-
Hypericum L. is one of the largest angiosperm genera, comprising more than 450 species distributed almost worldwide[1]. Members of the genus have been used in traditional remedies across diverse cultures and civilizations for centuries and have even been considered symbols of religion[2]. Traditionally, Hypericum plants have been prepared in olive oil or alcohol extracts or used as herbal teas to treat wounds, gastrointestinal problems, and mood disorders[3,4]. More recently, plants of the genus have been recognized for their antidepressant, antiviral, antibacterial, and anticancer properties[5−9]. Dr. Norman Robson, who conducted the extensive monograph on the genus's taxonomy, noted: "Hypericum has thus been associated with pharmacy and folklore for many centuries; so, its recent 'discovery' by Western medicine is not surprising, though it may be regarded as rather belated"[2]. This highlights the long-standing tradition associated with the genus Hypericum, confirmed by numerous ethnobotanical studies conducted worldwide over the past decades[3,4,10−12]. Today, dietary supplements and medicines made from Hypericum perforatum, also known as St. John's wort, are traded globally. St. John's wort is the most well-known species in the Hypericum genus. These products primarily treat mood disorders. In 2021, annual sales in the USA alone reached USD
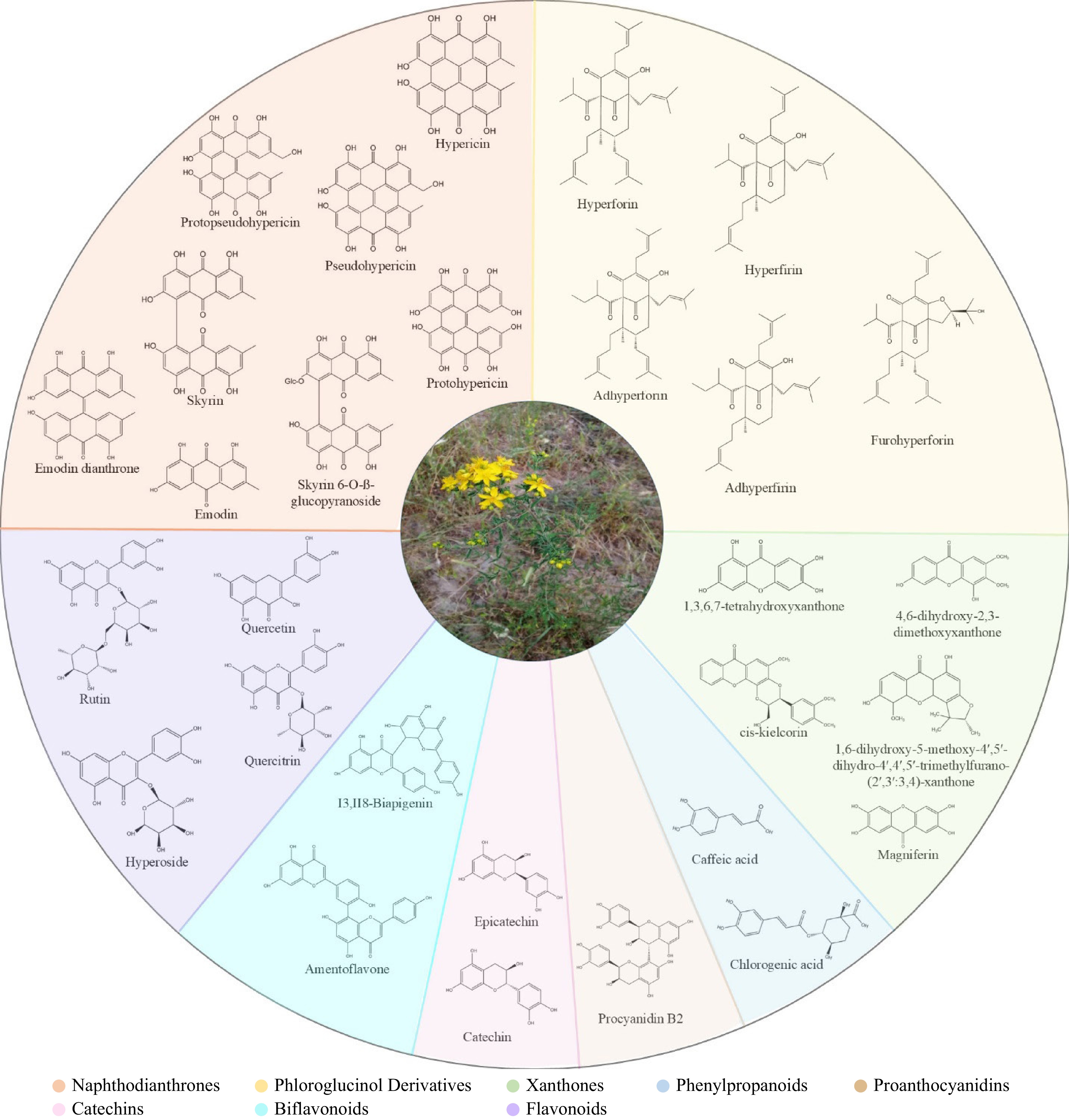
${\$} $ Hypericum plants produce diverse natural product classes (Table 1, Fig. 1), with variations observed among species. Among them, naphthodianthrones (hypericin, pseudohypericin, protohypericin, and protopseudohypericin) and prenylated phloroglucinols (hyperforin, adhyperforin, and their oxygenated derivatives) are of particular interest due to their unique activities and rarity in other organisms. The most common natural compounds include xanthones (e.g., 1,3,6,7-tetrahydroxyxanthone), flavonoids (e.g., hyperoside, rutin, quercitrin), biflavonoids (I3, II8-biapigenin, Amentoflavone), tannins, proanthocyanidins, and phenolic acids. The essential oil contains both hydrocarbons and terpenoids[14]. In recent years, there has been a notable increase in research focused on the Hypericum genus's secondary metabolites. The potential of Hypericum plants as a source of natural products for developing new medicines is currently being investigated, although this is still an ongoing process. Similarly, the study on the biosynthesis of secondary metabolites represents a significant area of continuing research. While new tools have emerged in transcriptomic and metabolomic research, facilitating research on plant secondary metabolism, further expansion of the current knowledge is necessary concerning biosynthetic pathways and genetic and environmental factors that influence the biosynthesis of the key chemical constituents present in Hypericum plants. The above insights can potentially improve medicinal plant cultivation and encourage bioactive compounds' production via more efficient and productive biotechnological applications. This review paper aims to synthesize the current understanding of secondary metabolism in Hypericum plants, integrating the latest research developments with a particular focus on the biosynthesis of hypericin and hyperforin.
Class Substance Naphodianthrones (precussors and derivatives) Hypericin Pseudohypericin Protohypericin Protopseudohypericin Skyrin Skyrin-6-O-ß-glucopyranoside Emodin Emodin dianthrone Phloroglucinol derivatives Hyperforin Adhyperforin Hyperfirin Adhyperfirin Furohyperforin Xanthones 1,6-dihydroxy-5-methoxy-4′,5′-dihydro-4′,4′,5′-trimethylfurano-(2′,3′:3,4)-xanthone 4,6-dihydroxy-2,3-dimethoxyxanthone cis-kielcorin Magniferin 1,3,6,7-tetrahydroxyxanthone Phenylpropanoids Caffeic acid Chlorogenic acid Flavonoids Quercetin Quercitrin Hyperoside Rutin Biflavonoids I3,II8-Biapigenin Amentoflavone Proanthocyanidins Procyanidin B2 Catechins Catechin Epicatechin -
Hypericum plants have a long history of traditional use and have been known for their medicinal applications since antiquity. In the modern era, they are acknowledged for their antidepressant, anti-inflammatory, neuroprotective, anti-cancer, and wound-healing properties. The antidepressant effects of Hypericum are primarily attributed to its active compound, hyperforin. Hyperforin inhibits the reuptake of neurotransmitters such as serotonin, dopamine, and norepinephrine, increasing their availability in the brain and consequently enhancing mood regulation[19,20]. In addition to its antidepressant properties, Hypericum species have been demonstrated to possess notable anti-inflammatory and antioxidant effects. The plant's flavonoids, including quercetin and rutin, have been demonstrated to inhibit Cyclooxygenase-2 (COX-2), a key enzyme in inflammatory processes[21]. Such anti-inflammatory effects are beneficial in conditions such as arthritis and dermatological inflammation. Furthermore, Hypericum demonstrates considerable antiviral and antibacterial properties. Extracts of H. perforatum plants have demonstrated efficacy against various SARS-CoV-2 variants, with the main active constituents being hypericin and pseudohypericin[22]. Additionally, these substances have demonstrated efficacy against herpes simplex virus (HSV) and other viruses by inhibiting viral replication[23]. Moreover, it is effective against Gram-positive bacteria, such as Staphylococcus aureus, making it a valuable therapeutic option for treating infections[24]. Moreover, Hypericum extracts have neuroprotective properties, protecting neurons from oxidative stress and potentially delaying neurodegeneration[25]. These effects make it a promising candidate for further investigation in conditions such as Alzheimer's disease. Additionally, Hypericum is historically used to promote wound healing due to its anti-inflammatory, antibacterial, and collagen-stimulating properties. Topical application of extracts has been demonstrated to accelerate tissue regeneration[26]. Furthermore, Hypericum has been identified as a promising agent for cancer treatment. The potential of hypericin as a photodynamic therapy (PDT) agent has been investigated. PDT is a treatment modality that employs light-sensitive compounds to eradicate malignant cells[5]. However, the use of Hypericum requires caution, particularly given its interaction with the cytochrome P450 enzyme system, notably CYP3A4, which can accelerate the metabolism of various drugs and decrease their efficacy[27].
-
Hypericin (1), (Fig. 2), its derivatives, and protoforms (pseudohypericin, protohypericin, and protopseudohypericin) are naturally occurring naphthodianthrones with a dark red coloration, which are found in the aerial parts of plants[14]. They are predominantly found in plants of the Hypericum genus. Although anthrone derivatives have been reported in other subfamilies, Hypericum is the only plant genus known to contain concentrated anthrones such as hypericins[15]. Hypericins accumulate in multicellular nodules with a black to reddish tint, typically called Dark Glands (DGs)[28] (Fig. 2a−c). These structures do not resemble any internal secretory structure described in other plants. They comprise a central core of large cells surrounded by an irregular sheath of flattened cells. In leaves, they do not span the entire height of the mesophyll, as they are separated from the adaxial epidermis by a layer of flattened palisade parenchyma cells[28]. DGs have been observed in approximately two-thirds of the taxonomic sections of the genus[2], and their size and number are positively correlated with naphthodianthrone content[1,29]. The arrangement and distribution of the DGs is also worthy of note, particularly in the leaves, sepals, and petals of the plants, as they serve as a key character for the taxonomic identification of different species[1].
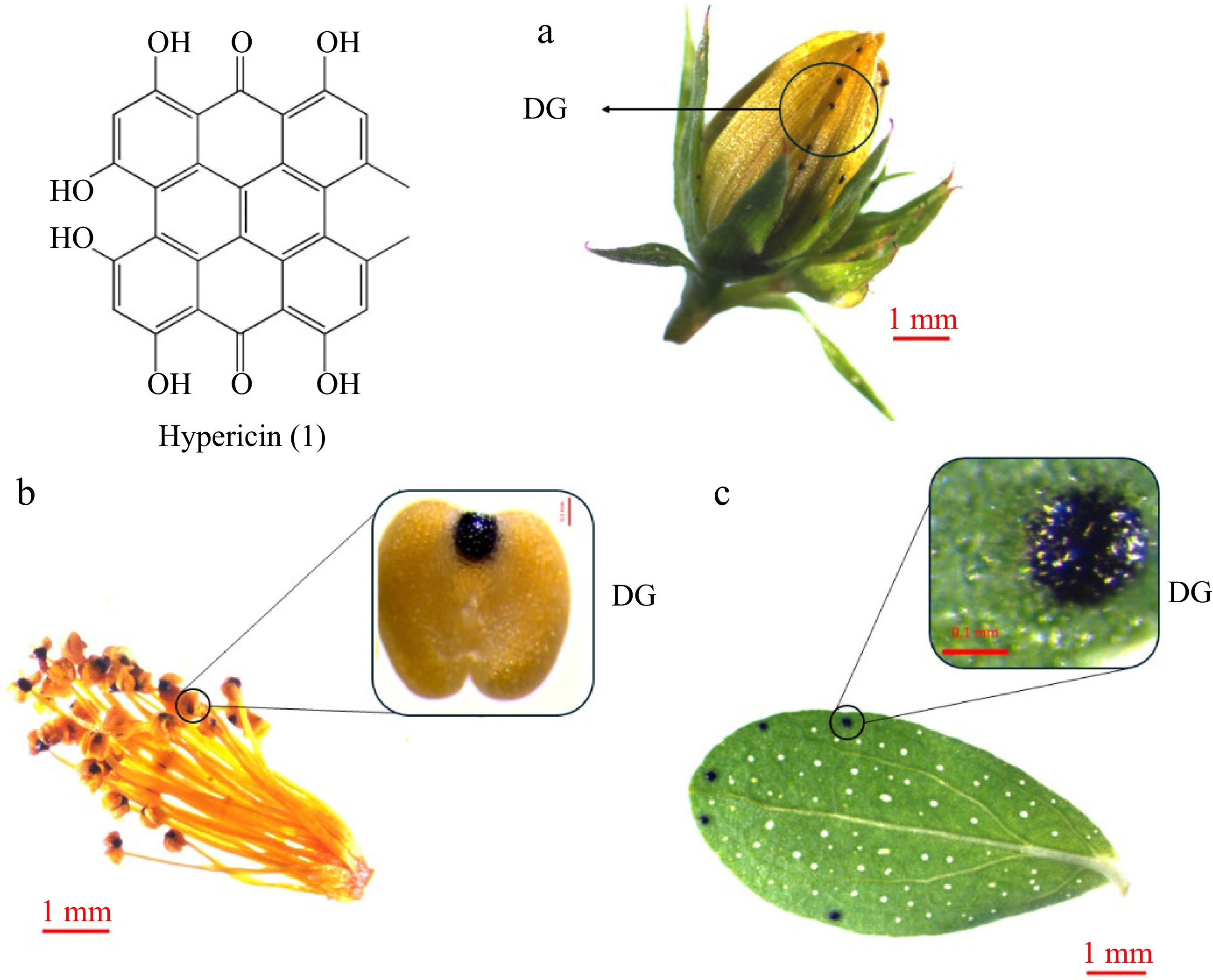
Figure 2.
Hypericin accumulation in Hypericum perforatum organs: (a) bud, (b) stamens, (c) leaf. Hypericin (1) is accumulated in Dark Glands (DGs) of the aerial parts of the plants like (a) sepals and petals, (b) stamens, and (c) leaves.
Medicinal importance of hypericin
-
A substantial body of evidence has accumulated over the past few decades indicating that hypericin can elicit a range of pharmacological effects[30]. Given its status as one of the most potent photosensitizers in nature, hypericin has been the subject of considerable investigation for its potential use in photodynamic therapy (PDT) for cancer treatment, mainly due to its ability to exhibit cytotoxic activity upon light activation[5,7]. The results thus far appear promising concerning the induction of cancer cell death. However, further high-quality clinical studies are required to establish hypericin's safety and clinical utility in cancer patients[30,31]. In addition to its potential anticancer properties, hypericin demonstrated robust antiviral activity[30] and has proven to ameliorate cognitive deficits in mouse models of Alzheimer's[32].
Biosynthesis
-
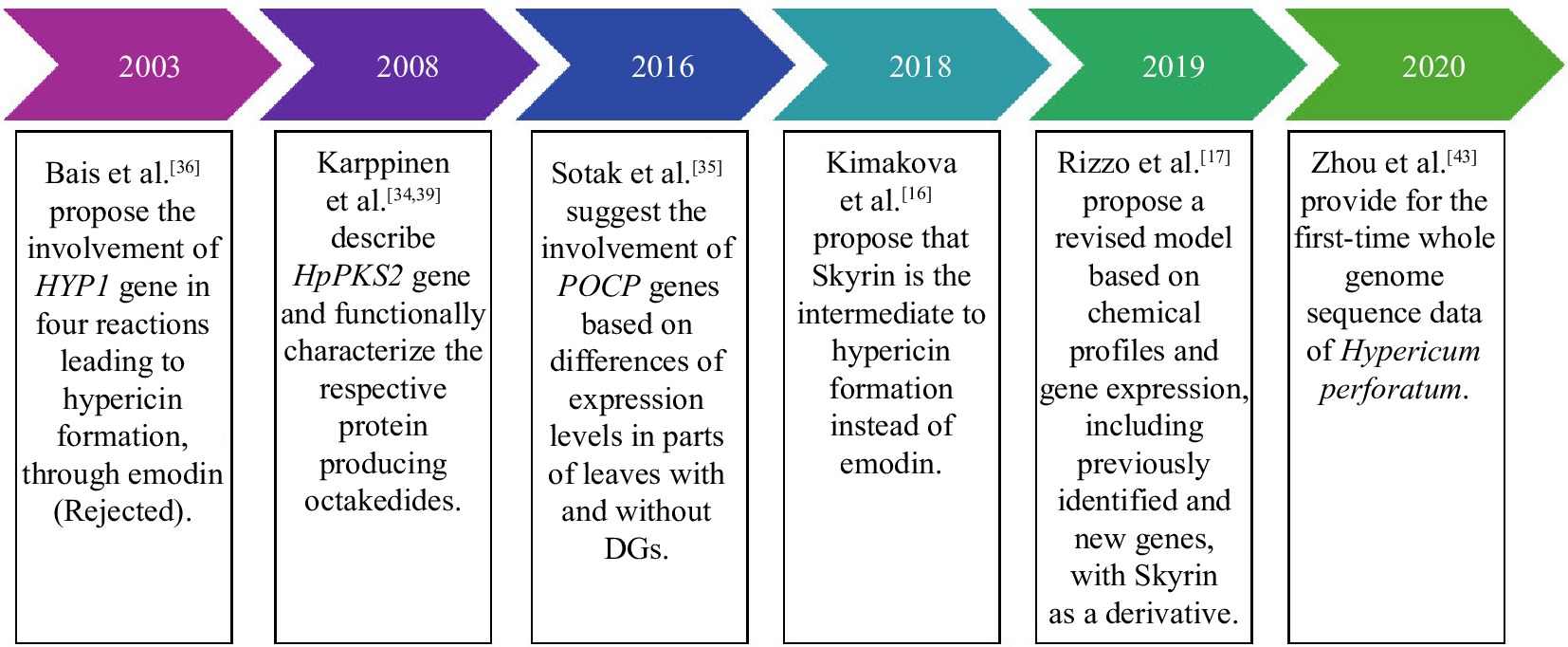
As the medicinal applications of hypericin are being supported by increasing data over recent years, there has been a growing need for a deeper understanding of its formation. Despite introducing new potential genes over the past decades, the biosynthetic pathway leading to hypericin formation in Hypericum plants remains unclear (Fig. 3, Table 2). Until 2003, there had been no suggestions about the genes or enzymes involved in the biosynthesis of hypericin; it was only known that its production was via the polyketide pathway, most likely via the anthrone emodin[33]. The first step in this pathway was considered to be the polymerization of acetyl-coenzyme A (acetyl-CoA) with malonyl-coenzyme A (malonyl-CoA) via a polyketide synthase (PKS) followed by a subsequent cyclization step to produce an anthrone derivative[33].
Table 2. Genes suggested to participate in hypericin and hyperforin biosynthesis.
Genes involved in hypericin biosynthesis Genes involved in hyperforin biosynthesis Confirmed by the functional characterization of the respective protein HpPKS2[34] BCKDH[37] CLL[37] PKS[37] PT1-4[38] Hypothetical based on transcriptional data POCP1-4[35] TER[17] BBE[17] Rejected by experiments HYP1[36] HpPKS1[39] In 2003, a gene and its encoded protein, potentially involved in hypericin biosynthesis, were reported and described for the first time[36]. The HYP1 gene encodes a phenolic oxidative coupling protein with high homology to Bet.v.1 class allergens[36]. The enzymatic activity attributed to this protein involved the catalysis of four reactions to produce hypericin: (i) a condensation reaction of a molecule of emodin and its anthrone, (ii) a dehydration reaction to produce the dianthrone of emodin, and (iii), (iv) two oxidative coupling reactions to produce first protohypericin and then hypericin. The involvement of the HYP1 protein in the biosynthesis of hypericin was questioned by several publications later on[40−42].
In 2008, an octaketide synthase (OKS) was described[34,39]. This synthase was named HpPKS2, and the corresponding gene was found to be explicitly expressed in the black glands of the plant[34]. HpPKS catalyzed the condensation of one acetyl-coenzyme A with seven molecules of malonyl-coenzyme A, yielding octaketide products but not in the expected cyclic forms. Based on this result, it was suggested that this OKS catalyzed the first reaction towards hypericin formation, possibly followed by the action of a Polyketide Cyclase[34]. Their analysis was based on mRNA expression data from leaf parts with and without DGs, Soták et al.[35] suggested the involvement of phenolic oxidative coupling proteins (POCPs) in some reactions previously associated with HYP1 and introduced four potential genes (POCP1-4) without providing sufficient functional data for the encoded proteins.
Two relatively recent publications have laid new foundations in the debate on hypericin biosynthesis. In a study published in 2018 by Kimáková et al.[16] demonstrated that the presence of emodin and its anthrone, on the one hand, and the presence of hypericin in Hypericum species, on the other hand, are not correlated. This finding prompted a re-evaluation of the hypothesis that emodin functions as an intermediate in hypericin production. Instead, the researchers put forth an alternative hypothesis involving skyrin. In 2019, Rizzo et al.[17] presented an integrated metabolomic and transcriptomic profile for Hypericum perforatum, utilizing tissue from two different phenotypes of the carpel placenta (with and without DGs). By integrating the chemical profiles of key species compounds (hypericins, endocrine glycosides, flavonoids) with gene expression and black gland development, the authors proposed a revised model for hypericin biosynthesis (Fig. 4). The suggested pathway incorporates previously identified genes/proteins (OKS, POCP) and introduces new candidate genes. This model retained the initial condensation step facilitated by the identified octaketide synthase HpPKS2[34]. The second step is cyclization, which is proposed to be catalyzed by a polyketide cyclase (PKC). This step may address the cyclization error exhibited by HpPKS2 in catalyzing the same reaction. The third step involves two genes designated as dihydrofolate reductases that may encode thioesterases (TERs), which release octaketides and tetraketides from coenzyme A. These octaketides and tetraketides were suggested to act as mediators in the biosynthesis of hypericin and flavonoids[17]. Phenolic oxidative coupling proteins (POCPs), which were previously reported by Soták et al.[35] to play a role in the biosynthesis of hypericin were proposed to facilitate the C-C bond formation of the naphthodiachronic halves of hypericin and its derivatives. An alternative or complementary function to that of POCPs in forming one of the three C-C bonds between the naphthodiachronic halves that make up hypericin is attributed to a Berberine Bridge Enzyme (BBE). This likely involves the formation of the last of the three bonds, for which enzymatic catalysis is required[17]. The initial bond proposed by the authors to be formed is C5-C5', which is catalyzed by POCPs. The second bond is the C10-C10' double bond, which is formed after catalysis by BBE. Finally, the C4-C4' bond is formed in the last step non-enzymatically. The intermediates for hypericin formation are proposed to be atrochrysone carboxylic acid, atrochrysone, atrovirin B, and peniciliopsin. This suggestion completely bypasses emodin while retaining the possibility of forming skyrin, in accordance with the findings of Kimáková et al.[16]. In addition to the hypericin biosynthesis model, Rizzo et al.[17] presented, for the first time, two transcriptional factors potentially involved in DG differentiation by relating gene expression to their development in the carpel placenta.
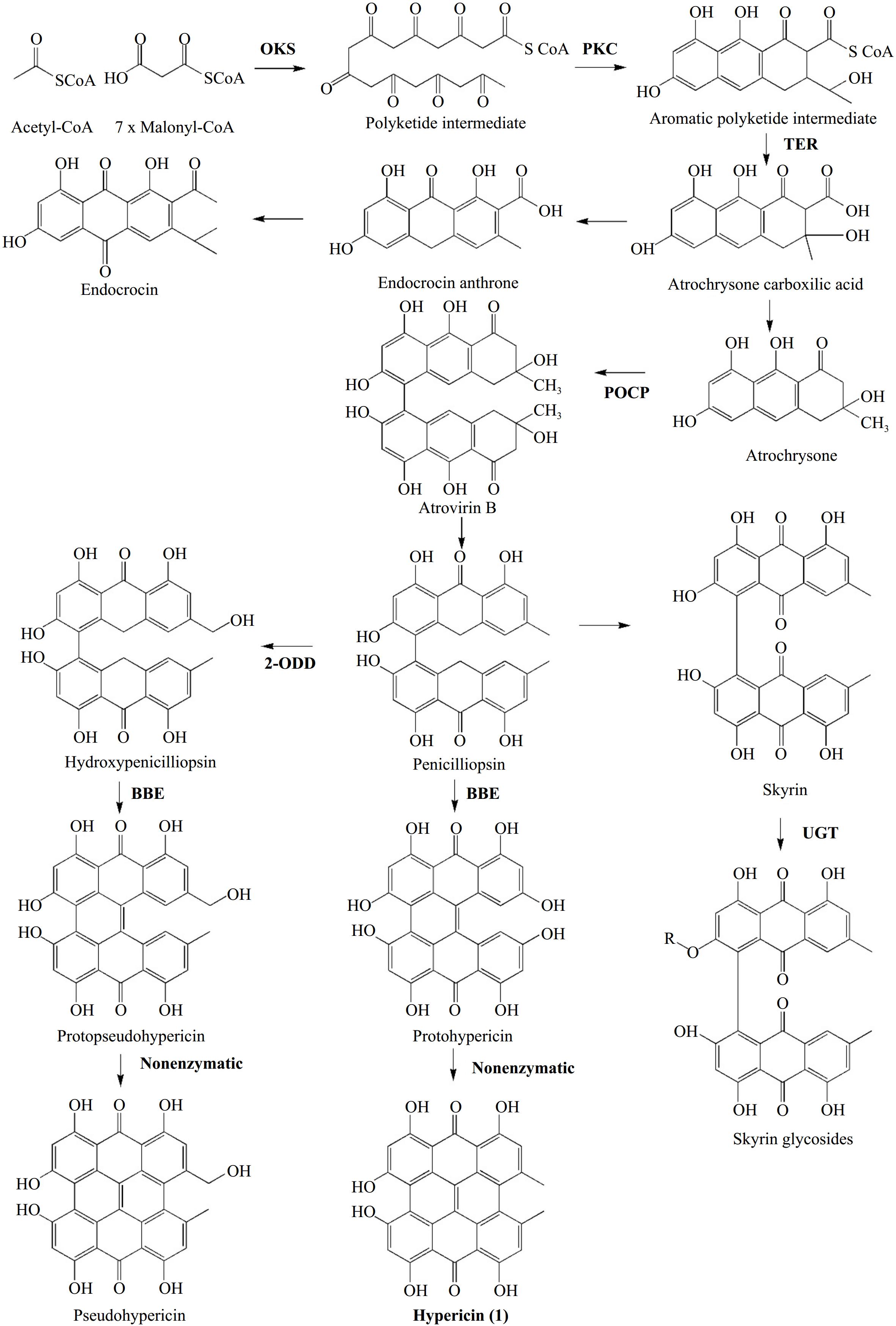
Figure 4.
Hypericin biosynthesis as presented by Rizzo et al.[17]. In bold: enzymes involved in reactions' catalysis. Abbreviations: OKS: Oktaketide Synthase, PKC: Polyketide Cyclase, TER: Thioesterase, POCP: Phenolic Oxidative Coupling Protein, BBE: Berberine Bridge Enzyme, 2-ODD: 2-oxoglutarate and Fe(II)-dependent oxygenase, UGT: UDP-glucosyltransferase.
In 2020, Zhou et al.[43] provided for the first-time whole genome sequence data of Hypericum perforatum. Following the annotation of the genome, the authors put forth two additional potential pathways for the formation of hypericin. The initial pathway was analogous to those previously proposed, commencing with the condensation of seven malonyl-CoA molecules and one acetyl-CoA, facilitated by a PKS that produces emodin anthrone. An aldolase is proposed to merge emodin and emodin anthrone, which subsequently undergoes phenolic oxidation to form protopseudohypericin and protohypericin. In the final step of the model, light catalyzes the transformation of hypericin and pseudohypericin precursors to the final form of the substances. The second probable pathway proposed by Zhou et al.[43] is consistent with a suggestion previously made by Kimáková et al.[16]. One molecule of 1,2,4,5-tetrahydroxy-7-(hydrymethyl)-9,10-anthraquinone and one molecule of 1,2,4,5-tetrahydroxy-7-methyl-9,10-anthraquinone-2-O-β-glycopyranoside merge through the enzymatic catalysis of a hydrogenase and a tyrosinase and produce Skyrin-6-O-glucopyranocide. Skyrin emerges from the activity of a glycosidase and, through some additional reactions not provided in the model, forms hypericin. While Zhou et al.[43] provided a valuable tool for research on novel biosynthetic genes, their proposed pathways for hypericin formation lacked sufficient evidence. This is particularly evident due to the lack of support from their tests on gene expression levels of the proposed genes in tissues with varying hypericin content.
To date, the work of Rizzo et al.[17] represents the most comprehensive and well-supported hypothesis regarding hypericin formation, including the well-characterized enzymatic activity of HpPKS2 and the POCP genes, which were previously found to be related to hypericin formation. Nevertheless, research on hypericin biosynthesis is ongoing. The novel genes introduced by Rizzo et al.[17] have yet to be confirmed by new gene expression experiments, and the functional characterization of the proteins translated by these genes remains to be done.
Regulation of hypericin biosynthesis
-
The research on the mechanisms controlling hypericin production and DG (dark gland) development is quite limited, with only a few studies published. Most of these studies focus on how H. perforatum responds to different conditions. For example, it has been shown that low temperatures help increase hypericin levels by activating related genes, though the exact mechanism behind this observation is still unclear[44,45]. UV-B light exposure has also been demonstrated to enhance hypericin production[46]. Plant growth regulators, including jasmonic acid, its derivatives, and select cytokinins, have also been shown to stimulate hypericin production[47−49]. To date, no transcription factors involved in hypericin production have been identified. However, Rizzo et al.[17] identified two transcription factors closely linked to the initial stages of dark gland formation and proposed their involvement in the differentiation process. One such factor is Agamous-like 6 (AGL6), a MADS-box transcription factor that regulates flower organ identity and meristem fate in Arabidopsis thaliana[50]. The other one is an R2R3-Myb transcription factor, which primarily regulates processes associated with secondary metabolism, cell fate, and organ identity[51].
-
Hyperforin (2) is a bicyclic polyprenylated acylphloroglucinol derivative[52] (Fig. 5). In contrast to hypericin, present in numerous genus taxa, hyperforin is primarily concentrated in H. perforatum[52,53]. The substance is concentrated mainly in the pistils and fruits of plants, where it may function as a defensive factor to safeguard the fruit from herbivores and microbes[52,54]. The pistil and capsule exhibit glandular streaks or patches on their walls. These are classified as vittae when flat or slightly swollen and vesicles when visibly swollen[1] (Fig. 5a). Additionally, hyperforin is accumulated in schizogenic extracellular spaces, along with essential oils[55,56]. This is the second type of gland observed in Hypericum plants, also called Pale Glands (PGs) (Fig. 5b). These structures are spherical or oblong glands comprising a sub-epidermal cavity surrounded by two layers of cells. The internal layer is composed of highly attenuated, thin-walled secretory cells. The external layer comprises parenchymatous cells with thicker walls[28,56]. PGs exhibit a color range from transparent to amber, and they are responsible for the perforated image of the leaves when observed against light[1].
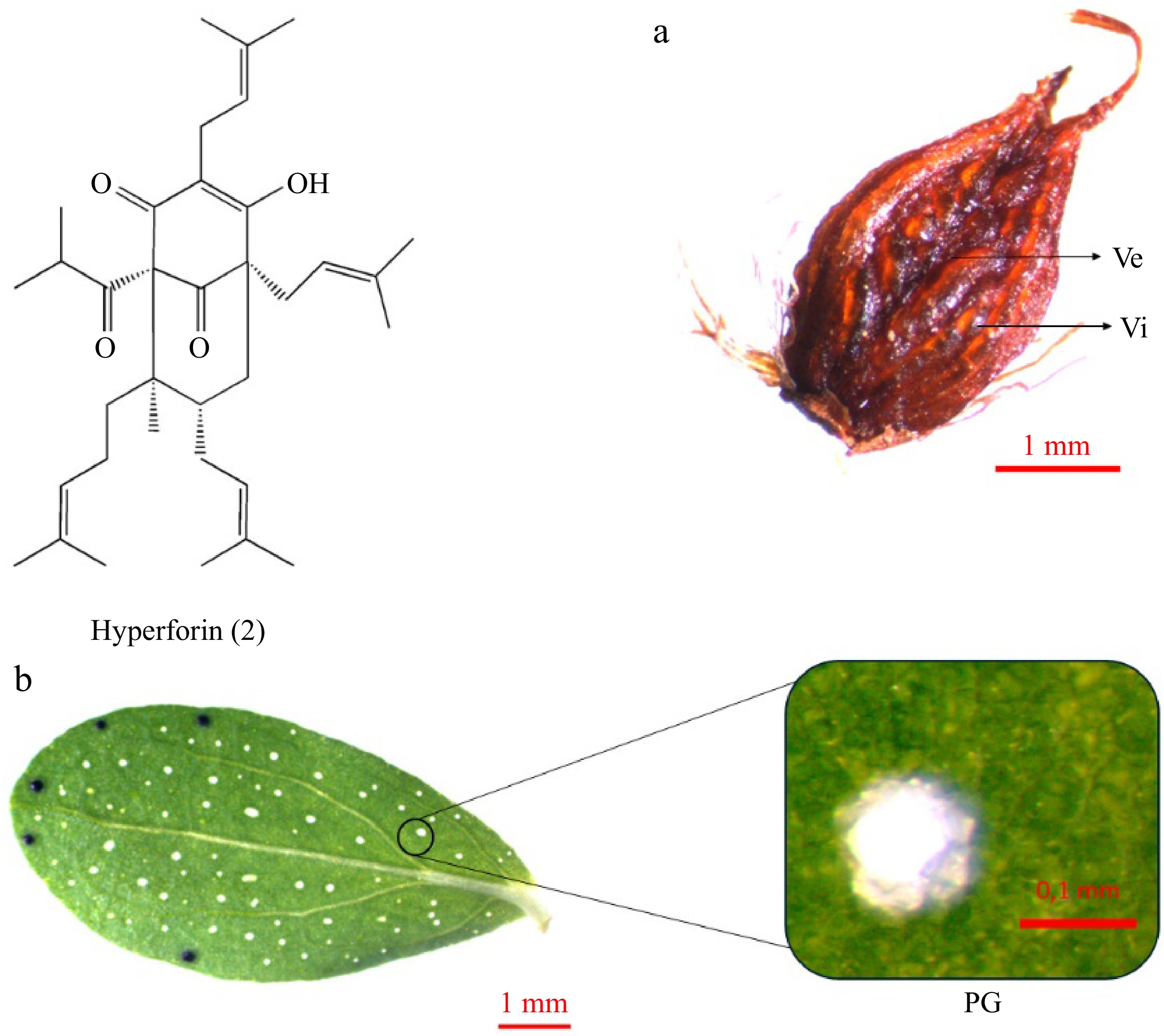
Figure 5.
Hyperforin accumulation in Hypericum perforatum organs: (a) fruit, (b) leaf. Hyperforin (2) is accumulated in formations like vittae (Vi) and vesicles (Ve) on the (a) fruit capsule, and in the (b) Pale Glands (PGs) of the leaves.
Medicinal importance of hyperforin
-
For an extended period, it was assumed that the antidepressant impact of H. perforatum was primarily attributable to the naphthodianthronic compounds, particularly hypericin. Nevertheless, after experimental and clinical studies, it is now accepted that hyperforin is the primary component responsible for the antidepressant effect[20,58]. Hyperforin acts as a broad-spectrum neurotransmitter reuptake inhibitor that affects the synaptosomal uptake of serotonin, dopamine, norepinephrine, glutamate, and gamma-aminobutyric acid (GABA) with similar efficiency[20]. Subsequently, adaptive alterations in the receptor system occur. The model of action of hyperforin is distinctive from other antidepressants in that it does not directly interact with transmitter transporters. Alternatively, it increases the intracellular sodium concentration, inhibiting gradient-driven neurotransmitter reuptake[57]. This effect on intracellular sodium concentration has been attributed to the activation of non-selective cation channels[58]. In contrast, synthetic antidepressants are competitive inhibitors of either one or, at most, two transporters at the transmitter binding sites. Therefore, hyperforin is not only structurally but also functionally a novel antidepressant[20,52,57]. Additionally, it has been demonstrated to possess antibiotic properties against gram-positive bacteria[24] and anti-cancer activity in vivo[59].
Biosynthesis
-
In contrast to hypericin, until recently, there was a lack of evidence regarding the biosynthesis of hyperforin, particularly at the level of the genes and enzymes involved, even though Adam et al.[55] described the chemical reactions leading to its formation two decades ago. The chemical structure of hyperforin, consisting of an acylphloroglucinol nucleus and five isoprenoid units, has led Adam et al.[55] to suggest that hyperforin formation is divided into two distinct phases. The initial stage of the process involves the formation of the hyperforin skeleton via a polyketide mechanism. This mechanism entails the condensation of one molecule of isobutyryl-coenzyme A with three molecules of malonyl-coenzyme A, producing phlorisobutyrophenone (hyperforin skeleton). The isobutyryl-CoA molecule acts as the starter molecule, and it is derived from valine[55]. Klingauf et al.[60] proposed later that the condensation is catalysed by an isobutyrophenone synthase (BUS). The second phase involves a series of prenylations of phlorisobutyrophenone (PIBP). A triple electrophilic substitution of the aromatic nucleus is necessary for the formation of hyperforin involving two dimethylallyl pyrophosphatase (DMAPP) units and one geranyl pyrophosphate (GPP) unit. In contrast, another DMAPP unit is involved in the closure of the second ring[55]. It is established that the five isoprenoid moieties are derived predominantly via the non-mevalonate (MEP) pathway[55].
Until recently, research on enzymes involved in these processes was quite limited. Karppinen & Hohotla[39] took an approach, presenting a cDNA encoding a polyketide synthase (HpPKS1) whose expression was found to be correlated with hyperforin content. Nevertheless, the HpPKS1 protein was observed to yield compounds that are beneficial for the biosynthesis of other secondary metabolites rather than hyperforin[61]. Boubakir et al.[62] described a dimethyl transferase as a participant in prenylation reactions. Subsequently, this enzyme was proposed to catalyze the initial prenylation reaction of the hyperforin skeleton.
However, two recent studies conducted by the same research team have yielded new insights regarding the enzymatic reactions involved in hyperforin formation (Fig. 6). The first study addressed the reactions mediating the hyperforin skeleton formation[37], while the second examined the prenylation reactions that lead to the final formation of hyperforin[38]. To investigate the enzymes involved in phlorisobutyrophenone formation, Wu et al.[37] employed genome mining on the genome data of H. perforatum, as published by Zhou et al.[43], to identify candidate Biosynthetic Gene Clusters (BGCs). Along with transcriptomic, phylogenetic, and metabolomic data, they were able to characterize two BGCs with branched-chain keto acid dehydrogenase E1 subunit alpha (BCKDHA), CoA ligase (CCL) and PKS genes, of which the translated enzymes catalyze the formation of hyperforin precursor, phlorisobutyrophenone. BCKDHA and CCL translated enzymes were attributed to facilitating the transformation of valine to isobutyryl-CoA, while PKS enzymes were proposed to act as catalysts in the condensation of isobutyryl-CoA with malonyl-coenzyme A, ultimately leading to the formation of phlorisobutyrophenone. The authors provided comprehensive data on the expression levels of the genes and the functional characterization of the respective proteins, thus establishing a well-defined model for phlorisobutyrophenone biosynthesis[37]. In their most recent research, the authors investigated the subsequent steps leading to hyperforin formation through a series of prenylation reactions of the phlorisobutyrophenone core[38]. The tetraploid genome of H. perforatum was sequenced, and single-cell atlases were generated for leaves and flowers. By mapping upregulated genes, previously characterized to be involved in phlorisobutyrophenone core formation (BCKDHA, CCL, PKS), the researchers identified a new type of cell, designated 'Hyper cells', in which de novo biosynthesis of hyperforin occurs. They identified and characterized four new prenyltransferases (PTs) belonging to the UbiA family, which are involved in the hyperforin biosynthesis pathway. Furthermore, by employing two synthetic biology platforms for heterologous expression, they were able to produce the protein molecules in yeast and tobacco and produce hyperforin by employing the necessary substrates[38]. The four prenyltransferases (PTs) introduced in this study addressed the knowledge gap regarding the enzymes involved in hyperforin biosynthesis. It is noteworthy that the final step in hyperforin biosynthesis involves an unconventional prenyltransferase that catalyzes the 1′-2 coupling (branching) between two isoprenyl units and facilitates the formation of a bond between C1 and C8, resulting in cyclization and biosynthesis[38]. Such cyclizations, which generate complex polycyclic structures, are a defining characteristic of natural product biosynthesis.
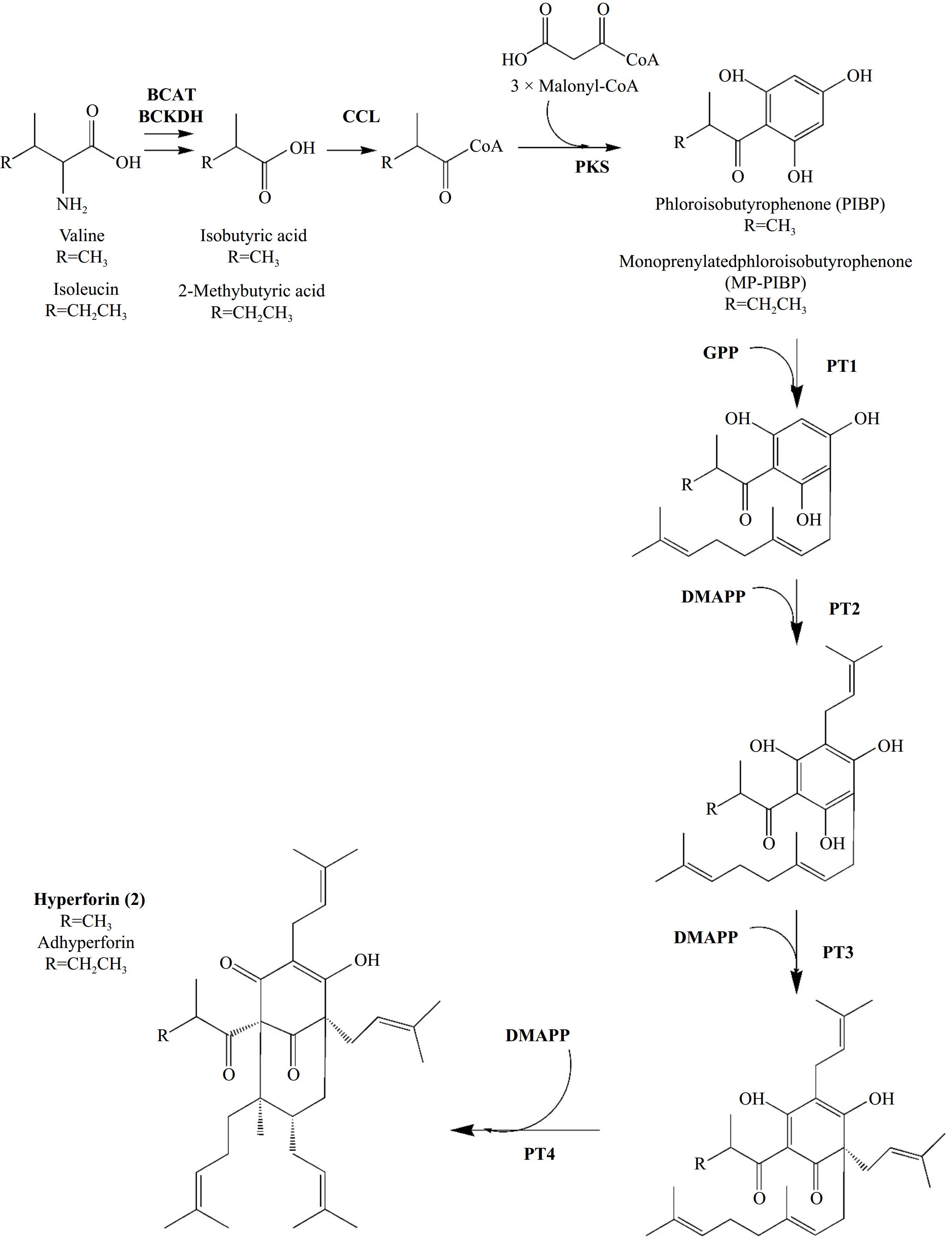
Figure 6.
Biosynthesis of Hyperforin (2) and its homologue Adhyperforin according to Wu et al.[37,38]. In bold: enzymes involved in reactions' catalysis and prenyl-groups. Abbreviations: BCAT: branched-chain amino acid aminotransferase, BCKDH: branched-chain alpha-keto acid dehydrogenase, CCL: CoA ligase, PKS: Polyketide synthase, PT: Prenyltransferase, DMAPP: dimethylallyl-diphosphate, GPP: geranyl-diphosphate.
These two studies fully elucidate hyperforin biosynthesis with an approach that can be regarded as complete and well-supported. However, they are still very recent and supporting those results from other experiments is necessary to confirm that hyperforin biosynthesis has now been established as a well-understood process at a chemical and molecular level.
-
Xanthones represent a diverse group of naturally occurring polyphenolic compounds extensively distributed among bacteria, fungi, lichens, and plants[63]. Simple oxygenated xanthones and prenylated xanthones are predominantly found in the roots of Hypericum species[18]. Over 100 xanthones have been isolated and identified from Hypericum species. These xanthones exhibit structural diversity, with variations in the patterns of hydroxyl, methoxy, prenyl, butenyl, and glycoside substitutions on the base structure. Notable xanthones present in H. perforatum include 1,3,6,7-tetrahydroxyxanthone (norathyriol), mangiferin, isomangiferin, and the xanthonolignoid kielcorin[18].
In recent decades, xanthones have emerged as a significant resource for developing new pharmaceutical agents. The pharmacological effects of plant xanthones are diverse, encompassing anticancer, antidiabetic, and antimicrobial activities[64].
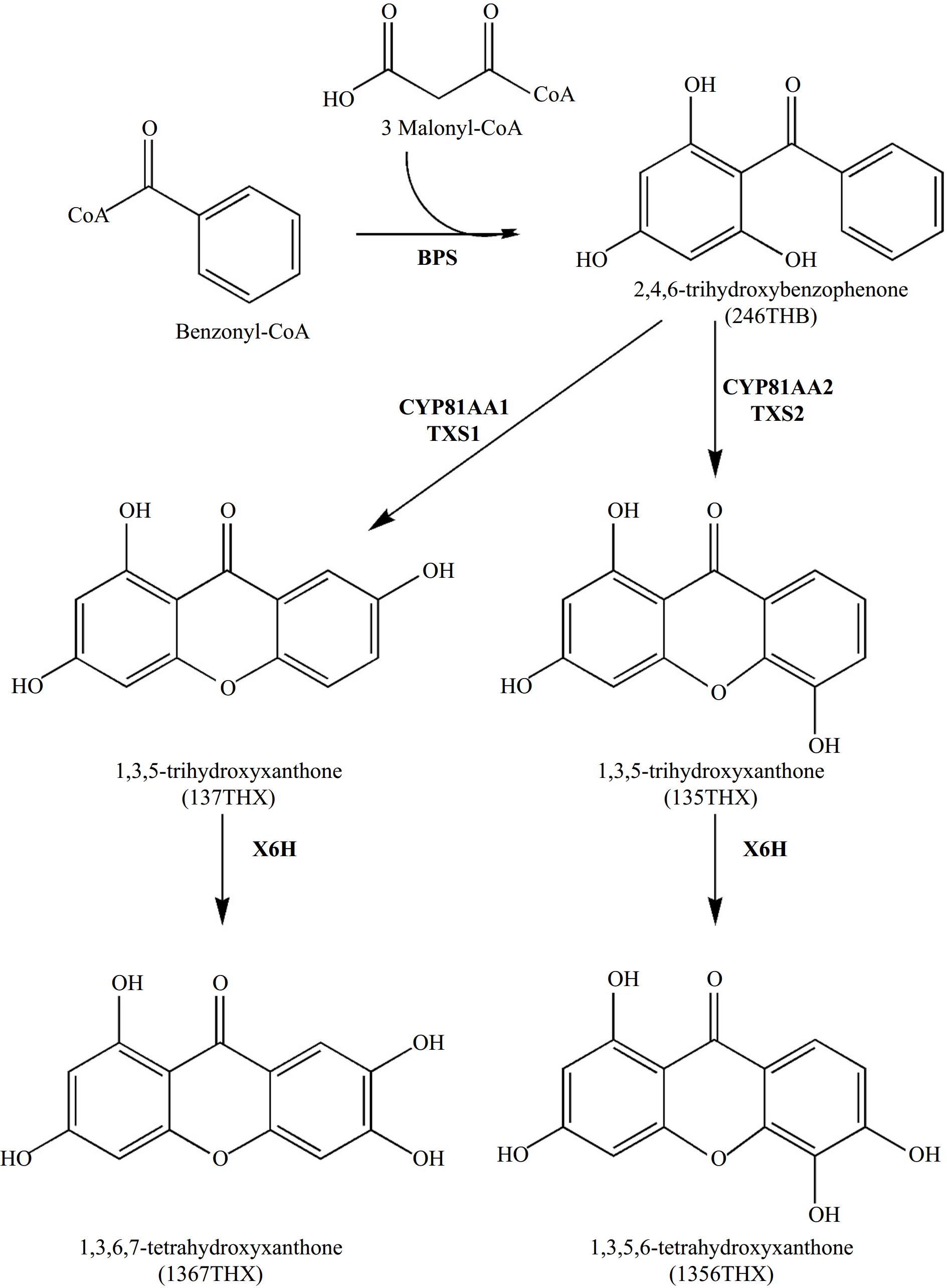
The biosynthesis of xanthones in Hypericum species has been thoroughly investigated. Analogous to hyperforin biosynthesis, the creation of the xanthone skeleton is followed by a series of reactions, such as hydroxylations and prenylations. Formation of the xanthone skeleton (Fig. 7) involves condensing one benzoyl-CoA molecule with three malonyl-CoA molecules to form 2,4,6-trihydroxybenzophenone (246THB). The reaction is catalyzed by a type III PKS known as benzophenone synthase (BPS)[65]. Subsequently, 246THB undergoes hydroxylation to give 2,3′,4,6-tetrahydroxybenzophenone (23′46THB), followed by regioselective oxidative phenol couplings to yield either 135THX or 137THX. The latter hydroxylation and coupling reactions are catalyzed by the bifunctional cytochrome P450 enzymes CYP81AA1 and CYP81AA2[66]. Finally, xanthone 6-hydroxylase (X6H) catalyzes the hydroxylation of 1,3,5- and 1,3,7-trihydroxylated xanthones to their corresponding tetrahydroxy derivatives, namely 1,3,5,6-tetrahydroxyxanthone (1356THX) and 1,3,6,7-tetrahydroxyxanthone (1367THX), respectively[67]. Recent advancements have led to characterizing genes encoding aromatic prenyltransferases that specifically prenylate xanthones from Hypericum perforatum at the C-4 position[63]. This discovery enhances our understanding of the formation of polyprenylated xanthone derivatives. Notably, the prenylation of xanthones at the C-4 position has been reported to improve the biological activities of the resulting prenylated derivatives.
-
Flavonoids represent a relatively diverse family of aromatic molecules derived from the amino acid phenylalanine and malonyl-coenzyme A[68]. Flavonoids are commonly involved in ultraviolet protection, flower pigmentation, and pathogen and herbivore resistance, and they also affect various developmental processes in plants[68]. Flavonoids derived from Hypericum plants have been demonstrated to possess antidepressant effects[69] by playing an important role in the modulation of the hypothalamic-pituitary-adrenal (HPA) axis[70]. The flavonoid biosynthetic pathway has been one of the most intensively studied metabolic systems in plants. Flavonoids are derived from the phenylpropanoid metabolic pathway and have a basic structure that comprises a C15 benzene ring structure of C6-C3-C6. Studies have highlighted the complexity of the biosynthesis of flavonoids in the plant kingdom, forming a wide network of reactions[71]. Enzymes identified to take part in flavonoid formation, specifically in Hypericum plants, include Chalcone Synthase[71] and, from more recent work, flavanone 3-hydroxylase, flavonol synthase, flavonoid 3-hydroxylase, and flavonoid 3-O-galactosyltransferase[72].
Essential oils (EOs) are complex blends of volatile compounds, well-known for their strong antimicrobial, antioxidant, and antiangiogenic effects. In species of the Hypericum, essential oils are stored in PGs and hyperforin (Fig. 5b). However, Hypericum species are often noted for their low essential oil content[73]. Key constituents of Hypericum essential oils include monoterpene hydrocarbons, sesquiterpene hydrocarbons, and oxygenated sesquiterpenes[74]. The essential oil and volatile constituents most frequently reported in Hypericum species include the aliphatic hydrocarbons n-nonane and n-undecane; the monoterpenes α-pinene and β-pinene; and the sesquiterpenes β-caryophyllene and caryophyllene oxide[73]. A recent review of the chemical composition and bioactivities of Hypericum essential oils has highlighted their medicinal potential[74]. Notably, essential oils from Hypericum species have demonstrated the ability to facilitate wound healing[75]. Given their likely presence in infused oils, these essential oils likely play a crucial role in enhancing the wound-healing effectiveness of traditional Hypericum preparations[75].
-
Research on the secondary metabolism of Hypericum species presents significant opportunities, driven by advances in biotechnology, pharmacology, cultivation techniques, and interdisciplinary collaboration. Using genomic and metabolomic tools is expected to enhance our understanding of the biosynthetic pathways leading to the formation of secondary metabolites in Hypericum. Techniques such as CRISPR may facilitate the targeted production of important compounds like hypericin and hyperforin, while synthetic biology offers the potential to replicate these pathways in microorganisms, supporting more sustainable large-scale production. In pharmacological research, well-designed clinical trials evaluating the efficacy and safety of Hypericum extracts and individual metabolites will be critical for establishing evidence-based practices and novel medicinal products. Moreover, exploring the potential synergistic effects of Hypericum compounds in combination therapies could provide new insights into treatment approaches. Collaboration among botanists, pharmacologists, chemists, and biotechnologists will be vital in advancing research and understanding the therapeutic potential of Hypericum. Continued work in these areas could lead to developing new therapeutic agents that address contemporary medical needs.
-
Hypericum is a highly diverse genus that has attracted the attention of researchers across various academic fields. Its complex taxonomy, ethnobotanical uses of different species worldwide, chemical diversity, and applications in modern medicine have made it a subject of extensive study for many years. This review aims to highlight the considerable pharmacological potential of Hypericum species, attributed to their diverse secondary metabolites while focusing on novel insights into their formation. Studying plant secondary metabolite biosynthesis can be challenging, requiring the integration of various pieces of knowledge to construct a comprehensive picture of a biosynthetic pathway. In Hypericum plants, the biosynthesis of two essential substances—hypericin and hyperforin—known for their significant medicinal value, has puzzled scientists for several years. However, recent advances in transcriptomics, metabolomics, and genomics, along with their increasing accessibility to researchers, have facilitated the identification of new enzymes and genes involved in their formation. As a result, elucidating the biosynthetic pathways of hypericin and hyperforin now seems closer than ever. Other substances present in Hypericum plants are more commonly found in other plant genera, and their biosynthesis is better understood due to studies conducted on those organisms. Despite these advancements, the field faces significant challenges, particularly in functional characterizing newly identified biosynthetic genes and enzymes. Further research is needed to confirm the roles of these genes and elucidate their regulatory mechanisms.
It is also important to note that most studies on hypericin biosynthesis primarily focus on H. perforatum. Investigating whether the genes expressed in H. perforatum are identical or similar to those in other Hypericum species would be valuable, as would understanding the extent to which gene expression is associated with hypericin content in these plants. Another intriguing question is whether these genes are present and expressed in taxa that do not produce hypericin, suggesting that these genes may serve additional functions or be unique to hypericin-synthesizing taxa. Furthermore, understanding the influence of genetic and environmental factors on metabolite synthesis will be essential for optimizing the production of these bioactive compounds in cultivation. Integrating biotechnological applications holds promise for enhancing the yield and efficiency of metabolite production, which could lead to the development of novel therapeutic agents derived from Hypericum species.
In conclusion, the ongoing investigation of Hypericum secondary metabolites not only reaffirm their historical significance in traditional medicine but also pave the way for new avenues of pharmaceutical innovation. By addressing current knowledge gaps and employing advanced research methodologies, future studies can fully elucidate the medicinal potential of Hypericum species, contributing to developing effective and sustainable therapeutic solutions.
We acknowledge the support of this work by the project 'Upgrading the plant capital' (MIS 5002803), which is implemented under the Action 'Reinforcement of the Research and Innovation Infrastructure', funded by the Operational Programme 'Competitiveness, Entrepreneurship and Innovation' (NSRF 2014-2020) and co-financed by Greece and the European Union (European Regional Development Fund); the National Strategic Reference Framework (NSRF), Research Funding Programme of the Action RESEARCH – CREATE – INNOVATE (AROMADISTIL – 95783).
-
The authors confirm contribution to the paper as follows: study conception and design: Poulaki S, Vlachonasios K; draft manuscript preparation: Poulaki S; manuscript review & editing: Vlachonasios K, Poulaki S; figures design, original photos in Figs 2 & 5: Poulaki S. Both authors reviewed and approved the final version of the manuscript.
-
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Poulaki S, Vlachonasios K. 2024. Secondary metabolites of medicinal use in Hypericum spp.: a rich history and a promising future. Medicinal Plant Biology 3: e025 doi: 10.48130/mpb-0024-0027
Secondary metabolites of medicinal use in Hypericum spp.: a rich history and a promising future
- Received: 12 August 2024
- Revised: 12 October 2024
- Accepted: 31 October 2024
- Published online: 29 November 2024
Abstract: The genus Hypericum, which comprises over 450 species worldwide, has a long history of use in traditional medicine. It is now known for its antidepressant, antiviral, antibacterial, and anticancer properties. This review summarizes the current knowledge on the biosynthesis of the main bioactive secondary metabolites responsible for the pharmaceutical applications of plants, particularly hypericin and hyperforin. In addition, this review highlights the importance of other chemical constituents in Hypericum, such as xanthones and flavonoids, which contribute to the pharmacological potential of the genus. Hypericin, a naphthodianthrone, has been shown to have remarkable pharmacological effects, particularly as a potential anticancer agent. On the other hand, hyperforin, a polyprenylated acylphloroglucinol, has been identified as a potent antidepressant. Recent advances in transcriptomics, metabolomics, and genomics have identified novel genes and enzymatic pathways that facilitate the biosynthesis of these compounds, providing valuable insights into their formation. Despite these advances, further research is essential to fully characterize the biosynthetic pathways and optimize the production of bioactive compounds in Hypericum species.
-
Key words:
- Hypericum /
- Hypericin /
- Hyperforin /
- Xanthone biosynthesis /
- Secondary metabolites