involved in metabolite synthesis have been reported in plants[22,106,109]. By influencing the synthesis of enzymes and substrates, it is possible to directly regulate the synthesis of metabolites. However, this is dependent on a particular chemical basis. Several genes involved in monoterpene synthesis were identified in tomato glandular trichomes. The cis-prenyltransferase gene, neryl diphosphate synthase 1 (NDPS1), was expressed in tomato type VI glandular trichomes. The terpene synthase gene, phellandrene synthase 1 (PHS1) was also identified which is capable of synthesizing β-phellandrene as well as various other monoterpenes[99]. In the tomato acyl sucrose synthesis pathway, two different conformations of acyl sucrose have been identified produced by ASAT, predominantly the 'P-type' in wild tomato and the 'F-type' in cultivated tomato. Biochemical analysis revealed that a small number of amino acid changes in two acyl sucrose acyltransferases (ASAT2 and ASAT3) altered their acyl acceptor preferences, leading to a reversal of their reaction order and facilitating the switch between F- and P-type acyl sucrose synthesis[118]. Similarly, two enzymes affecting the accumulation of medium-chain acyl-CoA, acylsugar acyl-CoA synthetase (AACS) and acylsugar enoyl-CoA hydratase (AECH), are present on tomato chromosome 7, and both enzymes are able to affect the substrate for acylose biosynthesis[119]. Type VI glandular trichomes are morphologically different between cultivated tomatoes and wild tomatoes. Recently, researchers discovered that the sesquiterpene synthase 2 (SsT2) gene cluster on chromosome 8 of the wild tomato relative LA1777 controls plastid-derived sesquiterpene synthesis. When SsT2 was transferred to cultivated tomato (cv. Micro-Tom, MT), the trichomes of the MT-Sst2 infiltrated strain were able to produce high levels of plastid-derived sesquiterpenes. However, the type VI glandular trichomes produced by the infiltrated strain did not revert to the same size as the round type VI glandular trichomes in LA1777, nor did they regain the same insect resistance. It has been speculated that the sesquiterpene resistance in wild tomato may be specific to certain insects[120].
Mode II
-
Although many enzymes and substrates involved in the synthesis of secondary metabolites have plants have been identified, while fewer transcription factors responsible for regulating this process have been reported. The R2R3-MYB transcription factor SlMYB75 and the terpene synthase gene are able to bind to the promoters of SlTPS12, SlTPS31, and SlTPS35, inhibiting the transcription of these TPSs and ultimately suppressing the accumulation of sesquiterpenes in tomato. The gene scarecrow-like 3 (SlSCL3) encoding GRAS family transcription factor promotes biosynthesis of volatile terpene by activating TPS genes, while up-regulating the expression of terpene precursor substance biosynthesis genes[121]. The WRKY transcription factor WRKY73, MYC1 and the zinc finger-like transcription factor Expression of Terpenoids 1 (SlEOT1) are capable of transactivating the terpene synthase promoters in Nicotiana benthamiana trichomes, respectively. And the latter two transcription factors can synergistically exert a stronger trans-activation effect[101]. In cucumber, genes involved in flavonoid metabolism are significantly down-regulated in mict mutants, and further experiments suggest that Mict regulates flavonoid biosynthesis by directly binding to downstream functional genes such as CsTT4, CsFLS1, CsCER26, and CsMYB36[74]. A recent study found that CsTBH is not only involved in trichome development, but is also a key regulator of flavonoid biosynthesis in cucumber glandular trichomes[75]. However, studies examining the regulatory of secondary metabolites produced by multicellular trichomes in vegetable crops are lacking. Therefore, researchers must conduct further in-depth studies to fill this gap.
Mode III
-
It appears that changes in trichomes can cause alterations in the associated metabolites. Modifying a specific trichome type can alter the production of the corresponding secondary metabolites. For example, in tomato type I and IV trichomes predominantly produce acyl sugars, and enhancing their production can be achieved by increasing the number of these trichomes. However, in practice, this approach may also affect multiple trichome types. In pepper, Hairiness regulates the production of non-glandular trichomes (type II, III, and V), but whether it can synthesize specific metabolites has not been reported. Meanwhile, certain genes involved in trichome formation can also affect metabolite production. Overexpression of SlMX1 increases trichome density, as well as enhances the expression of enzymes involved in the terpene synthesis pathway, which subsequently increases terpene levels[40]. The presence of the SlMYC1 gene leads to the low-density, smaller type VI glandular trichomes in tomato, and the absence of this gene results in the disappearance of type VI glandular trichomes. Additionally, SlMYC1 positively regulates the synthesis of monoterpenes in leaf and stem trichomes while negatively regulates the synthesis of sesquiterpenes in stem trichomes[39]. The transcription factor SlMYB75 negatively regulates the transcription of terpene synthase genes (such as SlTPS12, SlTPS31 and SlTPS35) hindering the formation of type II, V, and VI trichomes, as well as the accumulation of sesquiterpenes when overexpressed[50].
These three methods have been proved to effectively regulate the production of metabolites in multicellular trichomes. Mode I is currently the most extensively studied method, as it directly affects the production of metabolites by acting as key structural genes, while the second and third approaches require further in-depth study.
-
Vegetable crops are often subjected to biotic and abiotic stresses during their growth and development, among which pest and disease problems are essential factors affecting the yield and quality. However, conventional breeding often relies on pesticide spraying to control pest and disease, which not only affects the growth of vegetables but also causes harm to the environment. Fortunately, plant trichomes can produced terpenoids, acyl sugars, flavonoids, and other secondary metabolites that play an important role in pest and disease resistance. Therefore, these secondary metabolites can be used to develop natural resistance in vegetable crops, and breeding for insect-resistant varieties can also be considered. By moving away from the chemical pesticides, we can greatly decrease their usage and play a vital role in protecting the environment to a certain extent.
Trichome development affects not only trichome density but also secondary metabolite production[25]. By studying key transcription factors in trichome initiation and morphogenesis, we can better understand cell differentiation and development, while also improving agronomic traits. This paper provides the first comprehensive review of the synthesis and metabolism of multicellular trichomes in vegetable crops and aims to enlighten researchers in this field with a large number of recent publications.
Although this paper discusses many aspects related to trichome development, metabolite synthesis and regulations, our research on trichomes is still in its infancy. There are still many doubts regarding the study of multicellular trichomes in vegetable crops. Therefore, this section will present some research gaps and future research directions.
1. How is the identity of trichomes determined during their developmental stage of multicellular trichomes? What regulates the density and distribution of trichomes?
2. Why does each trichome divide into only a specific number of cells? What regulates this division?
3. What is the detailed process by which glandular trichomes transport metabolites and function?
4. Are there conserved genes or regulatory pathways that regulate trichome synthesis and metabolism in each species?
5. How do various environmental factors affect the density and distribution of trichomes?
6. If humans can produce specialized metabolites, how can we produce them, and what should be the criteria for production?
In conclusion, trichome development is the basis for studying the secondary metabolite synthesis, and how they are produced and regulated is crucial, particularly in vegetable crops. Trichomes' ability to resist biotic and abiotic stresses is an effective biocontrol measure that meets the criteria for green vegetables. However, the genes related to glandular trichome secondary metabolites and their regulatory mechanisms require further research to clarify.
This work was supported by grants from the National Science Foundation of China (32072578) and the National Key Research and Development Program of China (2021YFF1000104). Finally, we gratefully acknowledge the help of Ali from Peking University Institute of Advanced Agricultural Sciences in revising this article.
-
The authors declare that they have no conflict of interest. Changxian Yang is the Editorial Board member of Vegetable Research. He was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer-review handled independently of this Editorial Board member and his research groups.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Yuan S, Li Q, Shen H, Wang W, Wang T, et al. 2023. Advances in the regulatory mechanisms of multicellular trichome formation and its secondary metabolite synthesis in vegetable crops. Vegetable Research 3:24 doi: 10.48130/VR-2023-0024
Advances in the regulatory mechanisms of multicellular trichome formation and its secondary metabolite synthesis in vegetable crops
- Received: 02 June 2023
- Accepted: 18 July 2023
- Published online: 05 September 2023
Abstract: Trichomes are specialized epidermal appendages, which can be divided into glandular or non-glandular types based on their diverse morphology. The glands of glandular trichomes are responsible for the biosynthesis and storage of many natural metabolites. Recent progress has been made in characterizing the regulatory mechanisms of trichome formation and metabolite biosynthesis in the trichome. In this paper, we describe the structural and morphological features of glandular trichomes in vegetable crops, mainly focusing on tomato and cucumber. We discuss the developmental processes and regulatory mechanisms involved in trichome formation, including the roles of regulatory factors, phytohormones and environmental influences. We also highlight recent advances in the regulatory mechanisms underlying glandular trichome-related metabolites. This review provides a basis for understanding the formation of multicellular trichome and their secondary metabolites.
-
Key words:
- Trichome /
- Secondary metabolites /
- Vegetable /
- Regulatory mechanisms












