-
Agrobacterium transformation efficiency is influenced by the plant genotype or cultivar, the effectiveness of the selection system, the reporter gene used to identify the transgenic shoots and the meristematic nature of the plant cells. Highly embryogenic cell cultures and meristematic cells in organogenic explants yield the best results in plant transformation. Generally, cultivars and genotypes with high transformation efficiency will regenerate more transgenics and edited plants[1].
Agrobacterium-plant interactions are complex and only partially understood[2]. Recalcitrance among different species might be due to any number of reasons stemming from the physiology of the donor plant, in vitro explant manipulations, plant stress physiology[3], or other problems[4−11]. In mature citrus explants, cells in the cambial ring become competent for transformation and regeneration[12], but there is immense cultivar variation in Agrobacterium transformation efficiency (Zale unpublished).
Mature 'Valencia' (Citrus sinensis L. Osbeck) citrus scions form shoots in tissue culture, but few are transgenic, so it is considered recalcitrant to Agrobacterium. Transformation efficiency based on the number of shoots (TES) and based on the number of explants (TEE) is consistently less than 5% on semi-solid medium with 100 mg·L−1 kanamycin[13−15]. In practice, its transformation efficiency is considerably less than 5%, and higher transformation efficiencies depend upon other factors, such as the re-invigoration of the mature scion donor plant by budding[16].
'Florida Early Valencia 1' ('Florida EV1') is a somaclone derived from 'Valencia' in tissue culture[17,18]. 'Florida EV1' matures from December to January in central Florida (USA), while 'Valencia' matures from early to late March. The term somaclonal variant was introduced by Larkin & Scowcroft in 1981 to describe the genetic variation in plants regenerated from cell cultures[19]. These alterations might be genetic or epigenetic changes induced by the stress of tissue culture or due to auxins and cytokinins generating disorganized cell growth[11,20,21]. In wheat, three near-isogenic lines identified in tissue culture differed in regeneration ability and transformation ability, but the most amenable wheat line had two translocations[9]. Somaclonal variation can create unwanted variations but is also a potential source of useful genetic diversity[19].
Citrus biotechnology has traditionally used the neomycin phosphotransferase II (nptII) gene for selection, which, unfortunately, is riddled by escaped shoots[22]. Canton et al. determined the best concentrations of kanamycin in liquid vs. semi-solid selection medium to screen two mature citrus rootstocks by GUS staining[23]. They found that selection at stringent kanamycin concentrations (150 to 200 mg·L−1) in liquid culture was optimal. The present research aims to apply this same technology using the GFP reporter to mature sweet orange scions.
-
The TIPS-EPSPS vector has been previously described[24]. The Citrus sinensis EPSPS gene was mutated at two sites with site-directed mutagenesis and shown to confer tolerance to glyphosate in immature citrus[24]. In the present study, it was transformed into Agrobacterium tumefaciens, strain AGL1, with the appropriate antibiotics and kanamycin bacterial selection at the recommended concentrations[25]. The vector also contains the neomycin phosphotransferase II (nptII) gene as a secondary selectable marker which encodes an aminoglycoside phosphotransferase that confers resistance to kanamycin in plants, and the enhanced green fluorescent gene (egfp) as the reporter in plants (Fig. 1).
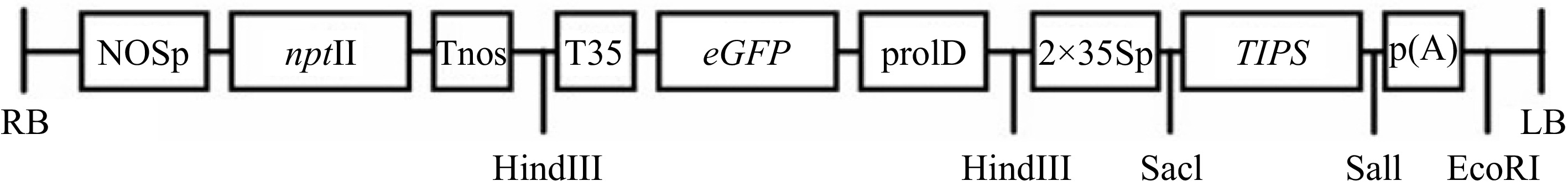
Figure 1.
Schematic map of the TIPS-EPSPS vector in pBI101 vector backbone for Agrobacterium-mediated genetic transformation. The nptII, eGFP, and TIPS-EPSPS are driven by the NOS promoter, prolD promoter, and double 35S promoter, respectively. LB, left border; RB, right border; NOSp, nopaline synthase promoter; prolD, A. rhizogenes rolD promoter; 2×35Sp, Enhanced Cauliflower Mosaic Virus promoter; nptII, neomycin phosphotransferase; p(A), CaMV polyadenylation signal; Tnos, nopaline synthase terminator; T35, Cauliflower Mosaic Virus terminator; eGFP, enhanced green fluorescence protein; TIPS, citrus shikimate pathway enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) modified to obtain glyphosate resistance[24].
Plant material and growing conditions
-
Mature internodal explants of 'Valencia SPB-1-14-19' and 'Florida EV1' were used in transformation experiments[18]. Mature scions were budded onto immature five-month-old Citrus volkameriana (L.) rootstocks[23]. Conditions in the growth room were maintained with a 12 h light cycle photoperiod provided by cool white, fluorescent lamps, with 50% humidity and a temperature of 26 ± 4 °C. Mature scion budsticks were collected at three months, and prepared accordingly[23].
Agrobacterium growth and tissue culture
-
Agrobacterium strain AGL1 was grown as described and used in transformations[15]. Plant transformations and tissue cultures were performed, and cultures were placed in a dark incubation with 100 mg·L−1 kanamycin selection for three weeks[23]. After three weeks, the explants were placed in liquid culture vessels with 3M paper or paper towels[23]. The explants were placed on top of this paper in the liquid culture vessels at 0, 100, and 200 mg·L−1 kanamycin and grown for four weeks[23]. A semi-solid medium with 100 mg·L−1 kanamycin was used as a control for three weeks in the dark incubation and four weeks in the light incubation[23]. Micrografting and secondary grafting of some but not all GFP positive shoots were performed[15,23].
Glyphosate assay of nodal budsticks
-
A glyphosate assay was developed to test whether the transgenic, nodal budsticks positive for the EPSPS transgene could sprout shoots on glyphosate medium. Roundup Super Concentrate (50.2% active ingredient, containing 3.7 lb glyphosate acid equivalent per US gallon or 2.6 M glyphosate) was used as a selection system in transgenic vs. wild-type (WT) nodal budsticks in semi-solid MT medium (Phytotechnology Labs, Lenexa, KS, USA) with 30 g·L−1 sucrose at pH 5.7 and different glyphosate concentrations (0, 2.6, 6.5, and 13.1 mM)[24,26]. Transgenic and WT budsticks were prepared[23], cut into nodal budsticks with one node per budstick, and plated onto the glyphosate medium. Three to six nodal budsticks per petri dish were plated onto the semi-solid medium and incubated in a chamber with a 12 h photoperiod, 45 μmol·m−2·s−1 light intensity, at 26 ± 2 °C. Sprouted shoots per nodal budstick were recorded after 28 days and analyzed using ANOVA.
Molecular analysis of transgenic plants
-
Shoots were examined for GFP fluorescence with a Nikon SMZ 745T stereoscope (Nikon, Melville, NY, USA) equipped with a NIGHTSEA fluorescence adapter (NIGHTSEA, Lexington, MA, USA) and a blue filter. These shoots were micrografted onto decapitated seedlings[15,23]. Transgenic lines were confirmed by PCR using primers for EPSPS, nptII, and gfp to amplify fragments of 455, 239 and 713 bp, respectively. The oligonucleotides used to amplify these genes were as follows: nptII (nptII-F: GTGGAGAGGCTATTCGGCTATGA and nptII-R: CTTCGCCCAATAGCAGCCAGT), gfp (gfp-F: CTGACCGGATCGGCACATTA, and gfp-R: CTTGTAGTTGCCGTCGTCCT) and EPSPS (EPSPS-F: AGAGGACACGCTGAAATCAC and EPSPS-R: AAGCATATGGTGAATATCTTCGC). Genomic DNA extractions were performed with the PowerPlant® DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA). The PCR reactions were set to a 15 μL volume total, containing 0.15 mM of each primer, 7.5 μL of Dream Taq PCR Master Mix (2X) (ThermoFisher Scientific®, Atlanta, GA, USA), and 20 ng of DNA template. The cycling parameters were programmed into an MJ Mini™ thermal cycler (Bio-Rad®, Hercules, CA, USA) with a preliminary denaturation at 95 °C for 3 min; followed by 35 cycles of 95 °C for 30 s; an annealing step at 62 °C for 30 s, and an extension step at 72 °C for 1 min, followed by a final 5 min extension at 72 °C. Polymerase chain reaction products were analyzed on 1.5% agarose gel electrophoresis with a 100 bp EZ Load molecular ruler (Bio-Rad®, Hercules, CA, USA).
Duplexed real-time TaqMan quantitative PCR (qPCR) for copy number
-
A duplex TaqMan real-time quantitative PCR (qPCR) assay determined the transgene copy number in some transgenic citrus lines vs a known, single copy, 'Hamlin' control[27,28]. Amplification of the nptII gene and the internal control gene CsLTP (Citrus sinensis lipid transfer protein) was analyzed in each transgenic line against the 'Hamlin' control known to contain only one copy of the nptII gene (supplied by the Mou lab, University of Florida, Microbiology and Cell Science, Gainesville, FL, USA). Primer probe combinations were standardized with the 'Hamlin' single copy nptII control reference line. A serial dilution was performed starting with 100 ng of total DNA, and Ct values were plotted vs the log nanograms of total DNA. The R-squared value (coefficient of determination) for the standard curves for both nptII and CsLTP genes was 0.99. The slope of the plot was −3.5 for nptII and −3.49 for CsLTP, giving an efficiency of 1.93 (93%) (Table 1). This is in the range (90%−97%) of good efficiency for a duplex qPCR.
Table 1. The linear range and standard curves for endogenous CsLTP and nptII genes.
Gene Linear range (CT) Regression equation Correlation coefficient nptII 21.07−26.16 f(x) = −3.505x + 21.987 0.999 CsLTP 21.92−25.32 f(x) = −3.491x + 21.126 0.999 Primers and probes for the duplexed real-time TaqMan qPCR reaction
-
Oligonucleotides and probes were designed and synthesized (Eurofins Genomics, Louisville, KY, USA) for the nptII transgene[29,30]. The oligonucleotide and probe sequences for the nptII gene were as follows: nptII-F: ATCATGGTGGAAAATGGCCG, nptII-R: GCCAACGCTATGTCCTGATAG and nptII-probe: [FAM]-TTCTGGATTCATCGACTGTGGC-[BHQ1], yielding an 86 bp product. The Citrus sinensis lipid transfer protein (CsLTP) gene was used as the endogenous single copy control[27,28]. The oligonucleotide and probe sequences for the CsLTP gene were as follows: CsLTP-F: CGGATCAATCCCTAACCTCAAC, CsLTP-R: GTCAGTGGAGATGCTGATCTTG and CsLTP-probe: [TxRed]-CGAGCTTGTGGAGTCAGCATTCCT-[BHQ2] and they yield a 94 bp PCR product.
Duplexed real-time TaqMan qPCR reactions and calculations
-
Duplexed TaqMan PCR reactions were performed in 12.5 μL volume in a 96-well format using a QuantStudio™ 3 Real-Time PCR system (Applied Biosystems, Foster, CA, USA). The reaction mixture contained 6.25 μL of 2x TaqPath™ master mix (Applied Biosystems, Waltham, MA, USA), 1 μL DNA (~50 ng), and 900 nM nptII-F/R primers, 900 nM CsLTP-F/R primers and 100 nM each gene-specific probe. The PCR program consisted of initial denaturation at 95 °C for 3 min, 40 cycles of 10 s at 95 °C, 20 s at 55 °C, and the fluorescence was collected at 55 °C. A no-template control and non-transgenic plant DNA were included. The instrument software determined the Ct values. The log10 of the DNA dilution series was plotted vs the Ct values obtained for each dilution. The PCR efficiency (E) was calculated using the following equation (E=10(−1/slope))[31]. This formula was used to calculate the copy number for each transgenic line and control as formulated by Pfaff[32].
$\rm Ratio = \dfrac{(E_{target})^{(DeltaCt(target)(control-sample))}}{(E_{reference})^{(deltaCT(reference)(control-sample))}} $ Data collection and statistical analyses
-
Transformation response variables included the total number of shoots greater than 2 mm (SL > 2) and the number of positive shoots (PS). The mean transformation efficiencies were based on the number of explants (TEE, the number of positive shoots/number of explants × 100), and transformation efficiencies based on the number of shoots screened (TES, the number of positive shoots/the number of shoots screened × 100). Data collection occurred after seven weeks. ANOVA was performed separately for each cultivar and comparisons were made within each cultivar. For 'Florida EV1' the statistics were calculated for 60 explants per vessel with ten replicates at each kanamycin concentration (0, 100, and 200 mg·L−1) in liquid medium and with ten replicates in semi-solid medium at the standard kanamycin concentration (100 mg·L−1) . For 'Valencia', the statistics were calculated based on 60 explants per vessel with seven replicates at each kanamycin concentration (0, 100, and 200 mg·L−1) in liquid medium and seven replicates in the semi-solid medium at the standard kanamycin concentration (100 mg·L−1). The square root transformation was used to transform the PS variable before ANOVA. Percentage data were divided by 100 and transformed into the arcsine transformation before ANOVAs. The Mann-Whitney non-parametric statistics test was used to compare the regeneration ability of each cultivar (SL > 2/number of explants) across all treatment levels and at 0 mg·L−1 kanamycin. Similarly, a non-parametric Mann-Whitney test compared the median number of positive shoots (PS) between 'Florida EV1' and 'Valencia'. Glyphosate tolerance was assessed after 28 d as the number of shoots that sprouted per nodal budstick at four different glyphosate concentrations (0, 2.6, 6.5, and 13.5 mM glyphosate) and used in ANOVAs. Descriptive statistics, ANOVAs, and multiple comparisons with Fisher's LSD were used to analyze the variables in the cultivars studied. All tests were calculated with Minitab Version 21.
-
In separate experiments, mature 'Florida EV1' and 'Valencia' sweet orange scions were transformed with the TIPS-EPSPS vector (Fig. 1), which possesses two selectable markers (P-Nos::nptII::T-Nos and 2x35S::TIPS-EPSPS::p(A)) in Agrobacterium strain AGL1. TIPS-EPSPS confers resistance to glyphosate[24]. A schematic representation of the process is shown in Fig. 2. A total of 74 'Florida EV1' transgenics and seven 'Valencia' transgenics were identified by GFP fluorescence in We-V™ liquid culture vessels with gravity wells using three levels of kanamycin selection (0, 100, and 200 mg·L−1 kanamycin) vs semi-solid medium at 100 mg·L−1 kanamycin (Table 2 & Fig. 2). The results of these experiments were analyzed separately using ANOVAs.
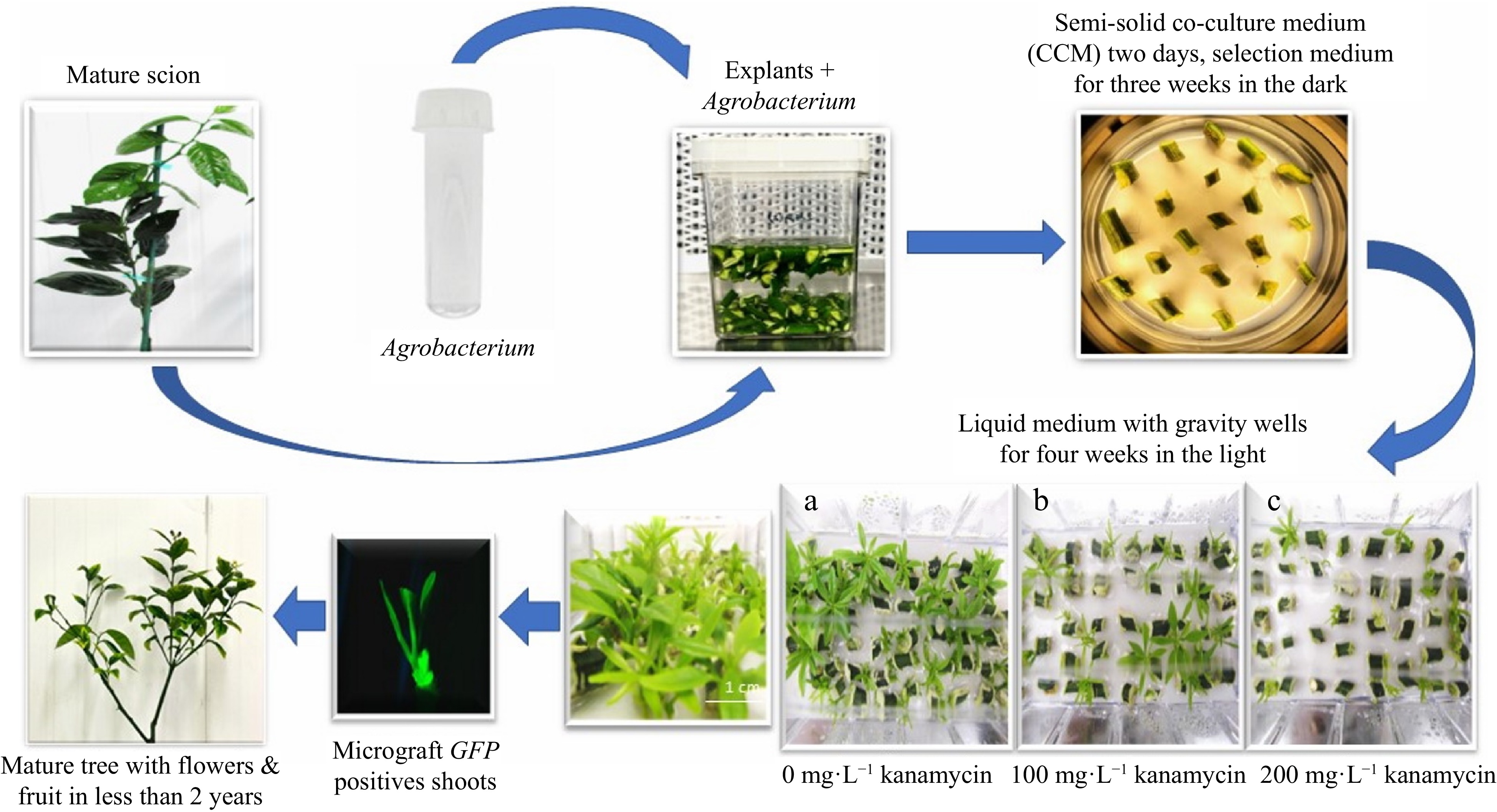
Figure 2.
Schematic representation of the transformation process using We-V™ liquid culture vessels with gravity wells with different kanamycin concentrations of (a) 0, (b) 100, and (c) 200 mg·L−1.
Table 2. 'Florida EV1' and 'Valencia' and the variables that were measured (shoots longer than 2 mm (SL > 2) and positive shoots (PS)) in liquid and semi-solid medium.
Cultivar1 Kanamycin
(mg·L−1)Medium Explants SL > 22 PS3 'Florida EV1' 0 Liquid 600 875 17 'Florida EV1' 100 Liquid 600 424 22 'Florida EV1' 200 Liquid 600 285 18 'Florida EV1' 100 Semi-solid 600 535 17 'Valencia' 0 Liquid 420 361 0 'Valencia' 100 Liquid 420 281 2 'Valencia' 200 Liquid 420 177 4 'Valencia' 100 Semi-solid 420 247 1 1The two cultivars were tested and analyzed separately. 2SL > 2, shoots longer than 2 mm. 3PS, GFP positive shoots. For 'Florida EV1', the mean shoot length greater than 2 mm (SL > 2) variable in the 100 and 200 mg·L−1 kanamycin treatments in liquid media and the 100 mg·L−1 control in the semi-solid medium was significantly less (p < 0.05) than the 0 mg·L−1 kanamycin liquid control treatment (Table 3 & Fig. 3a). Fewer shoots (n = 285) were screened at 200 mg·L−1 compared to the 0 mg·L−1 control (n = 875) (Table 2). The mean TES (transformation efficiency based on the number of positive shoots/total number of shoots × 100) variable at 200 mg·L−1 kanamycin in liquid medium was 7.9% ± 2.7; at 100 mg·L−1 kanamycin in liquid medium, it was 5.4% ± 1.4 (Table 3). However, the latter treatment generated more transgenics, although it was not statistically different from the mean TES value at 200 mg·L−1 (Fig. 3a). The mean TES value of 7.9% was significantly greater than the 0 mg·L−1 kanamycin liquid medium control and the 100 mg·L−1 kanamycin semi-solid control (p < 0.05), showing that liquid medium selection is superior (Table 3 & Fig. 3a). The mean TEE variable was not significant (Table 3 & Fig. 3b), and the number of positive shoots (PS) was not significant because transgenics were produced at every kanamycin level (Table 2). Transgenics readily regenerated in 'Florida EV1' at all kanamycin concentrations.
Table 3. The means for the number of positive shoots (PS), shoots with lengths greater than 2 mm (SL > 2), and transformation efficiencies (TES and TEE) at three concentrations of kanamycin in liquid and semi-solid medium for 'Florida EV1' and 'Valencia'.
Cultivar1 Kanamycin (mg·L−1) Medium Mean PS2 ± SE3 Mean SL > 24 ± SE Mean TES5 ± SE Mean TEE6 ± SE 'Florida EV1' 0 Liquid 1.7 ± 0.4 87.5a ± 14.3 2.4b ± 0.7 2.8 ± 0.7 'Florida EV1' 100 Liquid 2.2 ± 0.6 42.4b ± 3.7 5.4ab ± 1.4 3.7 ± 1.0 'Florida EV1' 200 Liquid 1.8 ± 0.5 28.5b ± 3.2 7.9a ± 2.7 3.0 ± 0.8 'Florida EV1' 100 Semi-solid 1.7 ± 0.4 53.5b ± 8.7 2.9b ± 0.7 3.2 ± 1.0 'Valencia' 0 Liquid 0 ± 0 51.6a ± 4.4 0 ± 0 0 ± 0 'Valencia' 100 Liquid 0.3 ± 0.2 40.1b ± 2.0 0.7 ± 0.4 0.5 ± 0.3 'Valencia' 200 Liquid 0.6 ± 0.4 25.3c ± 2.7 2.4 ± 1.7 0.9 ± 0.7 'Valencia' 100 Semi-solid 0.1 ± 0.1 35.3bc ± 5.5 0.3 ± 0.3 0.2 ± 0.2 1The two cultivars were tested and analyzed separately. 2PS, GFP positive shoots. 3SE, Standard error. 4SL > 2, the number of shoots longer than 2 mm. 5TES, transformation efficiency based on the number of shoots. 6TEE, transformation efficiency based on the number of explants. 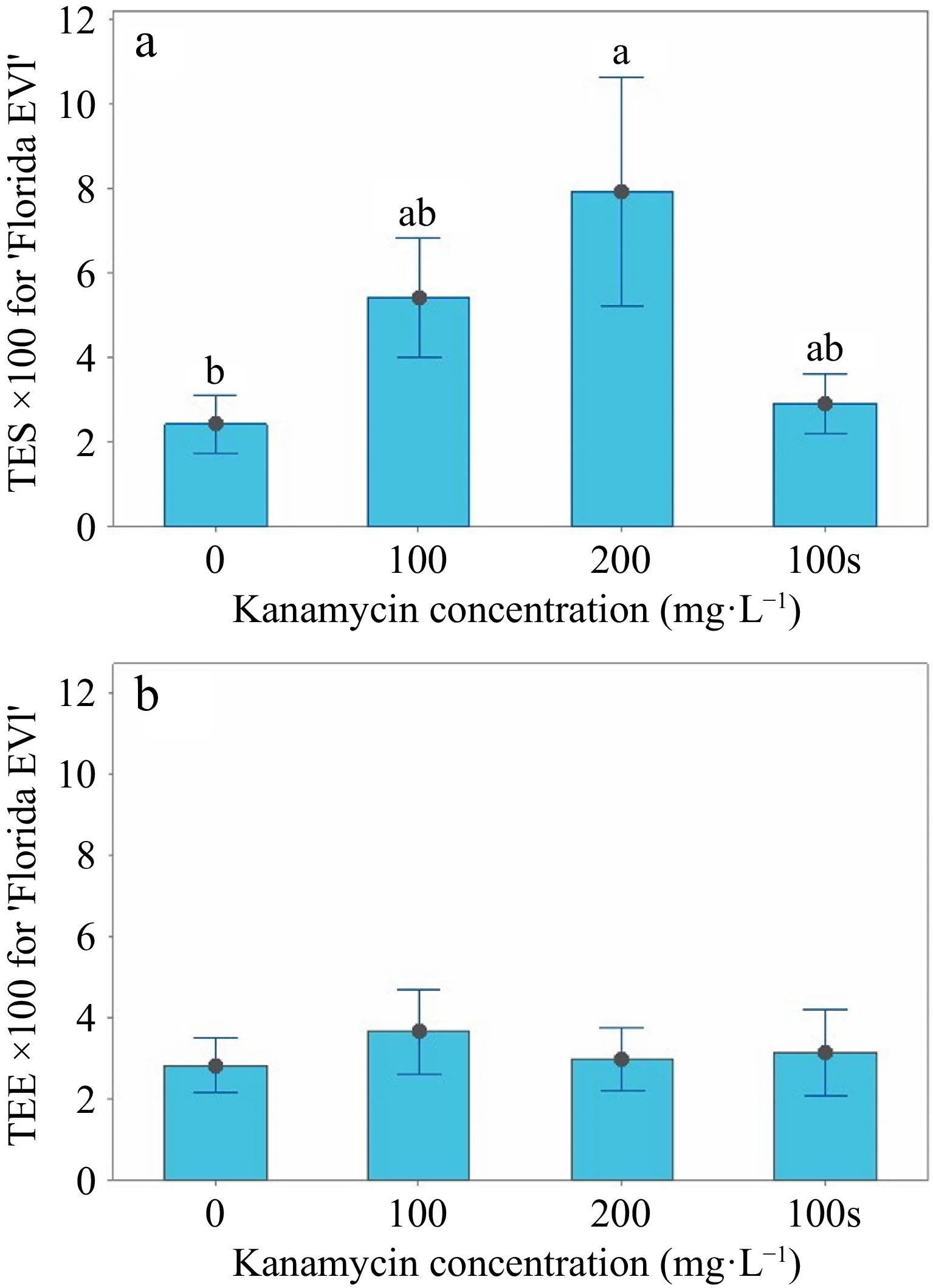
Figure 3.
TES and TEE interval graphs for 'Florida EV1'. (a) The means, standard errors (SE) for TES (transformation efficiency based on the number of positive shoots/total number of shoots × 100), and multiple comparisons are shown in liquid medium at 0, 100, and 200 mg·L−1 and in semi-solid medium at 100 mg·L−1. (b) The means and standard errors (SE) for TEE (transformation efficiency based on the number of positive shoots/explants × 100) are shown in liquid medium at 0, 100, and 200 mg·L−1 and in semi-solid medium at 100 mg·L−1.
For 'Valencia', the mean SL > 2 variable at 0 mg·L−1 was significantly greater (p < 0.05) than the other treatments (Table 3), but there were fewer shoots to screen (n = 177) at 200 mg·L−1 kanamycin (Table 2). In contrast, 361 shoots grew in the 0 mg·L−1 control. No significant differences existed for the TES or TEE variables (Table 3, Fig. 4a & b). Two transgenics were identified by GFP fluorescence at 100 mg·L−1, while four were identified at 200 mg·L−1 kanamycin in liquid medium, so selection was increased at higher kanamycin concentrations. Only one transgenic developed in the semi-solid control medium with 100 mg·L−1 (Table 2). No transgenics were produced without kanamycin selection in the liquid medium, and the PS variable was not significant (Table 3). One 'Valencia' transgenic died after grafting.
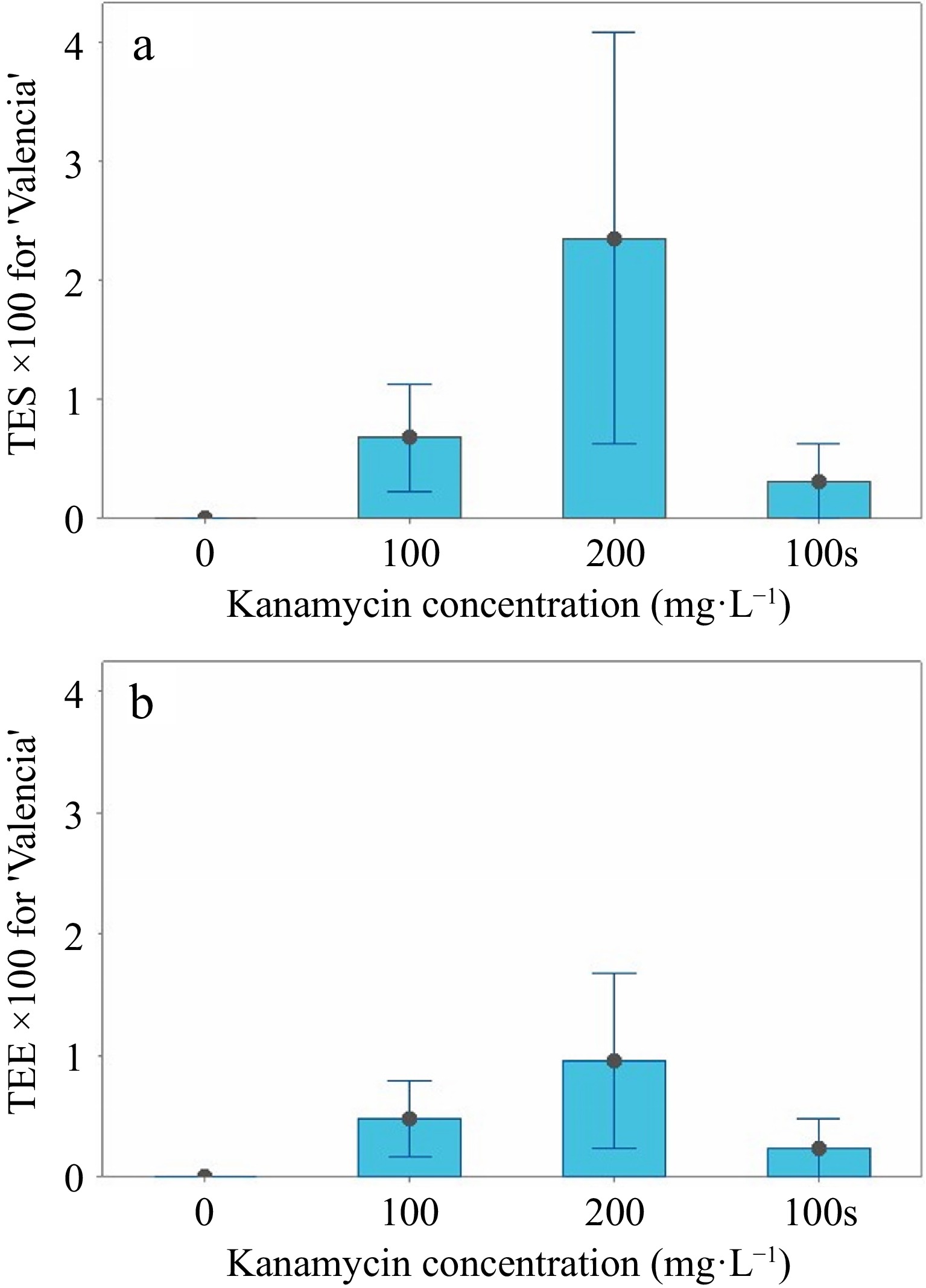
Figure 4.
TES and TEE interval graphs for 'Valencia'. (a) The means and standard errors for TES (transformation efficiency based on the number of positive shoots/number of shoots × 100) are shown in liquid medium at 0, 100, and 200 mg·L−1 and in semi-solid medium at 100 mg·L−1. (b) The means and standard errors (SE) for TEE (transformation efficiency based on the number of positive shoots/explants × 100) are shown in liquid medium at 0, 100, and 200 mg·L−1 and in semi-solid medium at 100 mg·L−1.
A nonparametric Mann-Whitney test was used to compare the median values for regeneration ability (the number of shoots per explant) between the two cultivars. Across all treatments, the median value for 'Florida EV1' was 0.71 (n = 40), which was significantly greater (p < 0.05) than the median value for 'Valencia' at 0.63 (n = 28). Yet in the control treatment at 0 mg·L−1 kanamycin in liquid medium, the median regeneration ability for 'Florida EV1' was 1.19 (n = 10) was not significantly different from 'Valencia's' median regeneration ability at 0.98 (n = 7). A Mann-Whitney test was also used to compare both cultivars' median values for GFP positive shoots (PS). The median value for 'Florida EV1' was 1.5 (n = 40), while the median value for 'Valencia' was 0 (n = 28), indicating a highly significant difference (p = 0.00) in the medium number of positive shoots between the two cultivars.
Glyphosate as a selectable marker in mature citrus
-
To test whether glyphosate can be used as a selectable marker in mature tissue transformation, an experiment to show glyphosate tolerance conferred by the EPSPS selectable marker was conducted with four levels of glyphosate (0, 2.6, 6.5, and 13.1 mM) with the transgenic lines and wild-type (WT). Nodal budsticks were plated on glyphosate medium, and the sprouting of shoots was measured after 28 d. An ANOVA of the data comparing transgenic and WT nodal budsticks for their ability to sprout shoots at these concentrations of glyphosate medium revealed significant differences (p < 0.05) between the two cultivars. Across all treatments and the control, 'Florida EV1' sprouted more shoots per node than 'Valencia' (Figs 5 & 6). At 0 mM glyphosate, 19 shoots sprouted from 12 nodal budsticks in 'Florida EV1', whereas in 'Valencia', 12 shoots sprouted from 12 nodal budsticks. Transgenic 'Florida EV1' had a mean of 0.9 (SE ± 0.1) sprouted shoots, while 'Valencia' had a mean of 0.5 (SE ± 0.1) shoots. Citrus lines 3 and 22 died on the glyphosate medium (photo not shown) (Fig. 5), whereas line 14 died after secondary grafting before this experiment. There was no significant difference among the different glyphosate concentrations, indicating that the lowest concentration (2.6 mM) was enough to select for herbicide resistance in transgenic mature citrus scions. All nodal budsticks of WT 'Florida EV1' and 'Valencia' died at all concentrations of glyphosate (Fig. 5, Supplementary Table S1).
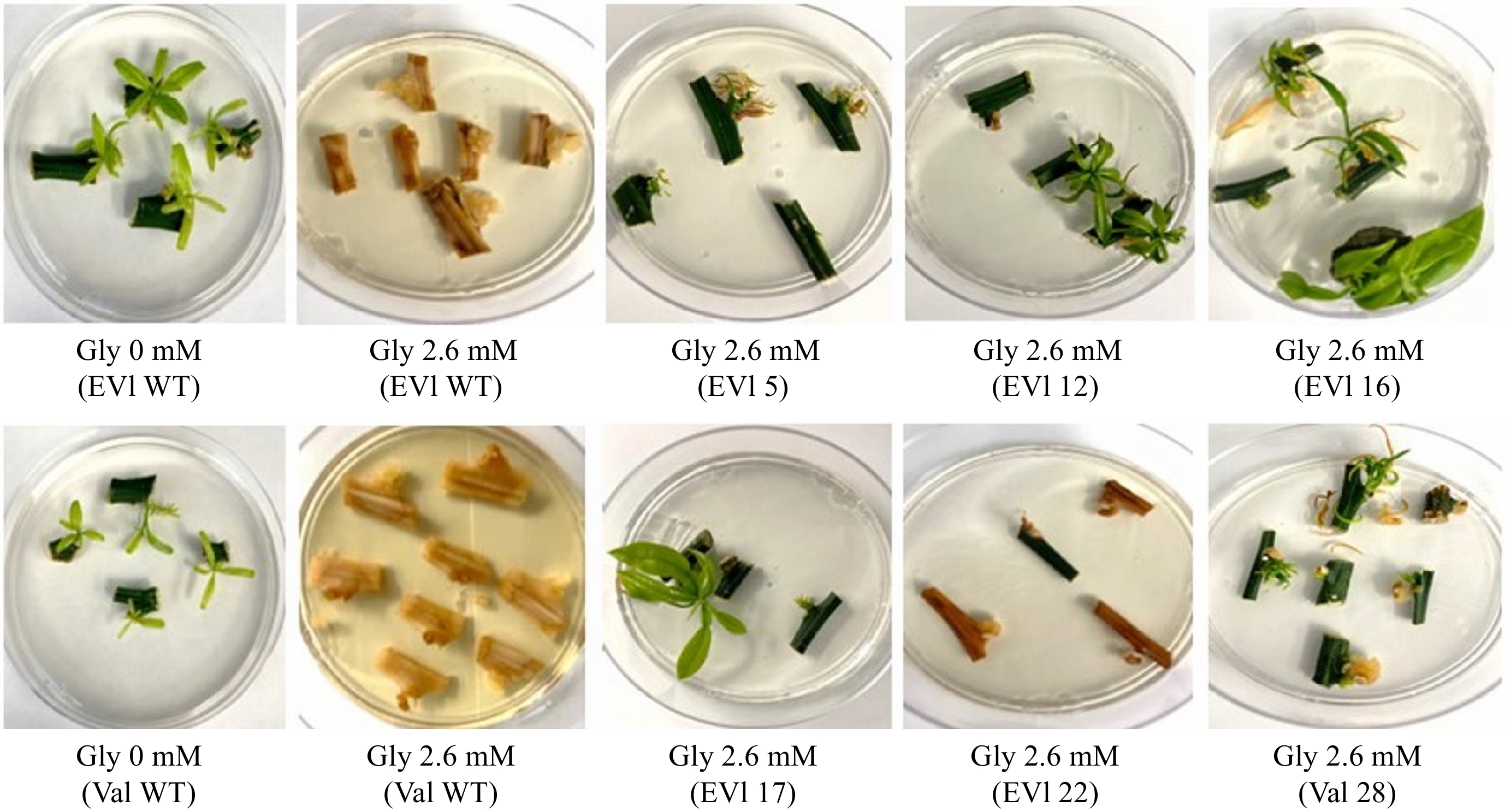
Figure 5.
Transgenic lines and wildtype (WT) nodal budsticks at two glyphosate concentrations (0 and 2.6 mM). Nodal budsticks of 'Florida EV1' (EV1) (samples 5, 12, 16, 17, 22, and WT) and 'Valencia' (Val) (samples 28 and WT) were plated on glyphosate (Gly) medium to show tolerance conferred by the TIPS-EPSPS transgene. Two additional concentrations of glyphosate were used but are not shown (Supplementary Table S1). 'Florida EV1' line 22 was later shown not to possess the EPSPS transgene.
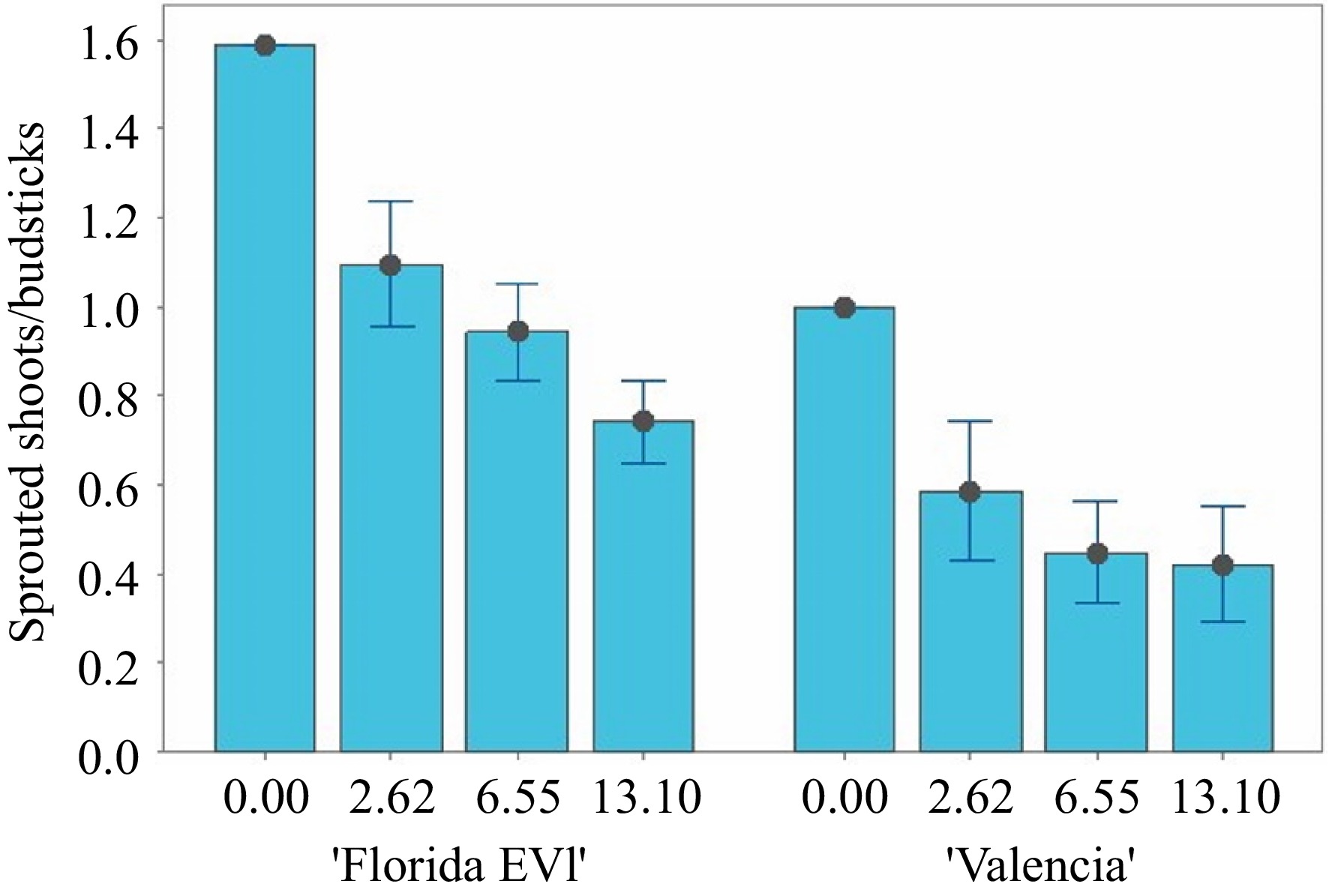
Figure 6.
Sprouted shoots per nodal budstick at four concentrations of glyphosate in semi-solid medium for 'Florida EV1' and 'Valencia'. In some treatments, 'Florida EV1' sprouted more than one shoot per nodal budstick.
Molecular confirmation and copy number
-
PCR tests were performed for the transgenes (nptII, egfp, and EPSPS) in 23 'Florida EV1' and six 'Valencia' transgenic lines. All transgenic lines analyzed carried the nptII and gfp genes at the correct sizes (nptII 239 bp; egfp 713 bp) (Fig. 7). The 455 bp EPSPS transgene was present in all samples except 'Florida EV1' lines 3, 10, 14, and 22. Line 23 of 'Florida EV1' could not be tested with the EPSPS primers because it died.
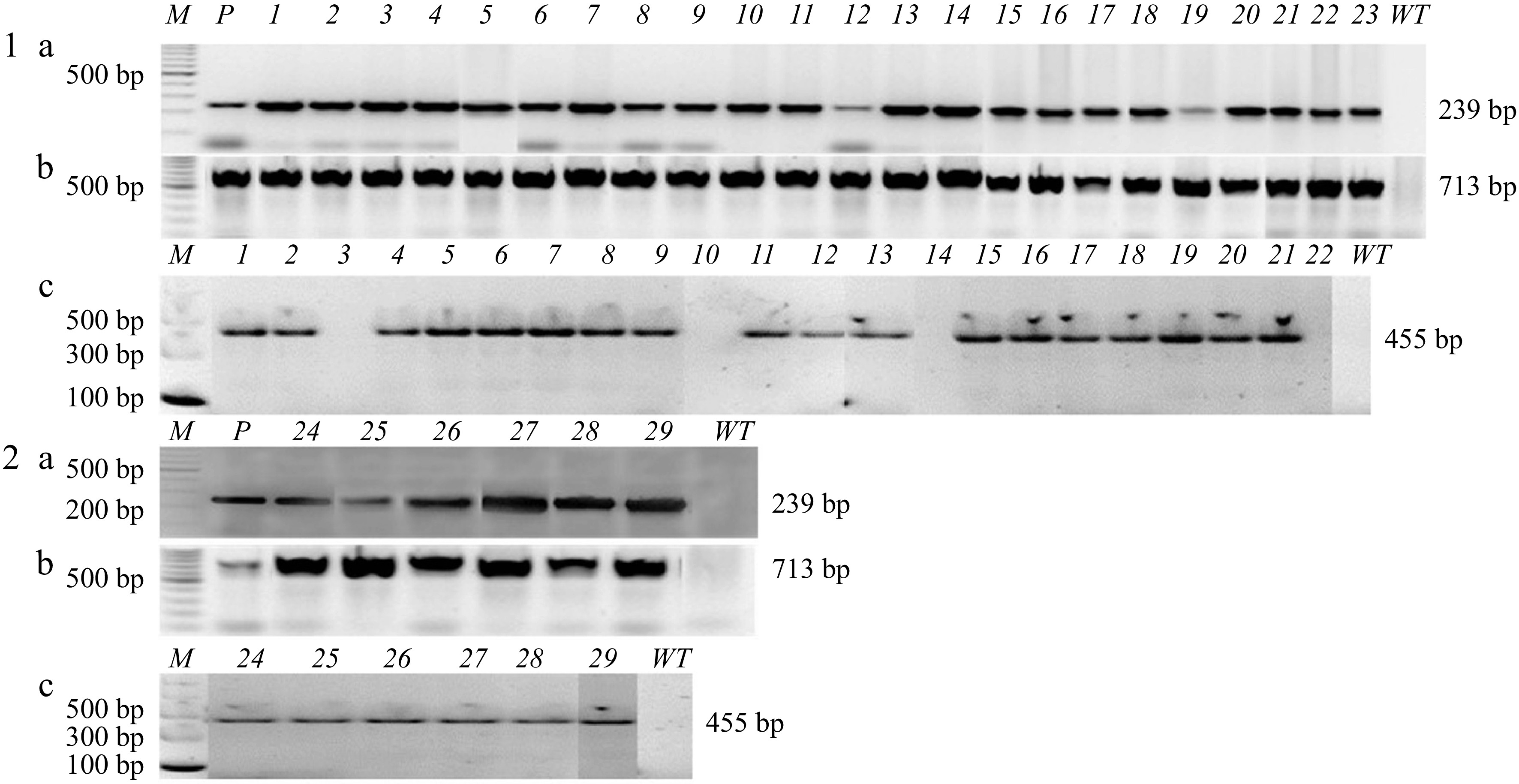
Figure 7.
PCR confirmation of transgenic lines transformed with the Mu-EPSPS vector and selected in liquid medium with gravity wells. (1a) 'Florida EV1' 1-23 samples, PCR for nptII 239 bp; (1b) egfp 713 bp; (1c) EPSPS 455 bp. Samples 3, 10, 14, and 22 contained the nptII gene but not the EPSPS gene. (2a) 'Valencia' 24-29 samples, PCR for nptII 239 bp; (2b) egfp 713 bp; (2c) EPSPS 455 bp. M: DNA ladder; P: plasmid; WT: wild-type.
The 'Florida EV1' plants selected for duplexed TaqMan real-time PCR for copy number analysis were the ones that had the greatest number of shoots on the glyphosate selection medium (Supplementary Table S1) as well as the 'Valencia' transgenics. Eight transgenic lines were analyzed: four from 'Florida EV1' and four from 'Valencia' (Table 3). The maximum copy number determined was two. For 'Florida EV1', there were three transgenic lines with a single nptII copy and one with two copies of this gene. In the case of 'Valencia', there were two lines with one copy and two lines with two copies.
-
There are no previous reports of transformation experiments with mature 'Florida EV1'. Earlier, we transformed 'Valencia' with the EHA101 Agrobacterium strain and the GFP reporter[15] and obtained similar transformation efficiency for TEE as reported here if you calculate the efficiency based on the same number of explants in semi-solid medium at 100 mg·L−1 kanamycin. From the present study, we can conclude that liquid medium doubles the transformation efficiency compared to semi-solid medium with same kanamycin concentration. One additional report cited a higher, but still low, mature 'Valencia' transformation efficiency[14] , but another report produced no transgenics in mature 'Valencia' whatsoever[13].
The most important measures in screening mature citrus shoots for transformation or gene editing in mature citrus are the number of shoots that must be examined that are long enough to be micrografted (SL > 2 mm) and the number of positive shoots (PS). Thus, the variable of transformation efficiency based on the number of shoots (TES) is the most valuable for our purposes[15,23,24,33]. In contrast, transformation efficiency based on the number of explants (TEE) better reflects the starting material (explants) required to initiate an experiment to find positive shoots. 'Florida EV1' is much more amenable to transformation and shoot regeneration, and it has repeatedly proven so in our transformation facility (unpublished results).
In both mature and immature citrus biotechnology, the standard for kanamycin selection is 100 mg·L−1 in a semi-solid medium, except nptII is a poor selectable marker because it permits many escaped non-transgenic shoots to grow[15,22]. However, it is useful in both immature and mature citrus transformation and gene editing because it does not kill the explants. At higher concentrations of kanamycin in liquid medium, fewer escaped shoots grew from mature rootstock explants as determined by GUS assays[23]. From that study and the present work, it can be concluded that screening shoots of mature citrus scion explants in liquid medium with high concentrations of kanamycin is superior to screening shoots on semi-solid medium with 100 mg·L−1 kanamycin. Transgenics were even produced at 0 mg·L−1 kanamycin, which is a normal occurrence for mature citrus[33].
'Florida EV1' lines 3, 10, 14, and 22 were positive for the nptII transgene but negative for the EPSPS transgene. EPSPS, on the 3' end of the T-DNA, was most likely truncated during T-DNA transfer because it is on the left border, which is not protected by the VirD2 protein[9,34,35].
The 5-enolpyruvylshikimate 3-phosphate synthase (EPSPS) genes in plants and microbes occur in nature and have evolved naturally in response to glyphosate selection pressure. Several glyphosate-resistant EPSPS variants have been successfully engineered to impart herbicide resistance in a variety of crop plants[36,37]. The highly efficient citrus TIPS-EPSPS glyphosate selection system can not only serve as an alternative to antibiotic-based selection methods in the genetic transformation of citrus, but it can also facilitate the production of intragenic citrus plants. This present work suggests that it can be used as a selectable marker in mature citrus.
Liquid culture maximizes the availability of water and nutrients and eliminates the adverse effects of contaminants in agar, which might contain impurities that limit plant growth[38−42]. The shoots in the medium avoid hyperhydricity and anoxia because the vessel is aerated[43]. A thin layer of 3M paper or paper towel is added to the vessel, allowing the explants to contact the liquid without being submerged and suffering hypoxia. The gravity wells gradually dispense liquid as the explants and shoots use it. Simple systems such as stationary thin films often yield high-quality plant growth without the complexities and cost of mechanized systems that require shakers and aeration tubes[44]. Liquid selection in mature citrus rootstock reduces labor and costs and increases transformation efficiency[23], and the results were confirmed here with mature citrus scions. The liquid medium allows for larger, more elongated shoots for micrografting, better multiplication, and easier subculturing.
-
'Valencia' is one of the most important citrus cultivars grown in the US, yet it is recalcitrant to Agrobacterium transformation. Mature 'Florida EV1', a 'Valencia' somaclone, has better transformation efficiency and regeneration ability than 'Valencia'. 'Florida EV1' performed remarkably well at all kanamycin concentrations in liquid and semi-solid medium. The TES variable in liquid selection in We-V™ vessels with gravity wells at stringent kanamycin concentrations (200 mg·L−1) was significantly better for identifying transgenics than in semi-solid medium at 100 mg·L−1 and the controls. A glyphosate assay with mature nodal budsticks suggests that the TIPS-EPSPS gene, which confers resistance to glyphosate, can be used as a selectable marker in mature citrus. 'Florida EV1' is being used in gene editing to alleviate disease problems afflicting the citrus industry in Florida. Transgenic and WT 'Florida EV1' trees produced fruit earlier than 'Valencia', which agrees with field performance data[18]. Future research will investigate the reasons for the differences in transformation efficiency and regeneration ability at the DNA sequence and chromosomal level between 'Florida EV1' and 'Valencia'.
-
The authors confirm contribution to the paper as follows: study conceptualization and design: Grosser J, Mou Z, Zale J; data curation: Canton M, Zale J; formal analysis: Canton M, Zale J; investigation: Canton M, Peraza-Guerra O, Wu H; methodology: Canton M, Peraza-Guerra O, Wu H; validation: Canton M, Zale J; draft manuscript preparation: Canton M, Peraza-Guerra O, Zale J; visualization: Canton M, Zale J; resources: Grosser J, Mou Z, Zale J; funding acquisition: Zale J; supervision: Zale J. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analyzed during this study are included in this published article and its supplementary information files.
We wish to acknowledge the University of Florida Institute of Food and Agricultural Sciences (IFAS) and the Citrus Research and Education Center (CREC) for Citrus Initiative Funding.
-
The authors declare that they have no conflict of interest.
- Supplementary Table S1 The number of nodal budsticks and shoots at each glyphosate concentration in the two cultivars.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Canton M, Peraza-Guerra O, Wu H, Grosser J, Mou Z, et al. 2024. A mature, sweet orange cultivar derived from 'Valencia' with high Agrobacterium transformation efficiency. Fruit Research 4: e036 doi: 10.48130/frures-0024-0030
A mature, sweet orange cultivar derived from 'Valencia' with high Agrobacterium transformation efficiency
- Received: 11 June 2024
- Revised: 07 August 2024
- Accepted: 19 September 2024
- Published online: 04 November 2024
Abstract: Transformation efficiencies of sweet orange cultivars 'Florida EV1' and 'Valencia', recalcitrant to Agrobacterium transformation, were investigated using liquid culture in We-V™ vessels. The two mature cultivars were transformed using Agrobacterium with a vector containing selectable markers, nptII and TIPS-EPSPS, and the GFP reporter. Transgenics were identified with GFP in liquid culture at 0, 100, and 200 mg·L−1 kanamycin or in the semi-solid control with 100 mg·L−1 kanamycin. For 'Florida EV1', there were significant differences in the mean transformation efficiency based on the number of shoots screened (TES) at all kanamycin concentrations. Selection at 200 mg·L−1 was better than at lower concentrations in liquid or semi-solid control medium with 100 mg·L−1 kanamycin. The variable TEE, transformation efficiency based on the number of explants, did not discern differences. The means ± standard errors for TES at 200 mg·L−1 were 7.9% ± 2.7% for 'Florida EV1' and 2.4% ± 1.7% for 'Valencia'. In total, 74 transgenics were produced in 'Florida EV1', whereas seven were generated in 'Valencia'. Obtaining transgenics in 'Florida EV1' was easy; fewer shoots were screened at 200 mg·L−1. 'Florida EV1' exhibited better regeneration ability, and all transgenics survived on glyphosate medium, suggesting the TIPS-EPSPS selectable marker could be useful in transformation. Molecular analyses confirmed their transgenic nature. 'Florida EV1' trees produced fruit earlier than 'Valencia' in less than two years. 'Florida EV1' could accelerate the production of HLB disease-resistant trees.













