-
For decades, researchers have made considerable efforts to study fruit quality traits to meet human preferences, with quality traits clearly impacting overall consumer acceptance, which is also the main factor affecting fruit price and market competitiveness[1−4]. Among these traits, color, as one of most eye-catching traits, directly affects consumer appeal. The color of many fruits results from the accumulation of various anthocyanins, a class of flavonoids, major water-soluble secondary metabolites[5−7]. Anthocyanins are generally considered to be one of the major sources of natural antioxidants and have a variety of biological functions[8−10]. Another important trait of fruit quality is acidity, which contributes to fruit taste[11,12]. Acidity is caused by the presence of organic acids, one of the main metabolites involved in the development of fruit flavor. It generally increases linearly with the amount of the dominant acid[13,14]. In fruits, the most common acids are malate, citrate, and ascorbate, and the dominant acid varies from species to species[15,16].
Recently, reports have revealed a possible relationship between the anthocyanin accumulation and acidity[17,18]. It has been shown that the accumulation of anthocyanins is not only accompanied by the accumulation of organic acids in the vacuoles, but also that a shift in the vacuolar pH causes a change in the absorption spectrum of anthocyanins[17,19]. Furthermore, a recent study revealed that organic acid accumulation enhances anthocyanin biosynthesis[20]. Moreover, extensive genetic studies at the level of structural genes and transcription factors suggested associations between these two metabolites, where a single gene may jointly regulate both traits[17,21,22]. Another fascinating example of this potential relationship is that the organic acid-related pathway provides the energy and primary metabolites of anthocyanin synthesis[23]. Therefore, understanding the regulation mechanism of the relationship between these two traits represents an important target for fruit crop improvement. Fruit quality traits are regulated by multiple TFs; however, there are few studies on the functions of individual TFs in the co-regulation of anthocyanin biosynthesis and acidity[17,18,21]. In this review, we mainly focus on recent advances in the identification of TFs as co-regulators of anthocyanin accumulation and acidity in fruit crops and discuss the regulatory mechanism networks that co-regulate these two quality traits.
-
Anthocyanins and organic acids, the two largest groups of plant metabolites, are derived from two distinct pathways. However, early studies revealed that the color of anthocyanin depends on the vacuolar pH[24]. This, together with studies of the structural genes and the level of TFs, suggested associations between the two metabolites. First, in plant cells, the vacuole stores a wide variety of compounds, such as secondary metabolites including anthocyanins, organic acids and sugars[13,25]; therefore, the accumulation of anthocyanins and organic acids is usually accompanied in vacuoles[17,26]. It is known that the color of anthocyanin depends on the vacuolar pH; when vacuoles become more acidic, the same anthocyanins that appear blue under alkaline conditions appear redder[24,27−29]. Notably, this co-relationship was initially discovered during the development of Ipomoea flowers, where a change in flower color from purple to blue is associated with an increase in vacuolar pH[24]. This co-relationship has also been recently described in fruits. For instance, in apple, the red-fleshed fruits accumulate more malate and have a lower pH than the non-red fleshed fruits (Fig. 1)[17]. Furthermore, a wild-type citron (Citrus medica) 'Poncire commun' has normal juice acidity around pH 2.5 and can synthesize anthocyanins. Correspondingly, 'Corsican' citron has very low juice acidity around pH 5.5 and unable to produce anthocyanins (Fig. 1)[18]. Moreover, Majia pummelo (Citrus maxima, 'Majiayou') has low citric acid content and does not accumulate anthocyanin in the pericarp (it was named as the low citric acid (LCA) cultivar)[30]. Recently, a new variety of Majia pummelo was identified in the same region; it accumulates higher contents of citric acid in the pulp (it was named as the high citric acid (HCA) variety) and accumulates anthocyanin in the fruit pericarp at the early developmental stage, indicating that HCA shows a phenomenon of tissue-specific accumulation of citric acid and anthocyanins in fruits (Fig. 1)[30]. Intriguingly, recent work also revealed that the accumulation of organic acid enhance anthocyanin biosynthesis. For example, an in vitro test exhibited that malic acid and citric acid could penetrate peach (Prunus persica) discs and then stimulate the activities of PAL (phenylalanine ammonia-lyase) and ANS (anthocyanidin synthase), further promoting anthocyanin biosynthesis during postharvest storage[20]. Moreover, the organic acid-related pathway was found to provide the energy and primary metabolites that enable anthocyanin synthesis. A study on plum fruit indicated that visible light regulated anthocyanin synthesis by controlling the malate metabolism via MDHs (Malate dehydrogenases), and then the ethylene signaling pathway[23]. During fruit ripening, the capacity of CO2 fixation decreased faster than that of the photochemical reaction and photosynthetic electron transport. This might result in more ATP and NADPH being accumulated in the chloroplasts of fruit peels. Once the produced NADPH is in excess, chMDH (encoding the malate dehydrogenase in chloroplasts) will consume NADPH to produce malate. Malate can be transported, via the malate/oxaloacetate shuttles, into the mitochondria, and then produce NADH catalyzed by MDHs[23]. In the mitochondria, NADH participates in respiration to produce ATP, which could be transported into the cytoplasm[31,32] to support the metabolisms, for instance, ethylene biosynthesis, and then induced the biosynthesis of anthocyanin[23].

Figure 1.
The morphological phenotypes of tissue-specific anthocyanin and organic acid accumulation of different fruits. Above: The phenotype differences between non-red-fleshed apple 'Jinshanyilamu' (left), a cultivar that accumulated less malate, and red-fleshed apple 'Yepingguo' (right), a cultivar that accumulated more malate[17]. Middle: 'LCA', a low citric acid pummelo cultivar (left) that is unable to produce anthocyanins, and 'HCA', a high citric acid pummelo cultivar (right) that is able to produce anthocyanins[30]. Below: Young leaves, flowers and seeds of 'Corsican Citron' (left) that has very low juice acidity around pH 5.5 and completely is unable to produce anthocyanins, and 'Poncire commun' (right) that has normal juice acidity around pH 2.5 and able to synthesize anthocyanins[18].
In addition, there are suggestions that structural genes may also link these two metabolites. For example, silencing of PsmMDH2 and PschMDH (encoding the malate dehydrogenase in mitochondria and chloroplasts, respectively) significantly reduced anthocyanin content in the peel of plum (Prunus salicina) fruits upon exposure to visible light[23]. Additionally, recent studies have revealed that vacuolar proton pumps and transporters also play important roles in the co-regulation processes of organic acid and anthocyanin accumulation. Over the past decades, genetic studies have proven that vacuolar proton pumps can simultaneously regulate the accumulation of both organic acids and anthocyanins in the vacuole. MdVHA-B2, a vacuolar proton pump subunit, in its overexpressed form, increased anthocyanin and malate accumulation in apple calli[17]. In grape (Vitis vinifera), VvVHP1; 2, a vacuolar H+-PPase, was highly expressed in grape berry skin, and its expression pattern was positively correlated with anthocyanin content of berry skin during berry ripening; the overexpression of VvVHP1; 2 promoted anthocyanin production in berry skins[33]. By analyzing the fruits of pummelo, Rangpur lime, orange, and sour lemon, Strazzer et al. found that CitPH1 and CitPH5 are the major downstream genes involved in vacuolar hyperacidification and anthocyanin accumulation[34].
At the level of TFs, MdMYB1 was shown to target the expression of the malate- and anthocyanin-related transporters genes, leading to anthocyanin and malate accumulation in apple[17]. Similarly, upregulation of MdbHLH3 was reported to result in the accumulation of both anthocyanins and organic acids[21,22]. We also found that overexpression of PpWRKY44 in pear (Pyrus spp.) fruit and calli leading to increased anthocyanin and malate accumulation[35,36]. Indeed, several studies have been carried out over the last years assessing the roles of TFs on the transcriptional co-regulation of several genes that are crucial for the accumulation of anthocyanins and organic acids in a wide range of plant species. The next paragraph summarizes the conclusions of these studies.
-
TFs play a crucial role in gene regulatory networks in higher plants by recognizing specific DNA-regulatory sequences within the genome[37,38]. In order for TFs to modulate the expression of their target genes, they must be present in the nucleus in an active state[39,40]. Transcriptional regulation often involves the synergistic action of multiple TFs from the same or different families that modulate the expression of the same target genes. It is now recognized that several TFs are involved in anthocyanin accumulation and acidity, but these studies have focused on single regulation rather than co-regulation[41−44]. In this review, we present four families of TFs that function independently, or cooperatively, to co-regulate the expression of different biosynthetic pathway genes associated with the accumulation of anthocyanins and organic acids, and thereby co-regulating anthocyanin accumulation and acidity in horticultural crops (Table 1).
Table 1. Upstream regulators involved in the co-regulation of anthocyanin accumulation and acidity.
Plant species TF family
classificationProtein name Negative/
positive regulatorFunctionality in trait regulation Reference Petunia (Petunia hybrida) MYB PH4 Positive Vacuolar acidification and flower color [45] Litchi (Litchi chinensis) LcMYB5 Positive Anthocyanin and malate accumulation [46] Citrus (Citrus spp.) CitTRL Negative Anthocyanin, PA and citric acid accumulation [47] Apple (Malus domestica) MdMYB1 Positive Anthocyanin and malate accumulation [17] Apple (Malus domestica) bHLH MdbHLH3 Positive Anthocyanin and malate accumulation [21,22] Citrus (Citrus spp.) CitAN1 Positive Anthocyanin and citric acid accumulation [34] Petunia (Petunia hybrida) AN1 Positive Vacuolar acidification and flower color [48] Grape (Vitis vinifera) WRKY VvWRKY26 Positive vacuolar acidification and flavonoid biosynthesis [49] Pear (Pyrus pyrifolia) PpWRKY44 Positive Anthocyanin and malate accumulation [35,36] Citrus (Citrus reticulata) ERF CitERF13 Positive Fruit color and citrate accumulation [50,51] MYB TFs, one of the largest groups of TFs in plants, contain a conserved DNA-binding domain (MYB domain) and extensive intrinsically disordered regions at the C terminus[52]. They can broadly classified into different types (1R, 2R and 3R) based on the number and placement of DNA-binding domains[53,54]. Multiple studies have revealed that MYB TFs play an important role in the transcriptional regulation of several genes that are crucial for the accumulation of anthocyanins and organic acids in a wide range of plant species[3,55−58]. In apple, MdMYB1 positively co-regulates the accumulation of malate and anthocyanin by promoting the malate- and anthocyanin-related transporters genes, MdVHA-E2, MdVHP1, MdMATE-LIKE1, and MdtDT[17]. In lychee (Litchi chinensis), LcMYB5, by directly activating the transcription of DFR (Dihydroflavonol 4-reductase), regulates anthocyanin biosynthesis. More interestingly, LcMYB5 has also been implicated pH regulation, where its expression pattern in the aril of lychee during the fruit development was generally consistent with malic acid content[46]. In addition, silencing of PH4, an R2R3-MYB that regulates vacuolar pH acidification in petunia flowers by activating vacuolar P-ATPase genes, causes a shift from purple to blue in petunia flowers[45]. Other recent studies have reported that MYB TFs also act as repressors of the accumulation of both malate and anthocyanin in Malus species. For example, In apple (Malus domestica), overexpression of MdMYB44 causes a repression in the expression of the malate-associated genes V-type ATPase A3 (MdVHA-A3), P-type ATPase 10 (Ma10), V-type ATPase D2 (MdVHA-D2), and Al-Activated Malate Transporter 9 (Ma1), resulting in a reduced malate accumulation[59]. In addition to controlling organic acid accumulation, the role of MYB44 has also been documented in regulating fruit coloration. In Malus 'Radiant', MrMYB44-like1/2/3, homologous to apple MYB44, inhibited anthocyanin accumulation and reduced pigment in leaf disks of Malus 'Radiant' and fruit peels of Malus domestica 'Fuji' when the three MrMYB44-likes was overexpressed, by downregulating the expression of anthocyanin biosynthesis genes, such as MrPAL, MrCHS, MrCHI, MrDFR, and MrANS[60].
Besides MYB TFs, bHLH (basic helix–loop–helix) TFs are also play crucial roles in the co-regulation of anthocyanin and organic acid accumulation[21,22] by binding the G-box and E-box in the promoter of their target genes[61]. In apple, overexpression of MdbHLH3 led to enhanced anthocyanin accumulation by targeting key genes, MdDFR and MdUFGT (flavonoid 3-O-glucosyltransferase), involved in anthocyanin biosynthesis[22]. In addition to anthocyanin accumulation, MdbHLH3 have been also implicated in controlling malate accumulation. MdbHLH3 overexpression leads to malate accumulation by directly activating the expression of MdcyMDH (cytosolic NAD-dependent malate dehydrogenase)[21]. In citrus (Citrus spp.), Butelli identified Noemi, which encodes a bHLH TF and controls anthocyanin accumulation and fruit acidity[18]. Moreover, CitAN1 is associated with pigmentation (anthocyanin and proanthocyanin) in flowers, seeds, and leaves and with citric acid accumulation in fruit pulp [34]. In petunia, ANTHOCYANIN1 (AN1; bHLH TF), that is required for anthocyanin biosynthesis, also controls vacuolar acidification in petal cells[48].
In addition to the MYB and bHLH TFs, other TFs, such as WRKY and ERF, are also implicated in co-regulation of the accumulation of organic acids and anthocyanins. For example, VvWRKY26 is involved in the vacuolar acidification pathway by regulating ATPase pump, and is also implicated in flavonoid biosynthesis by activating the expression of the flavonoid structural genes CHS-A, F3′H, F3′5′H and DFR-A[49]. Additional research on PpWRKY44 further elucidated how WRKY protein co-regulates anthocyanin and organic acid accumulation, with the overexpression of PpWRKY44 in pear fruit and calli leading to increased anthocyanin and malate accumulation. PpWRKY44 promotes anthocyanin biosynthesis by activating PpMYB10 expression[35], while it modulates malate accumulation by regulating the malate-associated gene PpALMT9[36]. In apple, MdWRKY40 positively regulates anthocyanin accumulation in red-fleshed apple callus by directly activating the expression of ANS[62]. In addition, PpWRKY40, homologous to apple WRKY40, positively regulates organic acid accumulation by directly activating the expression of VHA-B1[63]. An ethylene response factor, CitERF13, interacts with the vacuolar proton pump gene CitVHA-c4 at the protein level to regulate citrate accumulation[50]. Furthermore, the overexpression of CitERF13 resulted in rapid chlorophyll degradation by binding directly to the chlorophyll degradation-related gene CitPPH (pheophytin pheophorbide hydrolase), thereby changing fruit color from green to yellow and improving fruit quality[51].
Several recent studies have shown that some kind of posttranslational modifications also play an essential role in regulating TFs function and consequently affect the co-regulation processes of anthocyanins and organic acids. For example, both MdCOP1 and MdMIEL1 interact with MdMYB1 to regulate anthocyanin accumulation[17,64], but, whether MdCOP1 and MdMIEL1 affect fruit malate accumulation through MdMYB1 still needs further analysis. Recently, that MdBT2, a BTB-TAZ protein, was found to regulate the stability of MdMYB1 via ubiquitination and to negatively regulate anthocyanin and malate accumulation in apple[17,65]. In addition, MdBT2 also regulates the stability of MdMYB73 and MdCIbHLH1 via ubiquitination to negatively regulate malate accumulation[66,67]. Furthermore, MdbHLH3, a key co-regulator of anthocyanin and malate accumulation, was ubiquitinated and phosphorylated by the U-box-type E3 ubiquitin ligase, MdPUB29, and the glucose sensor, MdHXK1, respectively, affecting anthocyanin accumulation by regulating their downstream anthocyanin-associated genes in apple[22,68,69]. Moreover, MdSnRK1.1 (Sucrose-Nonfermenting1 (SNF1)-related protein kinases 1) interacted with and phosphorylated the MdJAZ18, which is a repressor in the jasmonate (JA) signalling pathway, to promote its degradation, which released MdbHLH3 thereby activating the expression of its target genes and consequently stimulating anthocyanin biosynthesis in apple[22,70]. However, further work is needed to investigate whether MdPUB29, MdHXK1 and MdSnRK1.1 regulate malate accumulation through MdbHLH3. In apple, MdSIZ1 (small ubiquitin-like modifier E3 ligase) targets MdbHLH104 to regulate the activity of the plasma membrane H+-ATPase[71] and, by sumoylating MdMYB1, stimulates anthocyanin biosynthesis in apple under low-temperature conditions[72]. -
Recent studies on the TFs listed in the Table 1 have confirmed that most of the TFs do not act independently, but are involved in complex regulatory mechanisms to coordinate in the co-regulation of anthocyanin accumulation and acidity in fruit and ornamental crops, in which a highly conserved transcriptional regulatory complex MYB-bHLH-WD40 (MBW) complex play a central role in this co-regulation (Fig. 2). In grape, VvMYC1 (bHLH) and VvWD1 (WDR) together with VvMYB5b activate flavonoid accumulation and together with VvMYB5a vacuolar acidification (Fig. 2b)[73]. Also, the VvMYC1-VvWD1-VvMYB5a complex directly activates the expression of VvWRKY26, and a VvMYC1-VvWD1-VvMYB5a-VvWRKY26 complex then activates the expression of P-type ATPases VvPH5 and VvPH1, which are involved in vacuolar hyper-acidification. VvWRKY26 can bind to VvWD1, the homolog of petunia AN11, and reactivate the PH3 target genes VvPH5 and VvPH1 (Fig. 2b)[49,73]. In apple, MdbHLH3 interacts with MdMYB1 to promote anthocyanin and malate accumulation in apple fruit[17]. In citrus, the homologues of the AN1-PH4 and AN1-Ruby1 (Ruby1 is a homologue of AN2) complexes are associated with anthocyanin accumulation and acidity (Fig. 2d)[47]. Recently, the MYB TF CitTRL (TRIPTYCHON-LIKE) was found to be a negative regulator of proanthocyanin and citric acid accumulation in citrus. CitTRL represses proanthocyanin and citric acid accumulation by competing with CitPH4 for binding to CitAN1, thereby alter the expression of leucoanthocyanidin reductase and the vacuolar proton-pump gene PH5, which are responsible for proanthocyanidins biosynthesis and vacuolar acidification, respectively (Fig. 2d). In addition, CitTRL expression is activated by CitPH4 to form a fine-tuned regulatory loop to balance the accumulation of proanthocyanin and citric acid. Furthermore, CitTRL competes with the R2R3-MYB CitRuby1 for binding to CitbHLH1 or CitAN1, thereby repressing the expression of downstream anthocyanin structural genes (Fig. 2d)[47]. Moreover, CitAN1 is associated with acidity and anthocyanin accumulation by forming a protein complex with CitPH4 or Ruby1[29, 49]. Taken together, these reports suggest that the transcriptional co-regulation of anthocyanin biosynthesis and organic acid accumulation is a complex regulatory process in plants.
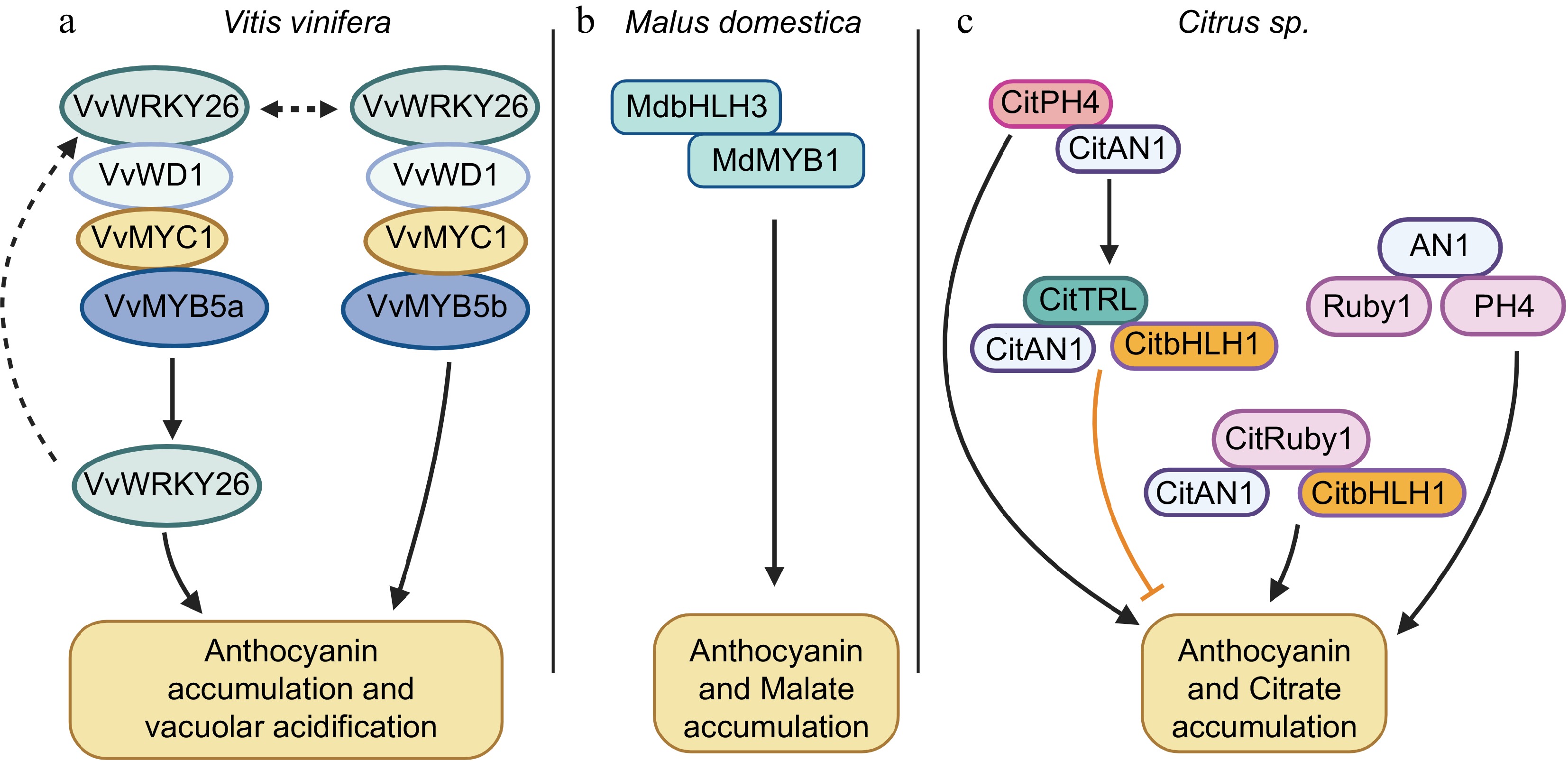
Figure 2.
Cooperation of TFs involved in co-regulation of anthocyanin and organic acid accumulation in fruit crops, including (a) grape (vitis vinifera), (b) apple (Malus domestica), and (c) citrus (Citrus sp.). Solid and dashed lines indicate proven and hypothetical (yet to be experimentally established) relationships, respectively. Arrows indicate positive regulators; T-bars indicate negative regulators.
-
Several stimuli (e.g., nutrient availability and temperature) are involved in the co-regulation of anthocyanins and organic acids via several signaling pathways, in which TFs play a critically essential role[74]. Recently, an important link between nitrate signaling and the accumulation of anthocyanins and malate has been identified in apple. Low nitrate leads to high levels of anthocyanins and malate, while moderately high nitrate results in lower levels of anthocyanins and malate, in which MdBT2, a nitrate-responsive protein, plays a critical role by physically interacting with the MdMYB1and MdMYB73. MdBT2 regulates the accumulation of MdMYB1-mediated anthocyanin and MdMYB73-mediated malate accumulation (Fig. 3)[17,65]. With respect to temperature, high temperature was found to reduce apple fruit color[75], and low temperature to promote anthocyanin synthesis in apple[76,77]. In apple, MdbHLH3, a cold-induced bHLH TF, was strongly induced by low temperature and enhanced the accumulation of anthocyanin biosynthesis and malate (Fig. 3)[21,22]. Other stimuli, including salinity, drought, and sucrose could also regulate anthocyanin biosynthesis and organic acid accumulation; but these studies have focused on single regulation rather than co-regulation.
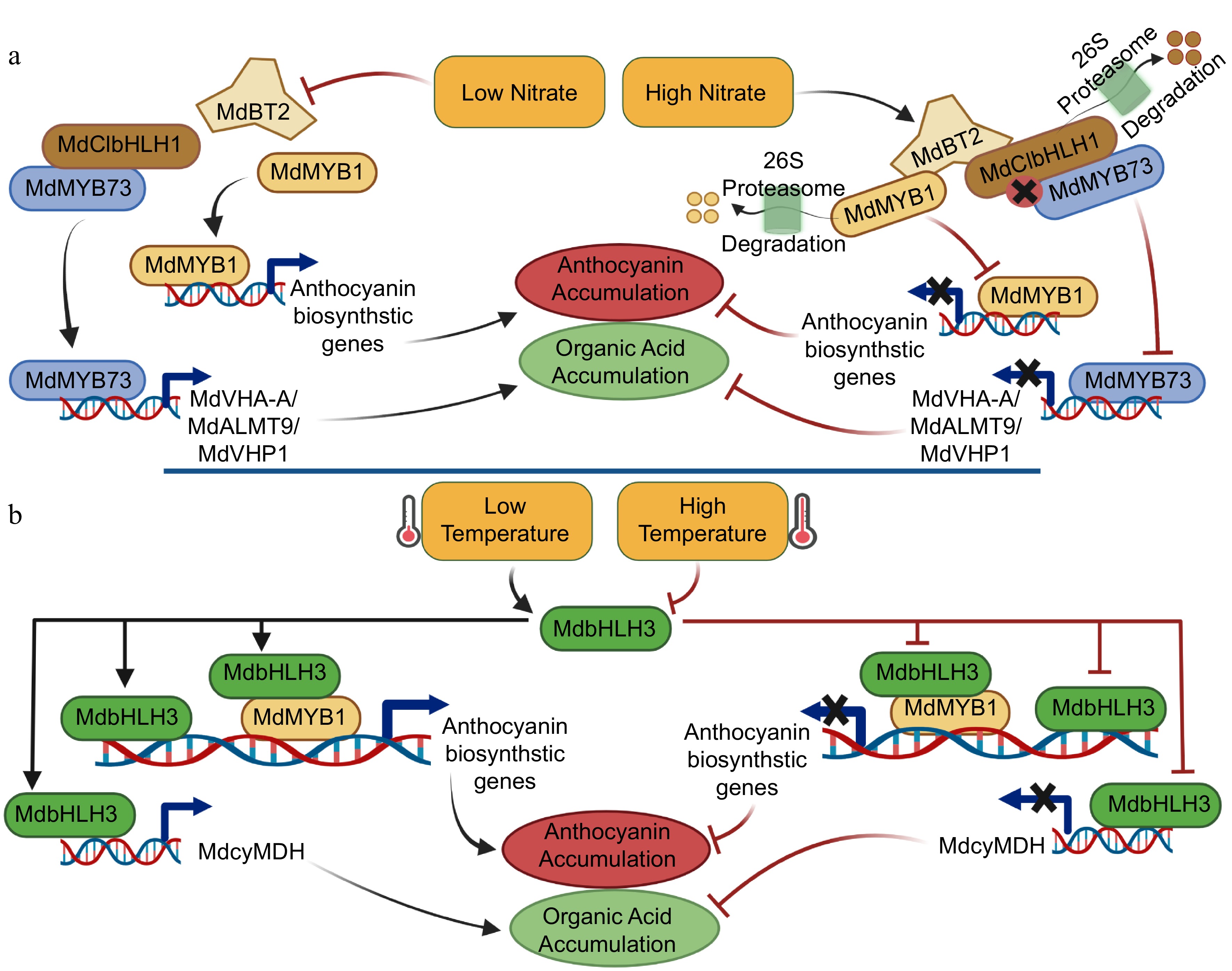
Figure 3.
A simplified model of environmental co-regulation of anthocyanins and organic acids in apple fruit. (a) High nitrate cues trigger MdBT2 to ubiquitinates MdMYB1 and the MYB-bHLH complex, MdCIbHLH1-MdMYB73, which are involved in the regulation of anthocyanin biosynthesis and organic acid accumulation in apple fruit, respectively; Low nitrate cues inhibit MdBT2, thus, MdMYB1 and the MYB-bHLH complex, MdCIbHLH1-MdMYB73, enhances the accumulation of anthocyanin and organic acid in apple fruit. (b) High and low temperature cues inhibit and trigger the MYB-bHLH complex, MdMYB1-MdbHLH3, which is involved in the co-regulation processes of anthocyanin biosynthesis and organic acid accumulation, respectively.
-
During the past decades, an important link between cell pH and flower color has been discovered, whereby a shift in vacuolar pH causes a change in flower color in plants. In contrast, this relation has been largely overlooked in fruits and is just now surfacing. The recent findings reviewed in this paper suggest that there is not only a genetic linkage between acidity and anthocyanin accumulation, but also a mechanism by which a single gene regulates both traits. These findings not only broaden our understanding of the genetic relationship of these traits, but also allow breeders to select appropriate targets for modulating agriculturally important traits. Although several transcription factors and structural genes co-regulate anthocyanin accumulation and acidity, further research is needed to know which specific transcription factor or structural gene plays a critical role and whether there are similarities across different species. One of the major challenges for the future will be functional identification of additional regulatory pathways involved in the co-regulation of fruit quality traits. In addition, with the completion of whole-genome sequencing of the important fruit trees, the analysis of large natural populations through high-throughput resequencing and genome-wide association studies (GWAS) will provide tremendous opportunities to explore key genes underlying important traits and their co-regulation, which may help to elucidate genetic mechanisms of these traits. Meanwhile, the accelerated development of genetic tools will support essential functional studies of transcription factors that co-regulate fruit quality traits. Currently, bud sports have become a powerful tool for studying functional genes, gene expression, and phenotypic differences[78−80]. Thus, more studies related to anthocyanin and organic acid accumulation in these mutants, especially red/green mutants, will further increase our understanding of the molecular mechanisms responsible for the co-regulation of these traits.
Although the co-regulation process of organic acid accumulation and anthocyanin biosynthesis by MYB-bHLH-WDR complex has been studied in several fruit trees, future studies are needed to identify other potential TFs involved in the co-regulation of these two traits in different fruit trees. The activity of the MYB-bHLH-WDR complex can be regulated by various post-translational modifications; however, current studies do not fully elucidate the roles of these post-translational modifications in the co-regulation processes of anthocyanin accumulation and acidity. In addition, the roles of non-coding RNAs, including miRNAs and lncRNAs, in the regulation of fruit traits have been uncovered in recent years[80,81]. However, much more work is needed to investigate their roles in the co-regulation processes of these two traits in different fruit trees.
-
The authors confirm contribution to the paper as follows: study conception and design: Teng Y, Alabd A; data collection: Alabd A; analysis and interpretation of results: Alabd A, Ni J, Bai S, Teng Y; draft manuscript preparation: Alabd A, Teng Y. All authors reviewed the results and approved the final version of the manuscript.
-
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
This study was supported by the National Natural Science Foundation of China (32072545), the Specialized Research Fund for Major Science and Technique of Zhejiang Province of China (2021C02066-5) and the National Key Research and Development Program (2018YFD1000202).
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Alabd A, Ni J, Bai S, Teng Y. 2024. Transcriptional co-regulation of anthocyanin accumulation and acidity in fruits. Fruit Research 4: e005 doi: 10.48130/frures-0023-0041
Transcriptional co-regulation of anthocyanin accumulation and acidity in fruits
- Received: 16 July 2023
- Revised: 18 October 2023
- Accepted: 23 October 2023
- Published online: 25 January 2024
Abstract: Color and acidity, two important fruit quality traits, greatly influence consumer choice and market competitiveness. They result from the accumulation of anthocyanins and organic acids in the vacuole. A shift in the vacuolar pH, caused by the accumulation of organic acids, leads to a change in the absorption spectrum of anthocyanins, and thus to changes in the color of tissues, suggesting a possible relationship between these two traits. Thus, the discovery of the molecular co-mechanism responsible for these processes is one of the most challenging for improving fruit quality traits and ultimately increasing market value. Here, we review current knowledge on the relationship between anthocyanin accumulation and acidity, and highlight recent advances in the roles of TFs in regulating these quality traits via transcriptional co-regulation of different genes associated with anthocyanin accumulation and acidity for fruit quality improvement.
-
Key words:
- Anthocyanin /
- Organic acid /
- Co-regulation /
- Color /
- Acidity /
- Transcriptional regulation












