-
Tanshinones are a group of abietane diterpenoid natural products synthesized in the root of Salvia miltiorrhiza Bunge, commonly called Danshen, a renowned traditional Chinese medicine. To date, over 40 tanshinones and structurally-related compounds have been identified from Danshen[1], such as Cryptotanshinone (CPT), Tanshinone IIA (Tan IIA), Dihydrotanshinone I (DHT), and Tanshinone I (Tan I), which reportedly provide clinical benefits for treating cardiovascular diseases[2]. The accumulation of tanshinones is frequently induced by the surrounding biotic and abiotic stresses, and is mediated by multi-tiered hormonal signaling pathways and transcription factors[3]. Among these, jasmonates (JAs), including 12-oxo-phytodienoic acid (OPDA), jasmonic acid (JA), methyl jasmonate (MeJA), and jasmonoyl-isoleucine (JA-Ile), play crucial roles in the regulation of tanshinone metabolism[4]. JA-Ile, as a core small molecule in JA signaling transduction, plays a predomiant role in connecting coronatine Insensitive 1 (COI1) with jasmonate-ZIM domain (JAZ) proteins. It promotes the ubiquitination and degradation of JAZ proteins, leading to the release of transcription factors and the regulation of downstream biosynthetic genes[5]. For instance, SmMYC2a/b and SmMYB39 interact with SmJAZ1/2, SmERF73 interacts with SmJAZ3, SmbHLH37 interacts with SmJAZ3/8, which ultimately regulate the biosynthesis of tanshinones and phenolic acids[4,6−8]. Additionally, in Artemisia annua, the interaction between AaWRKY9 and AaJAZ9 has also been discovered[9]. The types of transcription factors involved in JA signaling transduction through interactions with JAZ proteins are gradually being expanded.
As a vital environmental factor affecting plant growth and development, light is involved in various physiological processes such as photomorphogenesis, lateral root development, seed germination and secondary metabolites accumulation[10−12]. It was reported that white light treatment promotes content of DHT, red and blue light treatment significantly improves salvianolic acid content[13,14]. Further investigation into the molecular mechanisms has revealed that light can indirectly promote the generation of the JA-Ile signaling molecule through the photoreceptors CRY1/2 and phyA/B[10,15] and in turn, leads to the degradation of JAZ proteins and the subsequent release of MYCs transcription factors, which activate ELONGATED HYPOCOTYL 5 (HY5) transcription factor[16]. HY5 further regulates downstream target genes to achieve the transmission of light signaling. These findings shed light on the complex relationship between light and JA signaling pathway in regulating plant growth under environmental stress.
HY5, a bZIP transcription factor, can be activated by photoreceptors to promote downstream photomorphogenesis. As a core regulatory factor regulating plant development, HY5 also participates in the biosynthesis of active compounds[17]. It was reported that AtHY5 affects anthocyanin accumulation by interacting with AtPIF3[18], regulates artemisinin biosynthesis by binding the ACGT motif on the AaGSW1 promotor[19] and meanwhile directly binds with G-box on the promoter of AaQH6, which encodes monoterpene pinene biosynthesis[20]. BBX (B-box) transcription factor is a zinc finger protein extensively participating in plant life processes[21], and is confirmed as a rate-limiting cofactor of HY5[22]. BBX IV group PpBBX18 and PpBBX21 antagonistically regulates anthocyanin biosynthesis via competitive interaction with PpHY5[23], OsBBX14 promotes photomorphogenesis by activating expression of OsHY5L1[24], AtBBX11 and AtBBX21 regulates plant development by forming a positive feedback loop with AtHY5[25]. Both HY5 and BBX have been reported to be associated with multiple signaling pathways in a dominant way, including the abscisic acid (ABA)[26−28], gibberellin (GA)[29,30], and brassinosteroid (BR)[31] pathways. However, it remains unknown whether there is a crosstalk between light and JA signaling, which could potentially regulate the formation of the HY5-BBX complex.
In this study, the temporal changes in JAs and tanshinone content after dark-to-light transition were measured, confirming the positive regulatory effect of light on both components. Transcriptome analysis was conducted at corresponding time points to analyze the expression patterns of genes involved in JAs and tanshinone biosynthesis. A co-expression analysis network was then constructed based on the expression patterns of transcription factors and biosynthetic genes, along with the types of cis-regulatory elements in promoters, inferring potential regulatory relationships between SmHY5-SmBBXs and tanshinone. Using Y2H-seq technology, SmHY5 bait was employed to screen and identify SmBBX21 from a yeast library of Danshen, and strong interactions were also confirmed between SmBBX20/22/23/24 of BBX IV group and SmHY5, therefore suggesting the presence of a potential regulatory network from light-JA signaling to SmHY5-SmBBXs and finally to tanshinone in Danshen. However, further validation is required to elucidate the regulatory relationships between downstream target genes of SmHY5 and SmBBXs, which would ultimately reveal the molecular mechanisms of crosstalk regulation between light and JA signaling on tanshinone biosynthesis.
-
In order to investigate the effect of light signaling on the regulation of JAs and tanshinones in Danshen, we designed a light-induced experiment and treated 4-week-old plants with dark-to-light transition (Fig. 1a). The results showed that OPDA, as a substrate for JAs, exhibited a transient increase in both leaves and roots, reaching its peak at 4 h after treatment (Fig. 1b). In leaves, the content of OPDA increased dramatically to 98.7 ng/g, which was 3.0 times higher than that at 0 h (CK), while in roots, it increased significantly to 88.0 ng/g, which was 1.8 times higher than that at 0 h (CK), and then gradually decreased to a slightly higher level than that before treatment. JA was not detected in leaves at 0 h, but its content level gradually increased and reached its peak at 32 h with a gradient increase, reaching 270.3 ng/g; however, in roots, it reached its peak at 4 h, which was 3.4 times higher than that at 0 h (CK), with a content of 220.6 ng/g, and then gradually decreased. The signaling molecule of the JA pathway, JA-Ile, was not detected in leaves at 0 h, but its content level gently increased and reached its peak at 32 h, showing consistent trend with JA, with a content of 57.3 ng/g, while there was no obvious fluctuation in the content of JA-Ile in roots.
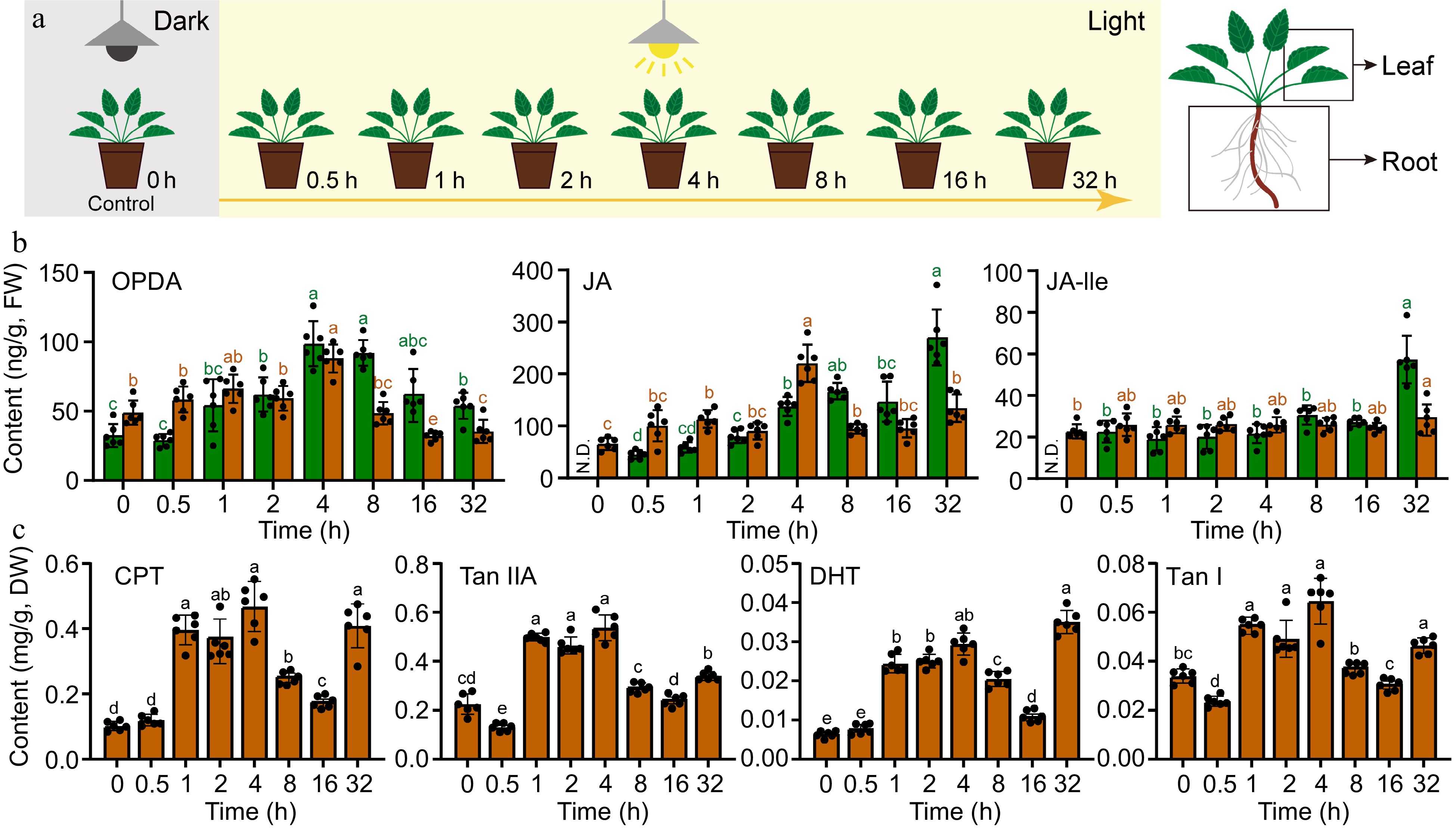
Figure 1.
Transient increase in the accumulation of JAs and tanshinones after dark-to-light transition in Danshen. (a) The schematic representation depicts the dark-to-light transition treatment. (b) Contents of 12-oxo-phytodienoic acid (OPDA), jasmonic acid (JA), jasmonic acid-isoleucine (JA-Ile) in root and leaf determined by UPLC-MS/MS. (c) Contents of Cryptotanshinone I (CPT), Tanshinone IIA (Tan IIA), Dihydrotanshinone (DHT), and Tanshinone I (Tan I) in root determined by UPLC-MS. Data are shown as the mean ± SD (n = 6). Variance test was performed for multigroup comparison, with different lowercase letters indicating significant difference between groups (p < 0.05).
The content of diterpenoids also showed a trend of transient changes in response to dark-to-light transition (Fig. 1c). CPT was at a low level at 0 and 0.5 h, and increased abruptly at 1 h, then reached its peak at 4 h with a content of 0.47 mg/g, which was 4.7 times higher than that at 0 h (CK), and then gradually decreased but still significantly higher than that at 0 h (CK). There was a significant accumulation at 32 h. The content of Tan IIA also increased sharply at 1 h, reaching its peak at 4 h with a content of 0.54 mg/g, which was 2.5 times higher than that at 0 h (CK). DHT accumulated in a similar manner as CPT, increasing sharply at 1h and reaching its peak at 4 h with a content of 0.029 mg/g, which was 4.5 times higher than that at 0 h (CK), and showed a significant accumulation at 32 h. Tan I accumulated in a similar way as Tan IIA, increasing sharply at 1 h and reached its peak at 4 h with a content of 0.065 mg/g, which was 1.9 times higher than that at 0 h (CK), and then gradually decreased. These results indicate that alteration in light can positively induce the elevation in content levels of JAs and diterpenoids, but the specific molecular mechanisms are still unclear.
Dark-to-light transition induces expression of JAs and tanshinone biosynthetic genes
-
To further investigate the effect of light on JA biosynthesis in Danshen, we measured the transcriptome after dark-to-light treatment (Fig. 2a). Among the six key genes involved in JA biosynthesis, the upstream SmLOX showed a decreasing trend in expression in both leaves and roots, while the two rate-limiting enzyme genes, SmAOS and SmAOC, were significantly upregulated in roots. SmAOS exhibited transient high expression in both leaves and roots within a short time, peaking at 0.5 h with an 8.7-fold and 1.8-fold increase, respectively. In leaves, SmAOC also reached its peak at 0.5 h with a 9.1-fold increase compared to the control, while in roots, it showed a continuous upward trend, ensuring the sustained supply of JA precursor OPDA. The expression of SmOPR3 was relatively stable in both leaves and roots, with a slight increase in roots (Supplemental Table S1). Among the downstream signaling molecule biosynthesis genes, the peak expression level of SmJAR1 was later than that of SmAOS and SmAOC, reaching its highest level at 2 h in leaves, which was 2.2 times higher than that of the control, but remained relatively stable in roots, possibly due to the stable content of JA-Ile in roots. In leaves, the expression level of SmJMT also peaked at 2 h, reaching 5.1 times that of the control, but was not detected in roots. More importantly, the light-responsive transcription factor HY5 was activated and expressed within a short period of time, reaching its peak at 1 h in leaves, which was 76.1 times higher than the control. Therefore, light promoted the expression of most JA biosynthesis genes in Danshen, especially SmAOS and SmAOC, but the causal relationship and the complex synergistic regulation mechanism on tanshinone accumulation still need further exploration.
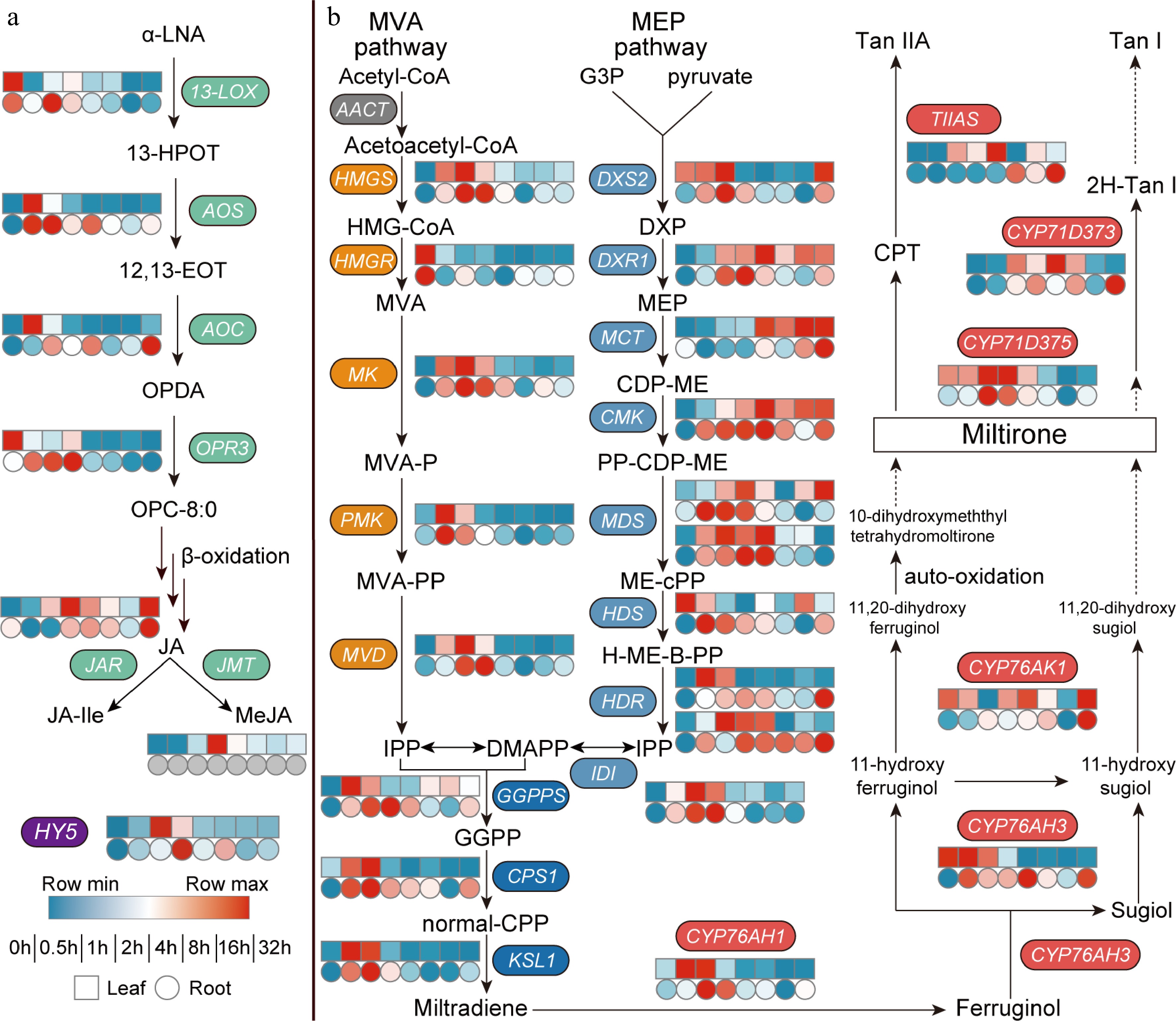
Figure 2.
Dark-to-light transition affects biosynthesis of both (a) JAs and (b) tanshinones. Heat map showing expression fold changes (log2 FC) of genes and FPKM values are shown in Supplemental Table S1. (a) α-LNA, α-Linolenic Acid; 13-HPOT, 13(s) -hydroperoxy-linolenic acid; 12,13-EOT, 12,13(S)-epoxy-9(Z),11,15(Z)-octadecatrienoic acid; OPDA, 12-oxo-phytodienoic acid; OPC-8:0, 3-oxo-2-(2-(Z)-pentenyl)-cyclopentane-1-octanoic acid; JA, jasmonic acid; JA-Ile, jasmonic acid-isoleucine; MeJA, methyl jasmonate; 13-LOX, 13-Lipoxygenase; AOS, allen oxidase; AOC, allene oxide cyclase; OPR, 12-oxophytodienoate reductase; JAR, JA-amino acid synthetase; JMT, jasmonate hydroxymethyl transferase; HY5, elongated hypocotyl 5. (b) MVA pathway, the mevalonate pathway; Acetyl-CoA, acetyl coenzyme A; Acetoacetyl-CoA, acetoacetyl coenzyme A; HMG-CoA, 3-hydroxy-3-methylglutaryl CoA; MVA, mevalonate; MVA-P, mevalonate-5-phosphate; MVA-PP, mevalonate-5-diphosphomevalonate; IPP, isopentenyl pyrophosphate; MEP pathway, the 2-C-methyl-d-erythritol 4-phosphate pathway; G3P, glyceraldehyde 3-phosphate; DXP, 1-deoxy-d-xylulose 5-phosphate; MEP, 2-C-methyl-d-erythritol 4-phosphate; CDP-ME, 4-(cytidine 5'-diphospho)-2-C-methyl-d-erythritol; PP-CDP-ME, 2-phospho-4-(cytidine 5'-diphospho)-2-C-methyl-d-erythritol; ME-cPP, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate; H-ME-B-PP, 1-hydroxy-2-methyl-2-butenyl 4-diphosphate; DMAPP, dimethylallyl pyrophosphate; GGPP, geranylgeranyl diphos-phate; normal-CPP, normal-copalyl diphosphate; AACT, acetoacetyl-CoA transferase; HMGS, hydroxymethyl-glutaryl-CoA synthase; HMGR, 3-hydroxy-3-methylglutaryl CoA reductase; MK, mevalonate kinase; PMK, phosphomevalonate kinase; MVD, mevalonate-5-pyrophosphate decarboxylase; DXS, 1-deoxy-d-xylulose-5-phosphate synthase; DXR, 1-deoxy-d-xylulose-5-phosphate reductoisomerase; MCT, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; CMK, 4-(cytidine 5'-diphospho)-2-C-methyl-d-erythritol kinase; MDS, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase; HDS, 4-hydroxy-3-methylbut-2-enyl diphosphate synthase; HDR: 4-hydroxy-3-methyl-but-2-enyl diphosphate reductase; IDI, Isopentenyl-diphosphate delta-isomerase; GGPPS, GGPP synthase; CPS, copalyl diphosphate synthase; KSL, kaurene synthase-like; TIIAS, tanshinone IIA synthase.
To investigate the effect of light on tanshinone biosynthesis, we evaluated the expression levels of key enzyme genes in the biosynthetic pathways (Fig. 2b). Regarding the genes involved in obtaining IPP and DMAPP via the MVA and MEP pathways, there was an overall increasing trend. However, in the MVA pathway, the rate-limiting enzyme gene SmHMGS reached its highest expression level in both leaves and roots at 1 h, while another rate-limiting enzyme gene SmHMGR showed a continuously decreased expression in both tissues. This opposite expression trend may limit the supply of precursors for diterpene biosynthesis through the MVA pathway. Nevertheless, the genes SmMK, SmPMK, and SmMVD still exhibited distinct expression peaks within a short time in both leaves and roots.
In the MEP pathway, the expression trends of the rate-limiting enzyme genes SmDXS2 and SmDXR1 were more consistent, reaching their highest levels at 1 and 2 h, respectively, ensuring the supply of precursors for subsequent diterpenoid compounds. Subsequently, the genes SmMCT, SmCMK, SmMDS, SmHDS, SmHDR, and SmIDI also showed varied degrees of upregulation. Of particular importance, during the formation of GGPP, the substrate for tanshinone biosynthesis, the gene GGPPS responded positively to light signaling in both leaves and roots, reaching its peak at 0.5 and 2 h, respectively. Subsequently, the genes involved in diterpene synthase and SmCYP450 enzymes exhibited specific high expression in roots, with SmCPS1, SmKSL1, and SmCYP76AH3 reaching their highest levels at 1 h. The expression peaks of CYP450 genes involved in late-stage modifications generally occurred later, with SmCYP76AK1, SmCYP71D373, and SmTIIAS gradually increasing and peaking at 32 h. These changes in gene expression ultimately reflected in the variation of tanshinone content. However, the molecular mechanism by which light signaling regulates tanshinone biosynthesis is still unclear and requires further research.
Multiple transcription factors potentially regulate the biosynthesis of JAs and tanshinones in the process of dark-to-light transition
-
As transcription factors play essential roles in plant secondary metabolite accumulation and growth, the upstream regulatory network of JAs and tanshinone accumulation was analyzed. Co-expression analysis revealed significant co-expression between genes involved in JAs (SmLOX, SmAOS, SmAOC, SmOPR3, SmJAR, SmJMT) and transcription factors such as WRKYs, ERFs, MYBs, bHLHs, bZIPs, BBXs (Fig. 3a). Similarly, genes related to tanshinone biosynthesis (SmDXS2, SmDXR1, SmGGPPS, SmCPS1, SmKSL1, SmCYP76AH1, SmCYP76AH3, SmCYP76AK1, SmCYP71D375, SmCYP71D373, SmTIIAS) exhibited co-expression with these transcription factors (Fig. 3b).
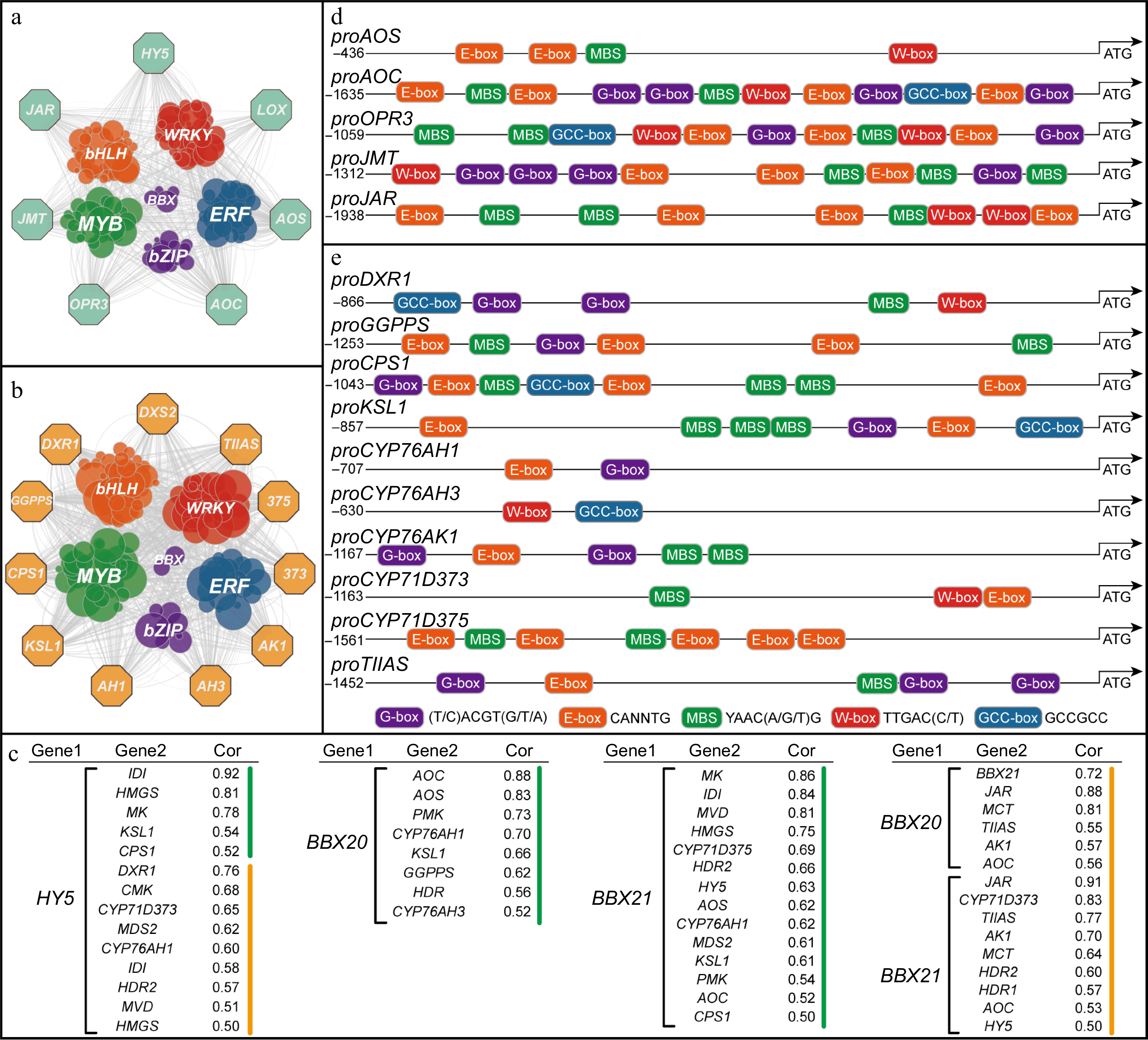
Figure 3.
JAs and tanshinone biosynthetic genes potentially regulated by transcription factors. (a) Co-expression analysis of JAs biosynthetic genes and transcription factors in leaf. (b) Co-expression analysis of tanshinone biosynthetic genes and transcription factors in root, while pathway genes are represented by octagons and transcription factors by circles. The screening criterion for selection was set as a correlation coefficient greater than 0.3. (c) Transcription factor-pathway genes with correlation coefficients, green represents genes in leaves and yellow in roots. (d) Cis-elements of the JAs biosynthetic gene promoters. (e) Cis-elements of the tanshinone biosynthetic gene promoters.
Further analysis revealed a high correlation (Cor > 0.75) between SmHY5 and key genes in the MVA pathway, such as SmHMGS, SmMK, as well as genes in the MEP pathway, including SmDXR1 and SmIDI (Fig. 3c). Furthermore, SmHY5 showed a moderate correlation (Cor > 0.5) with terpene synthase genes like SmCPS1 and SmKSL1, as well as CYP450 genes like SmCYP76AH1 and SmCYP71D373. However, no correlation was found between HY5 and genes involved in JA biosynthesis, suggesting that SmHY5 is more likely to regulate upstream terpenoid precursor forming related genes.
On the other hand, SmBBX20 exhibited higher expression level in leaves and showed strong correlations (Cor > 0.75) with JA biosynthesis genes such as SmAOS and SmAOC (Fig. 3c). The expression levels of these genes were consistent with BBX20 and all peaked at 0.5 h (Supplemental Table S1). SmBBX20 also displayed moderate correlations (Cor > 0.5) with MEP pathway genes like SmPMK, SmHDR, as well as diterpene synthase genes SmGGPPS, SmKSL1, and SmCYP76AH1, SmCYP76AH3. In roots, it showed a high correlation (Cor = 0.81) with MEP pathway gene SmMCT, and moderate correlations (Cor > 0.5) with SmCYP76AK1 and SmTIIAS. Interestingly, SmBBX20 also demonstrated a strong correlation with SmBBX21, indicating a possible regulatory relationship. In leaves, SmBBX21 displayed high correlations (Cor > 0.75) with MVA pathway genes like SmMK, SmIDI, SmMVD, and SmHMGS, as well as moderate correlations (Cor > 0.5) with MEP pathway genes like SmMDS2, SmHDR2, and diterpene synthase genes SmCPS1, SmKSL1, as well as SmCYP76AH1, SmCYP71D375. Notably, SmBBX21 also showed moderate correlations (Cor > 0.5) with JA biosynthesis genes like SmAOS and SmAOC, as well as SmHY5. However, SmBBX21 exhibited lower expression levels in leaves compared to roots, where it displayed strong correlations (Cor > 0.75) with SmCYP71D373, SmTIIAS, and moderate correlations (Cor > 0.5) with MEP pathway genes like SmMCT, SmHDR1/2. Additionally, the highest correlation of 0.91 was observed between SmBBX21 and SmJAR. Therefore, it is proposed that SmBBX20 and SmBBX21 potentially possess the ability to cooperatively regulate the biosynthesis of JAs and tanshinones.
Subsequently, based on the published genomic data, we cloned and analyzed the cis-elements in the promoter regions of JAs and tanshinone biosynthetic genes[32,33]. As shown in Fig. 3d & e, the promoter regions of genes involved in JAs and tanshinone biosynthesis contain multiple cis-elements that are binding sites for the above-mentioned transcription factors. These cis-elements include the G-box for HY5 (bZIP), T/G-box for BBX, E-box for bHLH, W-box for WRKY, MBS for MYB, and GCC-box for ERF, providing possibilities for direct or indirect signal transduction and transcriptional regulation.
SmHY5 specifically interacts with SmBBXs in the roots of Danshen
-
In order to uncover the proteins that interact with SmHY5, we built SmHY5 into a construct using pGBKT7 as back bone to screen Y2H library of Danshen root tissue. After ensuring no self-activation in pGBKT7-SmHY5, we collected 192 positive clones on SD-Trp/Leu/His on which we performed PCR single-cloning sequencing. Through BLAST analysis to eliminate duplicate sequences, we obtained 63 target genes (Supplemental Table S2), among which were several transcription factors, including SmWRKY1[34], SmWRKY41, SmMYB1R1, SmbHLH3[35], and SmBBX21. Further validation revealed that 13 clones, including SmbHLH3 and SmBBX21, were able to grow on SD-Trp/Leu/His/Ade plates and displayed blue color on SD-Trp/Leu/His/Ade + X-α-gal plates (Fig. 4a). Based on SmBBX21's classification in the IV subfamily of BBX, we proceeded to screen for potential interactions between proteins in this subfamily and SmHY5. Seven genes from this subfamily, SmBBX18, SmBBX19, SmBBX20, SmBBX21, SmBBX22, SmBBX23, SmBBX24, were cloned and then built into the constructs for yeast two-hybrid. From the results of -Trp/Leu and -Trp/Leu/His/Ade culture medium, the physical interactions between SmHY5 and SmBBX20, SmBBX21, SmBBX22, SmBBX23, SmBBX24 were proved, which can connect light pathway and the BBX gene family (Fig. 4b).
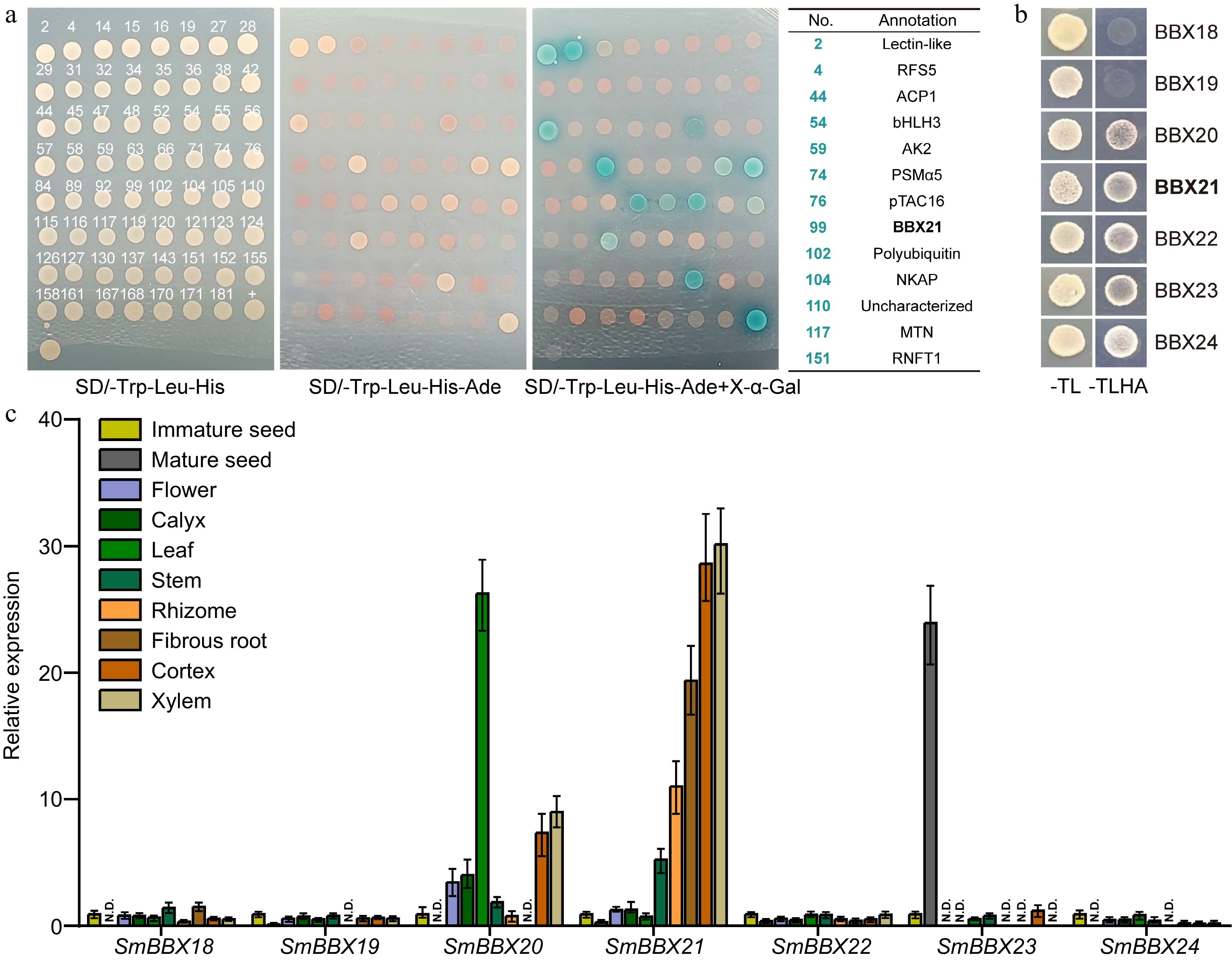
Figure 4.
Potential interaction between root-specific expressed SmBBX21 and SmHY5. (a) Yeast two-hybrid sequencing (Y2H-seq) screened potential interacting proteins of SmHY5 in root. Monoclonal sequencing results are shown in Supplemental Table S2. (b) Y2H confirmed the interaction between SmHY5 and SmBBXs. (c) The expression of the seven candidate SmBBX genes in different tissues of Danshen. ACTIN was used as an internal control; for each gene, the expression level relative to ACTIN in immature seed was taken as 1. Data are shown as the mean ± SD (n = 3).
In order to validate the potential role of SmBBXs in regulating tanshinone biosynthesis in root, we conducted tissue-specific expression analysis on Danshen. We found that SmBBX20 exhibits higher expression levels in leaves, stems, and flowers, while SmBBX21 shows specific expression in root tissues, including rhizome, fibrous roots, cortex, and xylem (Fig. 4c). This may be associated with the regulatory role of SmBBXs in different tissues.
-
Tanshinones, a class of valuable active components found in Danshen, are biosynthesized under the induction of various biotic and abiotic stresses. JAs serve as important hormone signaling molecules that respond to stress and act as 'brokers' linking the environment to secondary metabolism[3]. Among them, JA-Ile can promote the degradation of JAZ, release transcription factors, and activate downstream biosynthetic genes[4,6−9].
This study found that the important signaling molecule JA-Ile was not detected in leaves under dark conditions, but increased steadily after exposure to light, indicating that JA-Ile biosynthesis is light-dependent (Fig. 1b). Studies have reported that light can indirectly promote the generation of JA-Ile signaling molecules through photoreceptors CRY1/2 and phyA/B, and then regulate HY5 through MYCs transcription factors[10,15,16]. In this study, the two upstream key enzyme genes SmAOS and SmAOC in Danshen JAs biosynthetic pathway reached their peak at 0.5 h, and JA-Ile was detected and accumulated within the same time frame (Fig. 2a). This time point was prior to the high expression time point of SmHY5, which also suggests the possibility of indirect regulation of SmHY5 by JAs in Danshen.
Furthermore, HY5 can directly bind to the promoters of light-responsive transcription factors and biosynthetic genes, thereby regulating plant metabolism[17]. For example, AaHY5 activates the transcription of AaWRKY9/14, promoting artemisinin accumulation[9,36]. SmHY5 directly binds to SmC4H1 in hairy roots of Danshen, enhancing phenolic acid accumulation[37]. In this study, co-expression analysis revealed a strong correlation (cor > 0.5) between SmHY5 and various transcription factors, as well as tanshinone biosynthetic genes (Fig. 3c). Multiple G-box elements were identified in the promoters of these genes, suggesting their potential regulation by SmHY5 (Fig. 3e). However, it has been reported that the direct binding ability of HY5 to gene promoters is weak and requires the formation of heterodimers with BBX proteins for co-regulation with HY5-COP1 cascade module[38]. Among them, BBXs are believed to interact directly with HY5, exerting promoting or inhibitory effects on HY5 function[21]. The relationship between BBX IV group and HY5 is particularly close[39], with not only a direct interaction between AtBBX20 and AtHY5 but also a mutual effect on their protein abundance[40]. AtBBX21 interacts with AtHY5 and meanwhile exerts transcriptional regulation on AtHY5[41]. In this study, Y2H-seq was utilized to screen and identify SmBBX21 from a yeast library of Danshen roots using SmHY5 as bait (Fig. 4a). Furthermore, strong interactions were also confirmed between SmHY5 and members of BBX IV group, namely SmBBX20-24 (Fig. 4b). SmBBX20 showed a peak correlation with SmAOS, SmAOC, and SmJAR at 0.5 h (Fig. 2a, 3c), and it was specifically expressed in the aerial parts (Fig. 4c), suggesting its potential involvement in JA biosynthesis regulation. The peak of SmBBX21 expression occurred after 2 h (Supplemental Table S1), showing specific expression in the underground parts (Fig. 4c) and exhibiting strong correlations with MEP pathway and tanshinone biosynthetic genes (Fig. 3c). Based on the promoter G-box and T/G-box elements, it is speculated that there may be regulation of tanshinone biosynthesis by SmBBX21.
Therefore, a potential regulatory network from light-JA signaling to SmHY5-SmBBXs to tanshinone biosynthesis is constituted (Fig. 5). However, further experimental validation is needed for the downstream target genes of SmHY5 and SmBBXs, as well as the regulatory effects on tanshinone biosynthetic genes. Subsequent research is required to uncover the molecular mechanisms underlying the crosstalkregulation of tanshinone by the light and JA signaling. This study supplements the information on light and JA signaling in Danshen as a medicinal model plant and provides a basis for improving tanshinone production through the combination of light and JA.
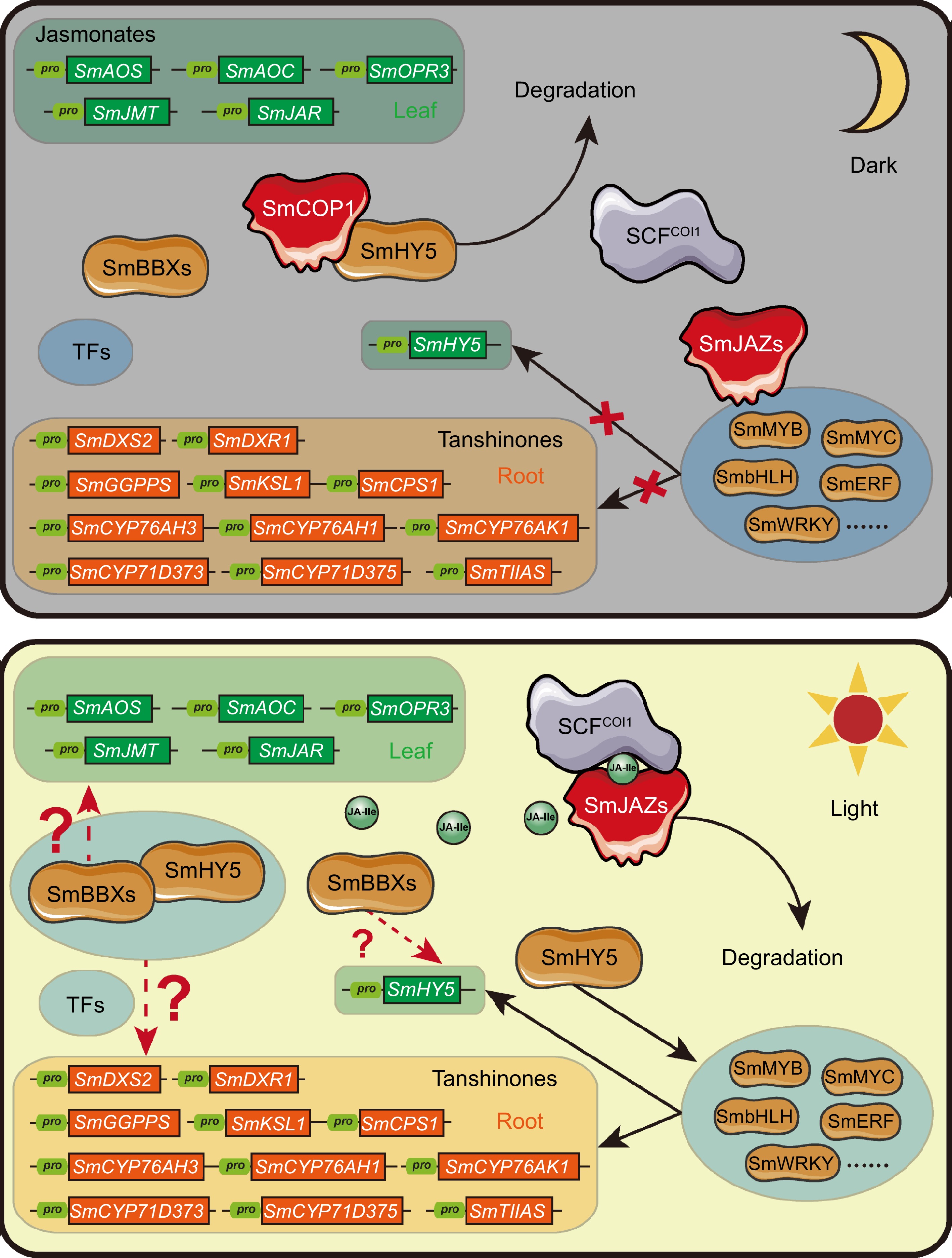
Figure 5.
Hypothesis for crosstalk between light and JA signaling regulating tanshinone biosynthesis through the SmHY5-SmBBXs complex. Under dark conditions, the absence of JA-Ile leads to the inhibition of multiple transcription factors by SmJAZ proteins. Simultaneously, SmCOP1 degrades SmHY5, preventing the formation of a regulatory complex between SmHY5 and SmBBXs. Consequently, the expression of downstream genes involved in tanshinone biosynthesis is at a lower level. In contrast, under light conditions, JA-Ile biosynthesis is enabled, leading to the degradation of SmJAZ proteins and the release of SmMYC and other transcription factors. Additionally, SmCOP1 releases SmHY5, allowing the formation of a regulatory complex between SmHY5 and SmBBXs. This coordination activates the expression of downstream genes involved in tanshinone biosynthesis. Finally, the crosstalk between light and JA signaling on tanshinone biosynthesis was realized through the formation of SmHY5 and SmBBXs complex or not.
-
Our research reports the positive regulatory effect of light on the accumulation of JAs and tanshinones, and analyzes the changing patterns of expression levels of genes involved in JAs and tanshinone biosynthesis after the transition from darkness to light. We identified transcription factors highly correlated with the aforementioned biosynthetic genes, including HY5 and BBXs. The interactions between HY5 and BBX IV group BBX20-24 were confirmed through Y2H-Seq and Y2H, and the tissue-specific expression of BBXs was also observed. Thus, we proposed a potential regulatory network in which the crosstalk between light and JA signaling regulates tanshinone biosynthesis through the SmHY5-SmBBXs complex.
-
The plants of Danshen were cultivated and stored in the greenhouse of National Resource Center for Chinese Materia Medica, China Academy of Chinese Medical Sciences (Beijing, China). The growth conditions were maintained at 25 °C with a 16-h light period each day for four weeks, followed by 48 h of darkness before the light treatment.
RNA-seq and analysis
-
Total RNAs treated with light (0−36 h) were extracted as described earlier[4]. RNA-seq library preparation and paired-end sequencing were both performed using an Illumina HiSeq 6000 platform (Novogene, Beijing, China). FeatureCounts was adopted to quantify total expressed transcripts with the FPKM (fragments per kilobase of exon model per million mapped reads) value[42]. Differentially expressed genes (DEGs) were obtained by comparing FPKM value among 0−36 h samples. DEGs with a log2 fold-change (|log2 FC|) ≥1 and an adjusted q-value ≤ 0.05 were identified using the DEGSEQ R package[43]. GENIE3 R package was used to calculate the Pearson correlation coefficients between transcription factors[44] and genes in batches and Cytoscape software (version 2.6.3) to draft the correlation map.
Phytohormone determination
-
Phytohormones including JA, OPDA, JA-Ile were quantitated using a previously reported method[45]. Fifty mg of ground plant material was subjected to extraction using an isopropanol-water-hydrochloric acid solution (2:1:0.002, V/V/V) for a duration of 30 min. Subsequently, the extract was further extracted with dichloromethane for another 30 min. The organic layer obtained was evaporated under a stream of nitrogen until dryness, resulting in a residue. This residue was then dissolved in 70% methanol to obtain a solution. To remove any insoluble particles, the solution was subjected to centrifugation at 12,000 rpm for 15 min, resulting in the collection of the supernatant. The quantitation of endogenous phytohormone was conducted using an ACQUITY UPLC I-Class system equipped with a 6500 QTRAP mass spectrometer. The mobile phase consisted of acetonitrile (A) and 0.05% formic acid-water (B), and the flow rate was set at 0.4 mL/min. The gradient elution was as follows: 0−3.5 min, 10% A; 3.5−4 min, 10%−95% A; 4−8 min, 95% A. For analysis, 1 µL of the sample solution was injected into the system. Mass spectrometry analysis was performed in both positive and negative modes using multiple reaction monitoring (MRM) mode.
Tanshinones determination
-
Leaves and roots were collected and lyophilized overnight using an FDU-1110 freezer dryer (Eyela, Tokyo, Japan). The samples were accurately weighed and sonicated in methanol at a ratio of 1:30 (w/v) for 30 min using an SB-800 DTD sonicator (Ningbo Xinzhi, China; power, 100 W; frequency, 40 kHz). The resulting supernatant was filtered through a 0.22 μm filter (Pall, Ann Arbor, MI, USA) for subsequent ultra-performance liquid chromatography coupled with triple-quadrupole tandem mass spectrometry (UPLC-TQ-MS) analysis. Experiments included six biological replicates and the statistical significance was evaluated using a variance analysis in SPSS v.16.0[46].
Quantitative real-time PCR analysis
-
To evaluate the expression profiles of the SmBBX genes in different tissues, a qPCR assay was conducted in a 10 μl reaction volume containing 5 μl of TB Green® Premix Ex Taq (TaKaRa RR820B, Japan), 0.5 μM of each oligonucleotide and 1 μl of cDNA template on a StepOnePlus PCR system (Roche LightCycler 480). The specific qPCR primers used in this study are listed in Supplemental Table S3. The β-actin genes were utilized as endogenous controls. Three technical replicates were performed for each sample, and quantitative analysis was performed using the 2−ΔΔCᴛ method[47].
Yeast two-hybrid assays
-
The yeast two-hybrid screening method was performed according to the manufacturer's user guide (Takara Bio Inc.). The full-length cDNA of SmHY5 was amplified and cloned into the pGBKT7 vector, which was then transformed into the yeast strain AH109. This construct served as the bait to screen a cDNA library prepared from equal amounts of poly(A)-containing RNA extracted from root tissue. The screening and plasmid extraction sequencing steps were performed following the manufacturer's instructions. The full-length cDNAs of SmBBXs were amplified and subcloned into the pGADT7 vector individually. The pGADT7 and pGBKT7 constructs were co-transformed into the yeast strain AH109, and the transformed cells were selected on SD/-Leu/-Trp medium. The resulting co-transformed cells were streaked on quadruple dropout medium (SD/-His/-Leu/-Trp/-Ade) and incubated at 30 °C for 72 h.
-
The authors confirm contribution to the paper as follows: study conception and design: Huang L, Zheng H; analysis and interpretation of results: Zheng H, Fu X, Yu M, Liu Q, Li C, Li L, Qian S, Chen K; draft manuscript preparation: Zheng H, Fu X, Yu M; manuscript editing: Zhang S, Tang K; project supervision: Huang L. All authors reviewed the results and approved the final version of the manuscript.
-
RNA-seq data have been deposited in the NCBI repository: SRR27169169- SRR27169184. Other data supporting the findings of this study are available within the paper and within its supplementary materials published online.
This research was funded by Natural Science Foundation of China (82104342), CACMS Innovation Fund (CI2021A041003, CI2023E002), National Key Research and Development Program of China (2023YFC3503900), Fundamental Research Funds for the Central public welfare research institutes (ZZ15-YQ-060), China Postdoctoral Science Foundation (2022M712099), key project at central government level: The ability establishment of sustainable use for valuable Chinese medicine resources (2060302).
-
The authors declare that they have no conflict of interest.
-
# Authors contributed equally: Han Zheng, Xueqing Fu, Muyao Yu
- Supplemental Table S1 The FPKM values of genes at different time points after light treatment.
- Supplemental Table S2 The results of yeast two-hybrid monoclonal sequencing.
- Supplemental Table S3 Primers used in this study.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Zheng H, Fu X, Yu M, Liu Q, Li C, et al. 2024. Transcriptome and yeast two-hybrid sequencing shed light on the crosstalk between light and jasmonate signaling in regulating tanshinone biosynthesis. Medicinal Plant Biology 3: e006 doi: 10.48130/mpb-0024-0006
Transcriptome and yeast two-hybrid sequencing shed light on the crosstalk between light and jasmonate signaling in regulating tanshinone biosynthesis
- Received: 31 December 2023
- Revised: 17 February 2024
- Accepted: 26 February 2024
- Published online: 03 April 2024
Abstract: As a high-value active component in Salvia miltiorrhiza (Danshen), the accumulation of tanshinones is under the influence of various environmental factors, yet it is unknown whether light, as an essential environmental factor for plant growth, regulates the biosynthesis of tanshinones. In this study, we first measured the temporal accumulation of jasmonates (JAs) and tanshinones under dark-to-light transition, and found that the accumulation of JA, JA-Ile and tanshinones is light-dependent. Transcriptome analysis revealed that SmAOS and SmAOC involved in JA biosynthesis responded rapidly under dark-to-light transition, followed by the accumulation of JA and JA-Ile. A light-responsive transcription factor SmHY5 was filtered indicating a potential regulatory relationship with tanshinone-related genes. Co-expression analysis identified multiple classes of transcription factors highly correlated with JAs and tanshinone biosynthetic genes, among which SmHY5 was confirmed to interact with SmBBX21 through Y2H-seq. Further confirmation of the interaction between SmHY5 and the IV subfamily of SmBBXs, including SmBBX20-24, was obtained through Y2H experiments. Specifically, SmBBX20 and SmBBX21 showed tissue-specific expression in aboveground and underground tissues, consistent with the accumulation sites of JAs and tanshinones. Taken together, this study contributes to the understanding of light and JA signaling in tanshinone biosynthesis in Danshen. We propose a crosstalk between light and JA signaling which regulates tanshinone biosynthesis through the SmHY5-SmBBXs complex and thus provide a basis for improving tanshinone production through the combination of light and JA.
-
Key words:
- Salvia miltiorrhiza /
- Light /
- Jasmonates /
- HY5 /
- BBXs /
- Tanshinones













