-
Plants are continuously exposed to pathogen attacks in natural environments. Through evolution, they have developed a sophisticated immune system to combat pathogens[1]. The first layer, Pathogen-associated Molecular Patterns (PAMPs)-triggered immunity (PTI), is initiated when pattern recognition receptors (PRRs) such as leucine-rich repeat receptor-like kinases (LRR-RLKs) detect conserved PAMPs. When pathogens bypass PTI via effector secretion, plants activate effector-triggered immunity (ETI), typically mediated by nucleotide-binding leucine-rich repeat (NLR) proteins that recognize specific effectors[2]. Both PTI and ETI can trigger the hypersensitive response (HR)[3], a form of programmed cell death (PCD) that restricts pathogen spread by forming a barrier of dead cells at infection sites. LRR-RLKs not only initiate PTI but also modulate HR through phosphorylation cascades regulating reactive oxygen species (ROS) burst and defense gene expression[4], playing a central role in early immune signaling and resistance responses. In Arabidopsis thaliana, LRR-RLK genes such as FLS2 and EFR detect bacterial flagellin and elongation factor EF-Tu, respectively[5]. However, the role of LRR-RLKs in resistance to Xanthomonas arboricola pv. pruni (Xap) in peach (Prunus persica) remains unclear. Xap causes bacterial spot disease, leading to leaf necrosis, fruit drop, and up to 30% yield loss[6]. Although chemical control can partially reduce symptoms, the lack of resistant cultivars remains a major limitation. Therefore, identifying resistance genes and elucidating their regulatory mechanisms is essential for peach breeding[7]. Here, an LRR-RLK gene, PpLRR1, that contributes to HR, was identified. Further analysis revealed that PpLIMYB transcription factor (TF) regulates PpLRR1 expression. These findings offer insights into PpLRR1-mediated resistance and provide valuable genetic resources for breeding disease-resistant peach varieties.
Healthy leaves of 'Guanghetao (GHT, Prunus mira)' and 'Qiubaitao (QBT, Prunus persica)' were inoculated with Xap. Disease symptoms appeared at 2 d post-inoculation (dpi) and became more pronounced by 4 dpi (Fig. 1a). Leaves were sampled at 0, 2, and 4 dpi for transcriptome sequencing, which identified ten upregulated LRR-RLK differentially expressed genes (DEGs) (Fig. 1b). Of these, four DEGs Prupe.1G168500, Prupe.4G069400, Prupe.4G069600, and Prupe.7G262300 were selected for preliminary functional validation based on their significant expression in both GHT and QBT (Supplementary Fig. S1). Among these, Prupe.4G069400 and Prupe.4G069600 encoded identical proteins; therefore, only one was used in subsequent analyses. Transient expression of the three candidate genes in Nicotiana benthamiana revealed that only PpLRR1 induced PCD (Fig. 1c), a response also observed in peach leaves (Fig. 1d; Supplementary Table S1). qRT-PCR analysis showed a significant upregulation of PpLRR1 at 4 dpi (Fig. 1e), suggesting its involvement in hypersensitive response (HR)-mediated cell death during pathogen infection. Therefore, PpLRR1 was chosen as the primary candidate for further study. Phylogenetic analysis (Supplementary Fig. S2) placed PpLRR1 within the LRR-RLK-IX subfamily, clustering closely with AtBARK1 from Arabidopsis and NbBIR1 from tobacco[8]. Additional analyses of gene structure and physicochemical properties of LRR-RLK-IX subfamily members were performed (Supplementary Table S2). The PpLRR1-encoded protein contains a canonical LRR-TM-PK (leucine-rich repeat–transmembrane–protein kinase) domain architecture, consistent with its predicted function as a receptor-like kinase and its homology to known plant resistance genes (Supplementary Fig. S3). Subcellular localization assays confirmed PpLRR1 is primarily localized to the plasma membrane (Fig. 1f), supporting its role as a pattern recognition receptor (PRR) involved in pathogen perception[9].
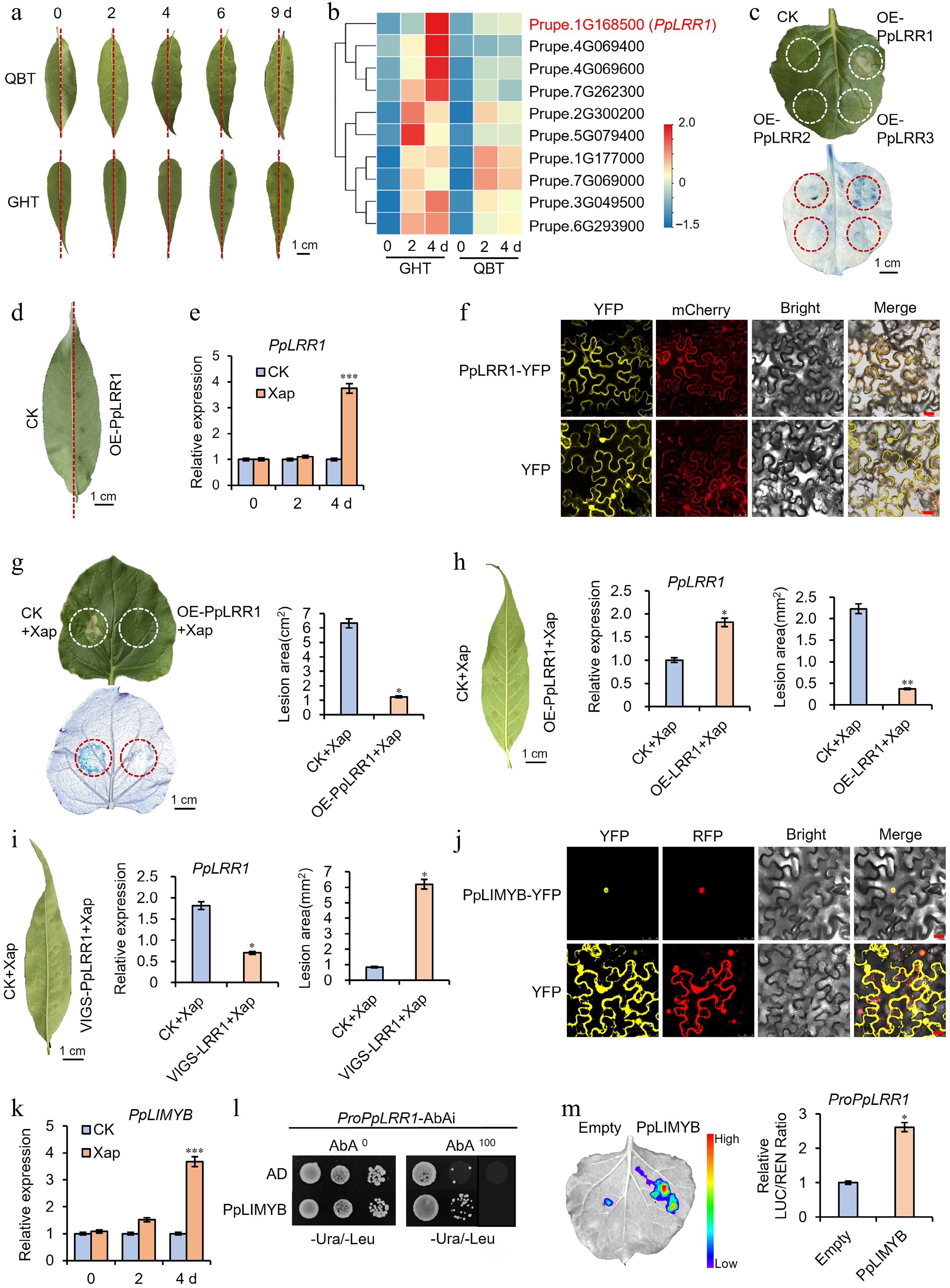
Figure 1.
Functional validation of PpLRR1 in resistance to peach bacterial spot disease. (a) Phenotypic comparison of peach leaves from 'Qiubaitao (QBT, Prunus persica)' and 'Guanghetao (GHT, Prunus mira)' cultivars inoculated with Xanthomonas arboricola pv. pruni (Xap) vs control at 4 dpi. Transcriptome sequencing was conducted on leaf samples at 0, 2, and 4 dpi. Scale bar = 1 cm. (b) Heatmap of differentially expressed genes (|log2FC| > 1, p < 0.05) identified from RNA-seq data. PpLRR1 is highlighted in red. (c) Trypan blue-stained tobacco leaves transiently overexpressing PpLRR1, PpLRR2, or PpLRR3 compared to the empty vector control at 3 dpi. (d) Hypersensitive response phenotype in peach leaves transiently overexpressing PpLRR1 at 4 dpi. (e) Relative expression of PpLRR1 in 'Mantianhong (MTH)' peach leaves at 0, 2, and 4 dpi, measured by qRT-PCR. Expression was normalized to the sterile water-injected control, set to 1. (f) Subcellular localization of PpLRR1-YFP fusion protein in Nicotiana benthamiana epidermal cells. Yellow fluorescence (YFP) co-localizes with the red fluorescent plasma membrane marker (mCherry). OD600 = 1.0. Cultivate for 48 h after injection. Scale bar = 25 μm. (g) Quantification of lesion area in tobacco leaves co-inoculated with Xap and overexpressing PpLRR1 at 3 dpi (trypan blue staining). (h) Lesion area and relative PpLRR1 expression in MTH peach leaves co-inoculated with Xap and overexpressing PpLRR1 at 4 dpi. (i) Lesion area and PpLRR1 expression in MTH peach leaves co-inoculated with Xap following PpLRR1 silencing. TRV, TRV1 + TRV2; PpLRR1-TRV, TRV1 + PpLRR1-TRV2. (j) Subcellular localization of PpLIMYB-YFP fusion protein in transgenic N. benthamiana epidermal cells. Yellow fluorescent signal (YFP) co-localizes with RFP (red). TM–NR: tonoplast marker and red fluorescent nuclear marker (Nucleus–RFP). OD600 = 1.0. Cultivate for 48 h after injection. Scale bar = 25 μm. (k) Relative expression levels of PpLIMYB in peach leaves at 0, 2, and 4 dpi assessed by qRT-PCR. Expression was normalized to the sterile water-injected control, set to 1. (l) Yeast one-hybrid assay showing direct binding of PpLIMYB to the PpLRR1 promoter. AbA0: medium without AbA; AbA100: medium with 100 ng/mL AbA. (m) Dual-luciferase reporter assay quantifying PpLIMYB-mediated activation of the PpLRR1 promoter. Data represent mean ± SD of three biological replicates (*p < 0.05). Each treatment was performed in triplicate, and each replicate contained three to five peach leaves. Error bars indicate standard deviation. Statistical significance was assessed by one-way ANOVA and t-test: * p < 0.05; ** p < 0.01; *** p < 0.001.
To further validate the functional role of PpLRR1 in disease resistance, the gene was co-expressed with the pathogen in tobacco leaves via Agrobacterium-mediated transformation. Trypan blue staining showed that PpLRR1 significantly reduced disease severity (Fig. 1g), consistent with the effects of LcPIP1 in Litchi chinensis[10], and Xa21 in Oryza sativa[3]. The overexpression of PpLRR1 significantly mitigated disease symptoms and reduced lesion area (Fig. 1h), while gene silencing led to enhanced susceptibility and increased lesion area (Fig. 1i), further supporting its role in resistance. Moreover, transgenic Arabidopsis plants overexpressing PpLRR1 exhibited significantly reduced lesion areas and milder disease symptoms upon Xap inoculation compared to wild-type plants (Supplementary Fig. S4), confirming its functional contribution to peach disease resistance by mediating HR and limiting pathogen spread.
To investigate how PpLRR1 regulates disease resistance, its promoter region was analyzed (Supplementary Fig. S5), and multiple stress-responsive cis-elements were identified, including binding sites for 14 MYB (C/TAACT/CG/A), three MYC (CAA/TTTG), two WRKY (TTGACC/T), and two bHLH (G-box: CACGTG) TFs. Notably, MYB-binding sites were the most abundant. This is significant given the established role of MYB TFs as central regulators of plant immunity, including antiviral defense[11,12] and antibacterial responses[13]. Although several TF families may regulate PpLRR1, focus was placed on MYB TFs. Transcriptome analysis identified 17 MYB-related TFs that were both pathogen-responsive and significantly upregulated. Among these, PpLIMYB was identified via yeast one-hybrid (Y1H) assays. Subcellular localization confirmed its nuclear function, and qRT-PCR revealed strong upregulation at 4 dpi, mirroring the PpLRR1 expression pattern (Fig. 1k). Additionally, co-overexpression of PpLIMYB and Xap in peach leaves led to simultaneous upregulation of PpLRR1 (Supplementary Fig. S6), suggesting a regulatory relationship. Y1H (Fig. 1l) and dual-luciferase reporter assays (Fig. 1m) confirmed that PpLIMYB directly binds to the PpLRR1 promoter and activates its transcription.
These findings reveal the important role of the PpLIMYB-PpLRR1 module in peach immunity against Xap (Supplementary Fig. S7). Similar to Arabidopsis LRR-RLKs (e.g., FLS), which activate MAPK-mediated defense signaling[5], PpLRR1 likely acts downstream of PpLIMYB to regulate defense genes (e.g., PR1 and PR5). The plant immune response is a complex process that may also trigger other immune responses, such as the activation of downstream immune genes, calcium ion influx, callose deposition, and the production of phytoalexins, among other cascading reactions. This mechanism resembles LcPIP1-mediated resistance in litchi. The membrane localization of PpLRR1 (Fig. 1e) suggests it perceives extracellular pathogen signals. Its activation of PpLRR1 by PpLIMYB provides new evidence for a 'receptor-TF' regulatory module in plant immunity. Future studies should examine interactions between PpLRR1 and co-receptors[14,15], and evaluate the conservation of this module across peach cultivars. PpLRR1 is strongly induced by Xap infection and confers resistance by activating downstream defenses. Therefore, these findings not only provide valuable genetic resources for breeding disease-resistant peach varieties but also offer an insight into the specific regulatory interactions in peach, a woody fruit tree with unique genetic and physiological characteristics. Further research will clarify the molecular functions of PpLRR1 and its applications in resistance breeding.
HTML
This work was financially supported by grants from the Chinese Academy of Sciences Talent Project (Grant No. E5299903), the Hubei Talent Project (Grant No. E4399901), the National Natural Science Foundation of China (Grant No. 32302497), and the China Agriculture Research System (Grant No. CARS-30).
-
The authors confirm their contributions to the paper as follows: study conception and design: An JP; data collection: Li MY, Zhao L, Di A, Li ZY; analysis and interpretation of results: Li MY, Liao L, Han Y, Luo CX; draft manuscript preparation: Li MY, An JP. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analyzed during this study are included in this published article and its supplementary information files.
-
The authors declare that they have no conflict of interest.
- Supplementary Table S1 Related PCR primer sequences.
- Supplementary Table S2 Comparative analysis of physicochemical properties of LRR-RLK-IX subfamily members in peach and Arabidopsis.
- Supplementary Fig. S1 Expression analysis of three selected genes based on transcriptomic data.
- Supplementary Fig. S2 Phylogenetic analysis of LRR-RLK-IX subfamily members across four species.
- Supplementary Fig. S3 Genomic characterization of LRR-RLK-IX subfamily members.
- Supplementary Fig. S4 DAB staining of Arabidopsis thaliana leaves after Xap infection, showing ROS accumulation (brown deposits).
- Supplementary Fig. S5 Cis-acting element analysis of the PpLRR1 promoter region.
- Supplementary Fig. S6 Quantitative RT-PCR analysis of PpLIMYB and PpLRR1 expression in peach leaves co-treated with PpLIMYB overexpression and pathogen infection.
- Supplementary Fig. S7 A working model of PpLRR1 functioning in disease resistance mechanism.
- Copyright: © 2025 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
| Li MY, Zhao L, Ai D, Li ZY, Liao L, et al. 2025. The leucine-rich repeat receptor-like kinase PpLRR1 enhances peach resistance to Xanthomonas arboricola pv. pruni. Fruit Research 5: e036 doi: 10.48130/frures-0025-0029 |













