-
Jasmine (Jasminum sambac) is a climbing shrub or upright plant, belonging to the Oleaceae family. Jasmine is widely distributed in tropical and subtropical regions and is renowned for its strong fragrance and extensive use in tea production, ornamentation, and medicine[1,2]. However, due to years of continuous cultivation, jasmine has exhibited signs of seed degradation, such as inferior flower quality, reduced resistance, and flower yield[3]. Crossbreeding is one of the most common and effective methods for germplasm innovation and variety improvement, having achieved significant success in plant breeding. However, sexual reproduction poses challenges in jasmine, and there have been no reports of successful hybrid breeding until now. Consequently, hybrid breeding significantly lags behind the requirements of planters in the field of jasmine[4]. A previous study revealed that pollen abortion is the main reason for the poor seed-setting in jasmine[5]. However, the precise reason behind pollen abortion in jasmine remains unclear.
Pollen plays a crucial biological function by transporting double sperm cells to the embryo sac[6]. The basic structure of a mature pollen wall comprises an outer exine layer filled with pollen coat and an intine layer[7]. The exine, primarily composed of sporopollenin, consists of the outer sexine that is made up of tectum and baculum and inner nexine. The intine, on the other hand, consists of cellulose, pectin polymers, hemicellulose, hydrolytic enzymes, and proteins. The synthesis of intine is exclusively controlled by microspores selves[8]. Proper pollen wall formation is necessary for pollen fertility, and any defects may result in abnormal pollen and male sterility[9]. Some genes related to exine or intine formation have been identified[10−12]. For example, overexpression of SiPKSA in Arabidopsis regulated exine layer development via sporopollenin biosynthesis, leading to male sterility[13]. In Triticum aestivum, SCULP1 was required for sporopollenin biosynthesis and exine integrity[14]. In rice, OsmiR528 regulated pollen intine formation by directly targeting the uclacyanin gene UCL23[15]. In A. thaliana, EFOP3 and EFOP4 indirectly regulated intine formation, resulting in pollen sterility[16].
Arabinogalactan proteins (AGPs) are a family of glycoproteins rich in hydroxyproline. They play a role in various aspects of plant development and reproduction, including pollen development and germination, pollen tube growth, tissue differentiation, response to biotic stresses, and cell expansion[17−19]. Based on the domain constituents of AGPs protein backbone sequences, AGPs are classified into six subfamilies, namely classical AGPs, AG peptides, fasciclin-like AGPs (FLAs), lysine-rich AGPs, non-classical AGPs, and chimeric AGPs[20]. FLAs are distinguished from other AGP subfamilies due to their typical fasciclin-like (FAS) domains[21]. The FAS domains comprise 110–150 amino acids, including two conserved regions (H1 and H2) separated by a highly conserved central YH motif[22].
FLAs have been characterized in numerous plant species, including 21 FLAs in A. thaliana[23], 27 in Oryza sativa[24], 30 in Musa acuminata[25], 19 in Corchorus olitorius[26], 17 in Cuscuta campestris[27], 40 in Populus deltoids[20], and 46 in Salix suchowensis[20]. FLAs play a crucial role in plant growth, development, and adaptation[28,29]. For instance, fascilin glycoprotein MTR1 controls the development of sporophytic cells, causing complete male sterility[30]. Arabidopsis FLA3 and FLA14 are pollen grain-specific genes that are essential for pollen wall development[31,32]. In poplar, the ptrfla40 ptrfla45 double mutant results in the enlargement of vessel and xylem fiber cell sizes, then subsequently increasing stem length and diameter[29]. At the same time, AtFLA11 and AtFLA12 are involved in second cell wall development and the regulation of lignin and cellulose synthesis under mechanical stimuli[33].
A previous study dynamically researched the mechanism of jasmine pollen and pistil interactions during crossing. The results showed significant changes in many genes, especially, the JsFLA3 gene exhibited significant up-regulation after pollination[34]. However, the biological function of JsFLA3 in jasmine has not been identified. In the present study, JsFLA3 was successfully cloned from jasmine. Subsequently, the expression patterns of the JsFLA3 in different jasmine tissues were evaluated. Overexpression vectors of the JsFLA3 gene were constructed, and the gene function was determined through its transformation into Arabidopsis. The findings suggest that the JsFLA3 gene may be involved in cellulose biosynthesis, subsequently affecting pollen wall development, and ultimately resulting in pollen sterility.
-
Single-petal J. sambac 'Danbanmoli' was obtained from the Jasmine Germplasm Resource Preservation Centre at Jiangsu Academy of Agricultural Sciences in Nanjing, China. The plants were maintained as described previously[35]. The A. thaliana ecotype plants were grown as previously described[36].
Isolation and sequencing of JsFLA3 cDNA
-
Total RNA was extracted from the jasmine 'Danbanmoli' leaves using MolPure® Plant RNA Kit (Yeasen, Shanghai, China). A total of 1 μg of total RNA was applied to synthesize the first strand cDNA using Hifair® III 1st Strand cDNA Synthesis Kit (Yeasen). The ORF of JsFLA3 was cloned using the specific primer pair JsFLA3-F/R (Supplementary Table S1), which was designed based on the sequence obtained from the J. sambac transcriptome database[34]. The amplified PCR products were purified using MolPure® Gel Extract Kit (Yeasen) and then inserted into the pMD19-T simple vector (TaKaRa, Tokyo, Japan) for sequencing.
Bioinformatic and phylogenetic analysis of JsFLA3
-
The composition of JsFLA3 amino acid was calculated using BioXM 2.6 software. The N-signal peptide cleavage site of JsFLA3 was predicted by SignalP 5.0 Server. The GPI (glycosylphosphatidylinositol) modification site of JsFLA3 was predicted by Big-PI Predictor. NetNGlyc-1.0 was used to predict JsFLA3 N-glycosylation sites. Euk-mPLoc2.0 was applied to predict JsFLA3 subcellular localization mode. Homologs of FLA3 in other species were identified through BLAST searches. Multiple sequence alignments of JsFLA3 and its homologs were conducted using software ClustalW[37]. The phylogenetic tree was generated using MEGA V6 software as described by Qi et al.[36].
Transcription profiling of JsFLA3
-
The fresh samples of leaves, stems, roots, petals, stamens, pistils, and sepals of 'Danbanmoli' were collected from 3-year-old plants for tissue expression analysis. The fresh samples were directly frozen with liquid nitrogen and subsequently stored at −80 °C for further usage. Total RNA isolation and cDNA synthesis were performed according to the manufacturer's instructions (Yeasen, Shanghai, China). The qRT–PCR reaction mixtures were prepared following the manuals provided by Hieff UNICON® Universal qPCR SYBR Green Master Mix (Yeasen, Shanghai, China). qRT–PCR was performed on a Roche Light Cycler 480 II Real-Time PCR System (Basel, Switzerland). The primer pair JsFLA3-QF/QR was designed using software Primer 5.0. JsEF1α was applied as the reference gene (Supplementary Table S1). Three biological and technical replicates were included for qRT-PCR analysis, respectively. The cycling procedures were performed as previously described[38]. The relative expression level was calculated by the 2−ΔΔCᴛ method[39].
Subcellular localization of JsFLA3
-
The primer pair JsFLA3 R4-F/R (Supplementary Table S1) was used to amplify the JsFLA3 ORF with a Phusion High-Fidelity PCR Kit (New England Biolabs, MA, USA). The restriction enzymes XhoI and SmaI were used to cleave both the empty pORE-R4 vector and the amplicon, and the resulting products were linked together using T4 DNA ligase (New England Biolabs). The pORE-R4-JsFLA3 fusion construct was subsequently sequenced for validation. A. tumefaciens strain GV3101, which harbored either pORE-R4 or pORE-R4-JsFLA3 was suspended in an infiltration buffer[36]. This suspension was then injected into the leaves of 6-week-old N. benthamiana according to the protocol made by Wang et al.[40]. At 72 h after injection, GFP fluorescence was detected by a confocal laser microscope (Zeiss LSM880, Germany).
A. thaliana transformation
-
The overexpression vectors, pORE-R4-JsFLA3, were transformed into the A. tumefaciens EHA105 and introduced into A. thaliana (Col-0) using the floral-dip method. The T0 seeds were screened on 1/2 MS medium containing 35 mg·L−1 kanamycin and T1 transgenic plants were confirmed using genomic PCR by the JsFLA3-F/R primer pair (Supplementary Table S1). Homozygous T3 transgenic lines were utilized for subsequent experiments. The transgene line was identified by a semi-quantitative RT-PCR assay by the JsFLA3-F/R primer pair (Supplementary Table S1).
Phenotype of JsFLA3 overexpressed plants
-
The silique phenotype and seeds in the siliques of eight-week-old wild-type (WT) and JsFLA3 overexpressed (OE) plants were photographed using a Canon EOS M50 digital camera. The phenotype of the florets was observed under a stereomicroscope (SZX10, Olympus, Japan), and the length of mature silique was measured using software Image J (version 1.8.0). All measured values represent the average of 30 replicates.
Sequencing of transcriptome libraries and RNA-Seq analysis
-
The fully extended young leaves of four-week-old WT and JsFLA3 overexpressed plants were sampled and immediately frozen in liquid nitrogen. Then total RNA was extracted and verified as previously described[34]. The RNA was saved at −80 °C for further usage. The mRNA of each library was generated using the Illumina TruSeq RNA Sample Preparation Kit (San Diego, CA, USA), and the libraries were sequenced on the Illumina HiSeq 2,000 platform. Three biological replicates were used for RNA-Seq analysis.
High-quality clean reads were gained by eliminating low-quality reads and adapters from the raw data. The RNA-seq reads were mapped to the Arabidopsis thaliana reference genome (TAIR10.dna.toplevel.fa) using software HISAT2. Fragments Per Kilobase of exon model per Million mapped fragments (FPKM) was applied to estimate the expression levels of genes. DEGs between WT and OE samples were identified using an algorithm developed by Audic and Claverie. The standard used as the thresholds for notable differences in gene expression were |log2Ratio| ≥ 1.0 and p-values < 0.05[41]. Blast2Go software was applied to annotate the GO functions of DEGs[42].
Pollen viability analysis and crosses
-
Alexander stain was used to observe pollen viability[43]. In vitro pollen germination was performed following the protocol described by Chen & McCormick[44], with subsequent counting of the pollen germination frequencies. For cross-breeding, florets were pre-emasculated and pollinated one day before anthesis. Pistils were collected at 4 h after pollination, aniline blue staining was performed as described by Miao et al.[32]. Three biological replicates were carried out for each cross. The samples were observed under a fluorescence microscope (Zeiss Axioskop40, Carl Zeiss, Jena, Germany).
Ultrastructure observation
-
Anthers were collected from freshly dehisced WT and JsFLA3 overexpressed plants for scanning electron microscopy (SEM) observation. The samples were dehydrated, dried, and photographed as described by Deng et al.[45], and the pollen abortion frequencies were counted.
Anthers at the dehisced stage were sampled from WT and JsFLA3 overexpressed plants for transmission electron microscopy (TEM) observation. The samples were fixed, embedded, sectioned, and photographed as described by Deng et al.[35].
Determination of cellulose content
-
One week after the first anthesis, the stems (between the first and the second branch), the fully extended young leaves, and the initial opening flowers were collected from the WT and OE plants. The samples were quickly frozen in liquid nitrogen and saved at −80 °C for future use. All the samples were collected in quadruplicate. The contents of cellulose were determined using the Cellulose Content Detection Kit (Solarbio, Beijing, China).
Expression pattern analysis of cellulose synthase genes
-
Total RNA was extracted from the stems, leaves, and flowers (by the method previously described) from the WT and OE plants, using the MolPure® Plant RNA Kit (Yeasen). Primer 5.0 software was used to design primers based on the sequences from the WT and OE transcriptome, and AtActin was applied as the reference gene (Supplementary Table S1). Changes in cellulose synthase genes (CesAs) expression level were detected by qRT-PCR. Three biological and technical replicates were performed for qRT-PCR analysis, respectively.
Statistical analysis
-
The data were analyzed by analysis of variance (ANOVA). The significance with means separated by Duncan's Multiple Range tests at the 5% level using SPSS version 20.0 software (SPSS Inc., Chicago, IL, USA). All results were shown as means ± standard errors (SE).
-
A JsFLA3 transcript with an 876 nt open reading frame (ORF) was isolated from J. sambac, which encoded a 291 amino acids (Supplementary Table S2). The molecular weight of the JsFLA3 protein was 30,305.94 Da, and its isoelectric point was 4.69. Its amino acid composition was typical of AGPs, rich in alanine (Ala, 13.4%), serine (Ser, 10.3%), proline (Pro, 8.2%), and threonine (Thr, 6.5%) (Supplementary Table S3). It exhibited high expression at 1 h after pollination (T1) during jasmine crossing (Fig. 1a). The JsFLA3 protein harbored a FAS domain, which had H1, [Y/F]-H and H2 three conserved domains (Fig. 1b). SignalP predicated a signal peptide cleavage site at the N-terminal between amino acids 24 and 25 (Fig. 1b). A potential GPI modification site was predicated at amino acid 267 (Ser), and three N-glycosylation sites were detected at amino acid 26, 103, and 159, respectively (Fig. 1b). The phylogenetic tree indicated that JsFLA3 was most similar to OeFLA3 (Fig. 1c).
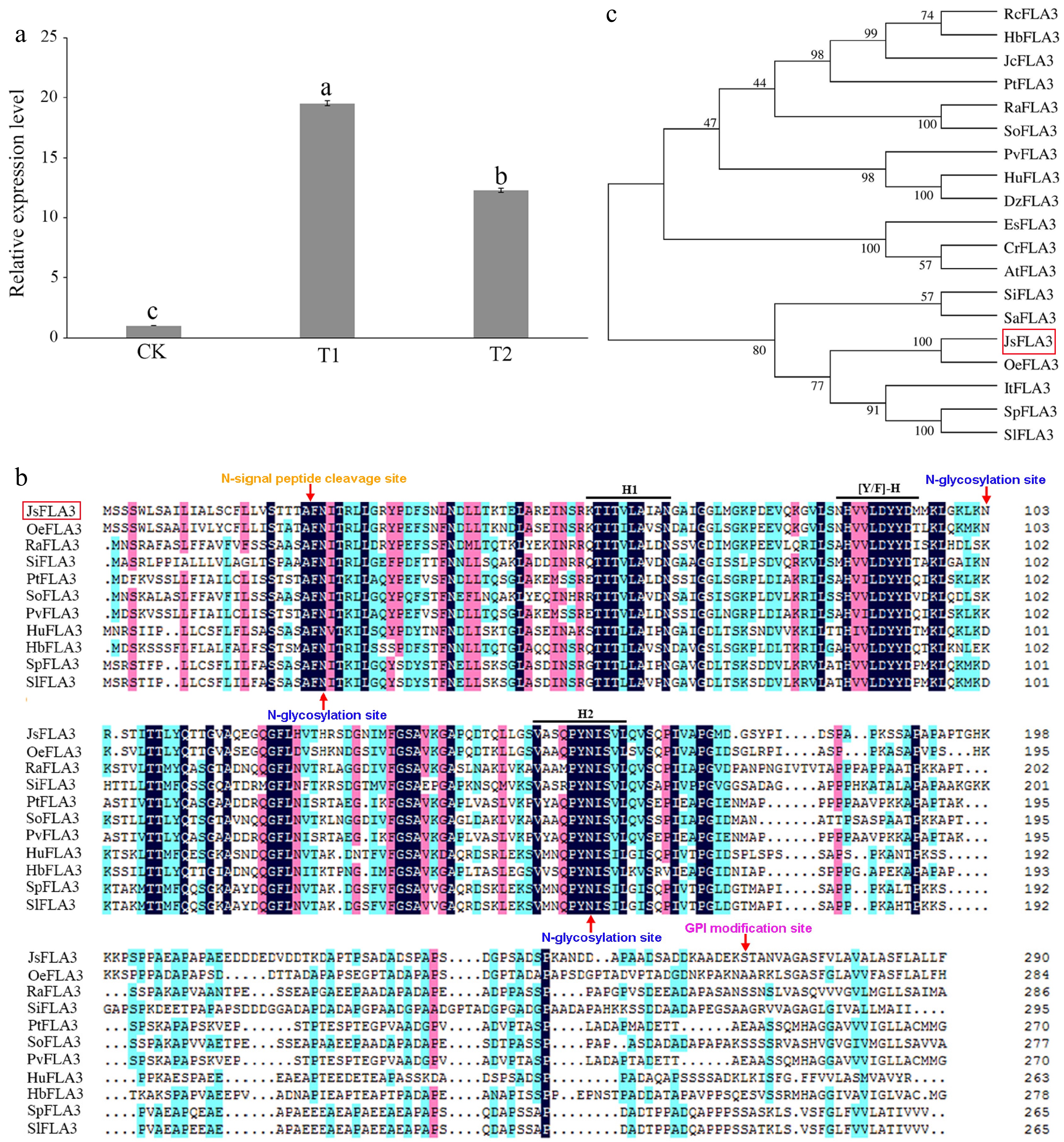
Figure 1.
Characterization of the JsFLA3. (a) Expression pattern analysis of JsFLA3 in stigma among T1, T2, and CK. CK, T1, and T2 represented stigmas at 0, 1, and 6 h after pollination, respectively. Values are shown as mean ± SE (n = 3). Columns headed by a different letter indicate significantly different JsFLA3 transcript abundances (p < 0.05). (b) Amino acid comparison between JsFLA3 and FLA3 homologs from other species. H1, [Y/F]-H, and H2 indicated three conserved domains of JsFLA3 and FLA3 homologs. (c) Phylogenetic tree of FLA3s. Oe, Olea europaea var. sylvestris; Ra, Rhodamnia argentea; Si, Setaria italic; Pt, Populus trichocarpa; So, Syzygium oleosum; Pv, Pistacia vera; Hu, Herrania umbratica; Hb, Hevea brasiliensis; Sp, Solanum pennellii; Sl, Solanum lycopersicum; Rc, Ricinus communis; Jc Jatropha curcas; Dz, Durio zibethinus; Es, Eutrema salsugineum; Cr, Capsella rubella; At, Arabidopsis thaliana; Sa, Striga asiatica; It, Ipomoea triloba. The bootstrap values were shown at the nodes. The red box indicated JsFLA3.
qRT-PCR revealed that JsFLA3 expression was specifically highly expressed in the stamen but barely detectable in the root, stem, leaf, petal, and pistil (Fig. 2a). The subcellular localization of JsFLA3 in the leaves of N. benthamiana showed that it was observed only in the plasma membrane, while empty vector pORE-R4 GFP fluorescence was distributed throughout the tobacco cells (Fig. 2b).
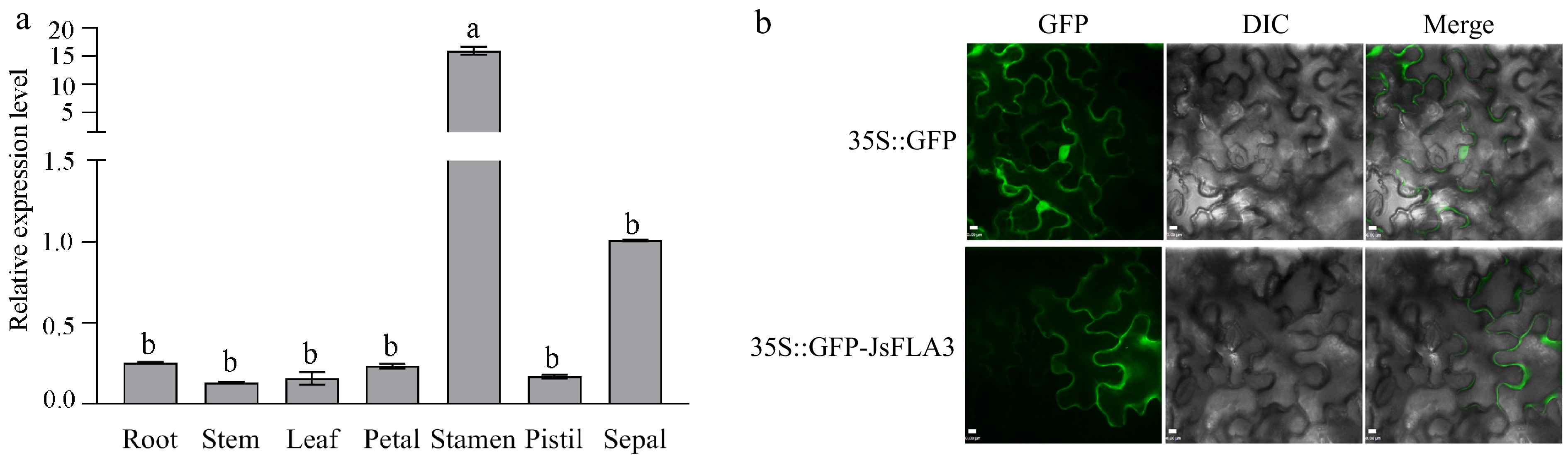
Figure 2.
The expression pattern analysis of JsFLA3. (a) Differential expression pattern analysis of JsFLA3 in various tissues of J. sambac 'Danbanmoli'. Values are shown as mean ± SE (n = 3). Columns headed by a different letter indicate significantly different JsFLA3 transcript abundances (p < 0.05). (b) Subcellular location of JsFLA3 in N. benthamiana epidermal cells. GFP, images photographed in the green fluorescence channel; DIC, images photographed in bright light channel; Merge, overlay plots. 35S::GFP, empty vector pORE-R4; 35S::GFP-JsFLA3, vector pORE-R4-JsFLA3. Bar: 50 μm.
JsFLA3 overexpressed transgenic plants caused severe sterility in A. thaliana
-
To investigate the function of JsFLA3, overexpressed (OE) transgenic plants were generated and confirmed by semi-quantitative RT-PCR. JsFLA3 transcript level was enhanced in OE lines, and three independent OE lines from the T3 progeny bred were selected for further analysis based on their transcript levels (Fig. 3a). The stigmas of OE transgenic lines were longer than the anthers during the anthesis stage (Fig. 3b). Compared to WT plants, the OE transgenic lines showed severe sterility (Fig. 3c). The small size of mature siliques of OE transgenic lines (1.16 ± 0.11 cm) was significantly shorter than WT plants (1.67 ± 0.14 cm) (Fig. 3d & f). In addition, the number of seeds per mature silique in the OE transgenic lines (21.6 ± 3.6) was less than WT plants (52.9 ± 6.0) (Fig. 3e & g).
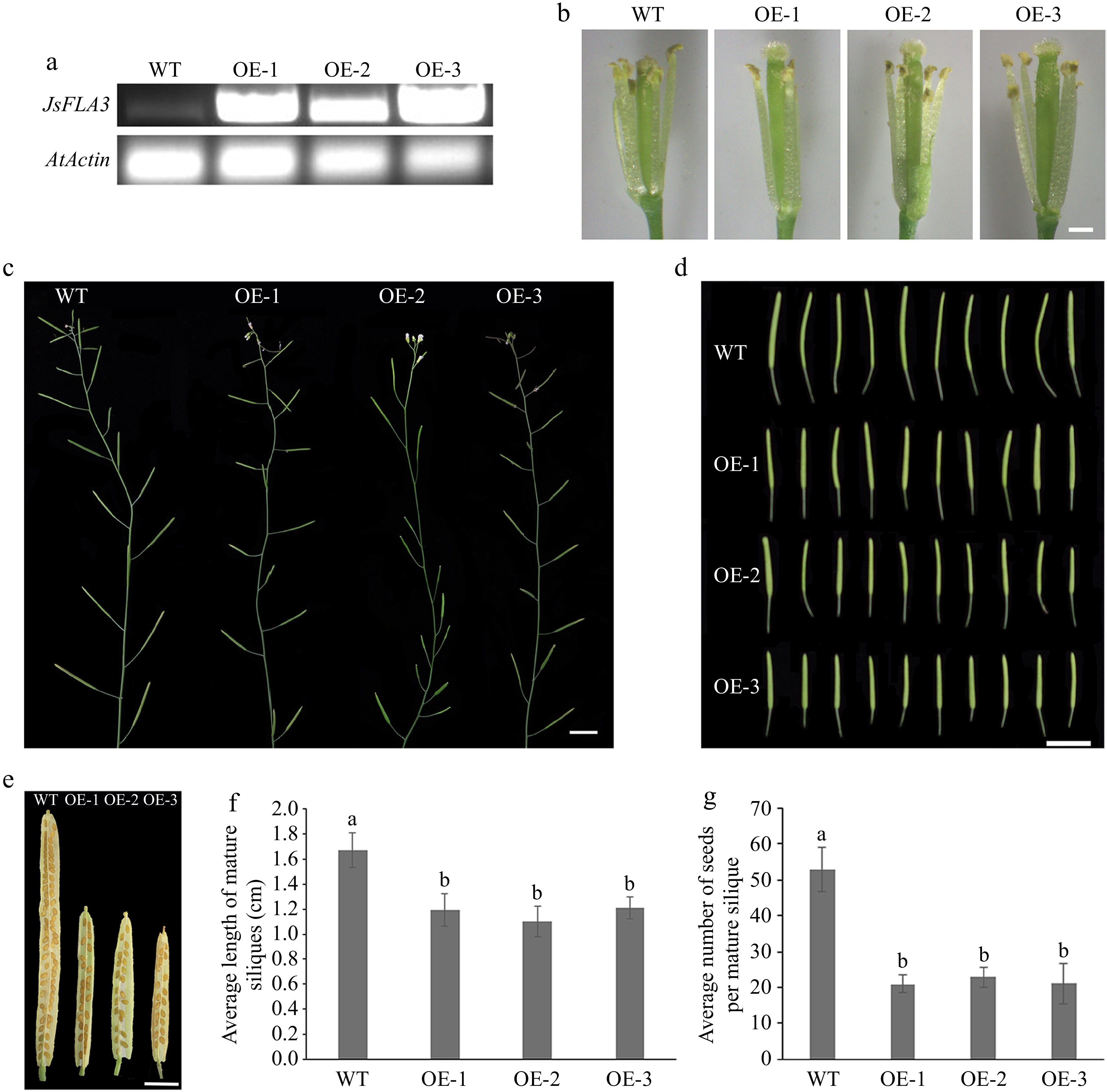
Figure 3.
Fertility analysis of ectopic expression of JsFLA3 in A. thaliana. (a) RT-PCR identification of the transgenic lines OE-1, 2, 3. (b) The appearance of stamen filament of WT and OE florets at anthesis stage. Bar: 0.5 mm. (c) The silique development of WT and OE plants. Bar: 1 cm. (d) The phenotype of the fifth silique. Bar: 1 cm. (e) The siliques of WT and OE plants. Bar: 1 mm. (f) Mature siliques length in WT and OE plants. Columns headed by a different letter indicate significantly different mature siliques length (p < 0.05). (g) Seed numbers per silique in WT and OE plants. Values are shown as mean ± SE (n = 30). Columns headed by a different letter indicate significantly different seed numbers per silique (p < 0.05). WT, wild-type A. thaliana; OE, overexpression transgenic A. thaliana.
Illumina sequencing and differentially expressed genes in JsFLA3 transgenic plants
-
To better understand the mechanisms through which JsFLA3 caused severe sterility, a large-scale screening of DEGs was performed in the WT and OE-1 transgenic line using RNA-Seq. As a result, 24,672 genes were obtained from the six libraries (WT-1, 2, 3, and OE-1, 2, 3) (Supplementary Table S4). A total of 23,539 genes were co-expressed in WT and OE, while 551 and 582 genes were expressed specifically in WT and OE, respectively (Supplementary Fig. S1a). A total of 87 DEGs were identified, including 66 up- and 21 down-regulated genes (Supplementary Fig. S1b & Supplementary Table S5). It was found that the transcription of AtFLA3 did not change. Functionally enriched these DEGs in the GO database. The result showed that 68, 68, and 71 DEGs were divided into molecular function (MF), cellular component (CC), and biological process (BP), respectively (Supplementary Fig. S2 & Supplementary Table S6). The most enriched terms of molecular function were oxidoreductase activity, acting on peroxide as an acceptor, and peroxidase activity, whereas the extracellular region-related genes were dominant in the cellular component category. The DEGs in the biological process category were enriched in detoxification, and cellular oxidant detoxification.
Some DEGs were identified to be involved in reproductive development and cell wall formation processes (Supplementary Table S7). For example, AT5G55020 (ATMYB120), AT3G50310 (MAPKKK20), AT3G18000 (NMT1), AT5G35770 (SAP), and AT4G12920 (UND) were involved in pollen sperm cell differentiation, gametophyte development, male gamete generation, pollen development, pollination, pollen tube growth, and development (Supplementary Table S7). AT5G57550 (XTH25) and AT3G30720 (QQS) participated in plant-type cell wall biogenesis, the cell wall polysaccharide metabolic process, cell wall organization, the hemicellulose metabolic process, and the polysaccharide metabolic process (Supplementary Table S7). AT5G57520 (ZFP2) and AT5G65080 (MAF5) were involved in the regulation of post-embryonic development (Supplementary Table S7).
Abnormal pollen wall development in JsFLA3 transgenic Arabidopsis plants
-
Observations from Alexander staining revealed that OE transgenic plant pollen grains displayed abnormal pollen viability (Fig. 4a). Pollen germination experiment showed that the frequency of pollen germination in OE plants (17.9%) was significantly lower than in WT plants (35.8%) (Figs 4b & 5n). Aniline blue staining experiment revealed that when WT pollens were used as the male donor, the number of pollen tubes that grew on WT or OE pistils was significantly higher than the OE pollen tubes on WT or OE pistils (Fig. 4c). In comparison to WT, the pollen morphology of OE lines was abnormal, with a large number of shrunken and wrinkled pollen grains appeared (Fig. 5a−l). OE lines displayed severe sterility with approximately 69% pollen abortion frequency, while only 4.6% was detected in WT plants (Fig. 5m). Furthermore, the ultrastructure of pollen wall structure showed that the intine layer of OE abnormal pollen disappeared, while the structure of exine was almost normal compared to WT pollen (Fig. 6). Therefore, it was suggested from the results that the sterility in OE lines was due to the abnormalities in the pollen wall.
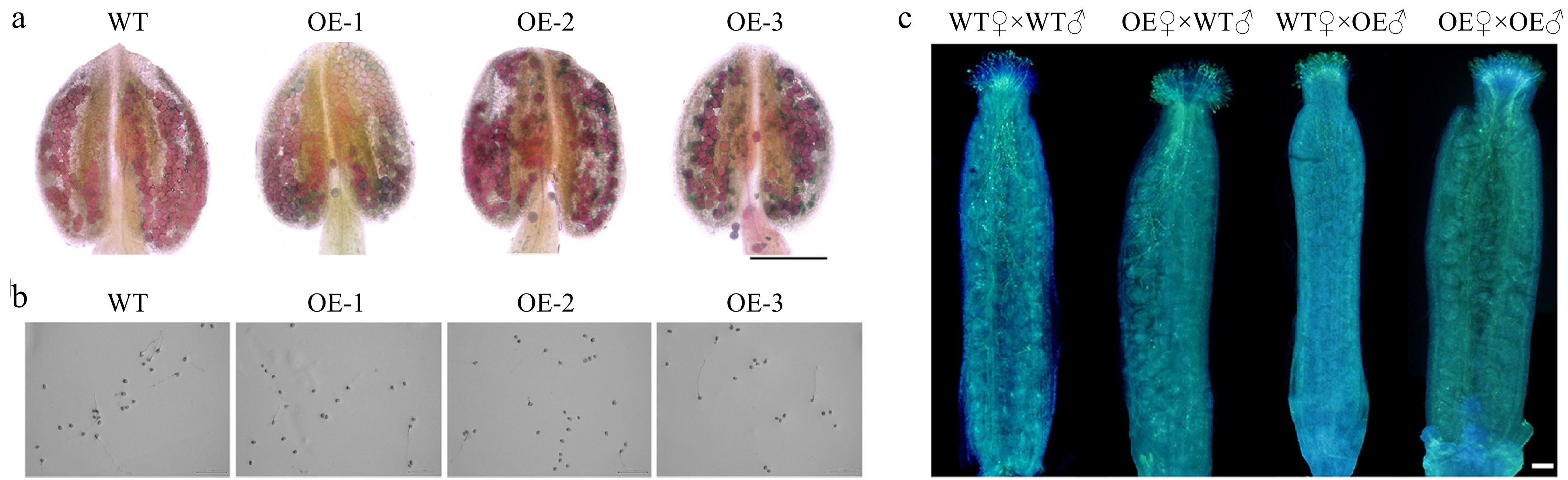
Figure 4.
Analysis of mature pollen in wild-type (WT) and overexpression (OE) transgenic A. thaliana. (a) Alexander staining of pollen grains in WT and OE plants. Bar: 200 μm. (b) In vitro germination of WT and OE pollen grains for 4 h. Bar: 200 μm. (c) Reciprocal crosses between WT and OE plants of A. thaliana, showing the pollen tube growth at 4 h after pollination. WT♀ × WT♂, WT plants were used as both the female receptors and male donors; OE♀ × WT♂, OE and WT plants were used as the female receptors and male donors, respectively; WT♀ × OE♂, WT and OE plants were used as the female receptors and male donors, respectively; OE♀ × OE♂, OE plants were used as both the female receptors and male donors. Bar: 100 μm.
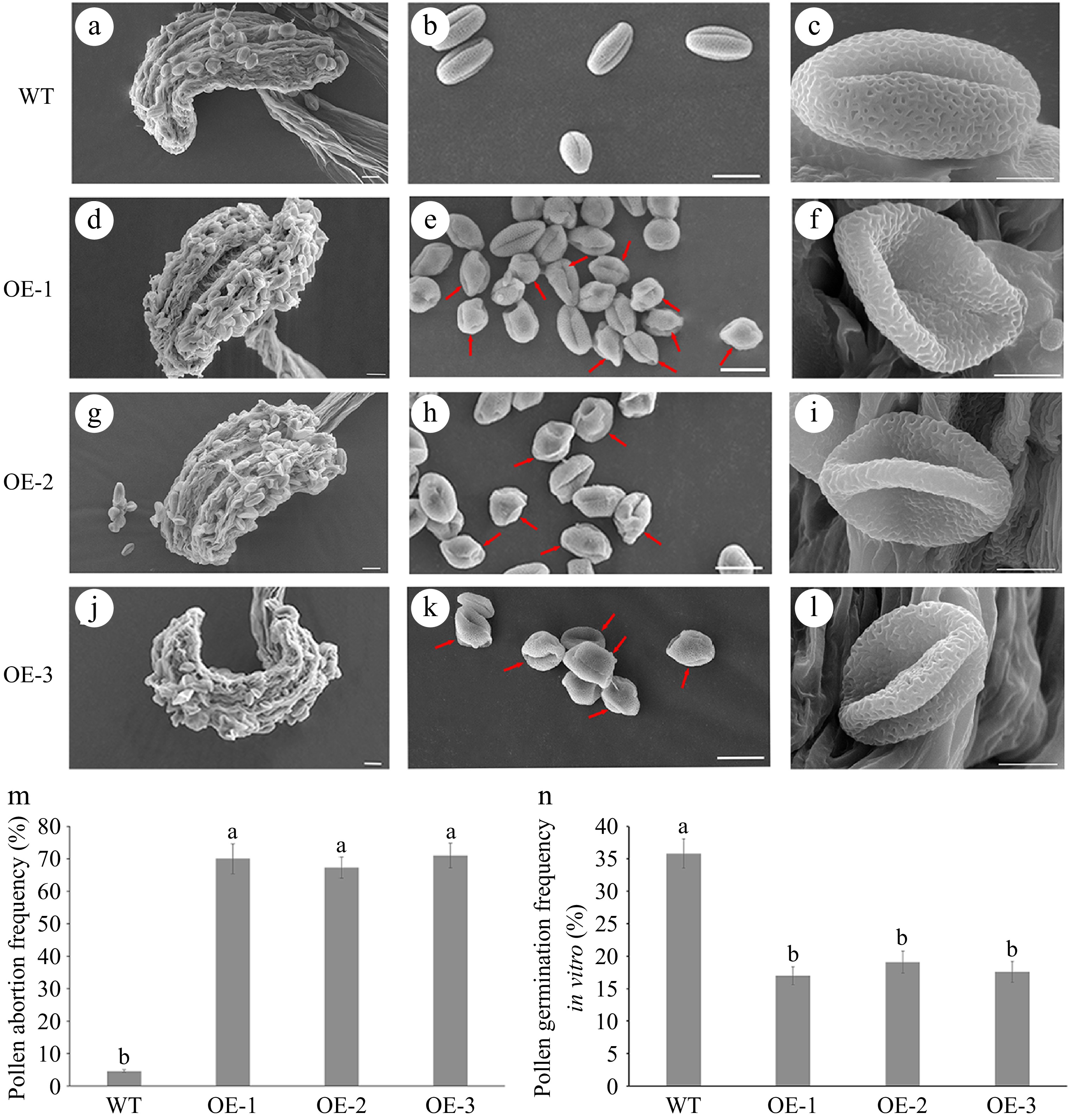
Figure 5.
Observation under scanning electron microscope (SEM) of mature pollen in wild-type (WT) and overexpressed JsFLA3 (OE) A. thaliana. (a), (d), (g), (j) SEM of anthers from WT and OE plants. Bar: 20 μm. (b), (e), (h), (k) SEM of mature pollen in WT and OE plants. Bar: 20 μm. (c), (f), (i), (l) Magnified pictures of b, e, h, k. Bar: 5 μm. (m) The frequency of pollen abortion in WT and OE plants. Columns headed by a different letter indicate significantly different pollen abortion frequency (p < 0.05). (n) The frequency of pollen germination of WT and OE plants in vitro. Values are shown as mean ± SE (n = 1,000). Columns headed by a different letter indicate significantly different pollen germination frequency (p < 0.05).
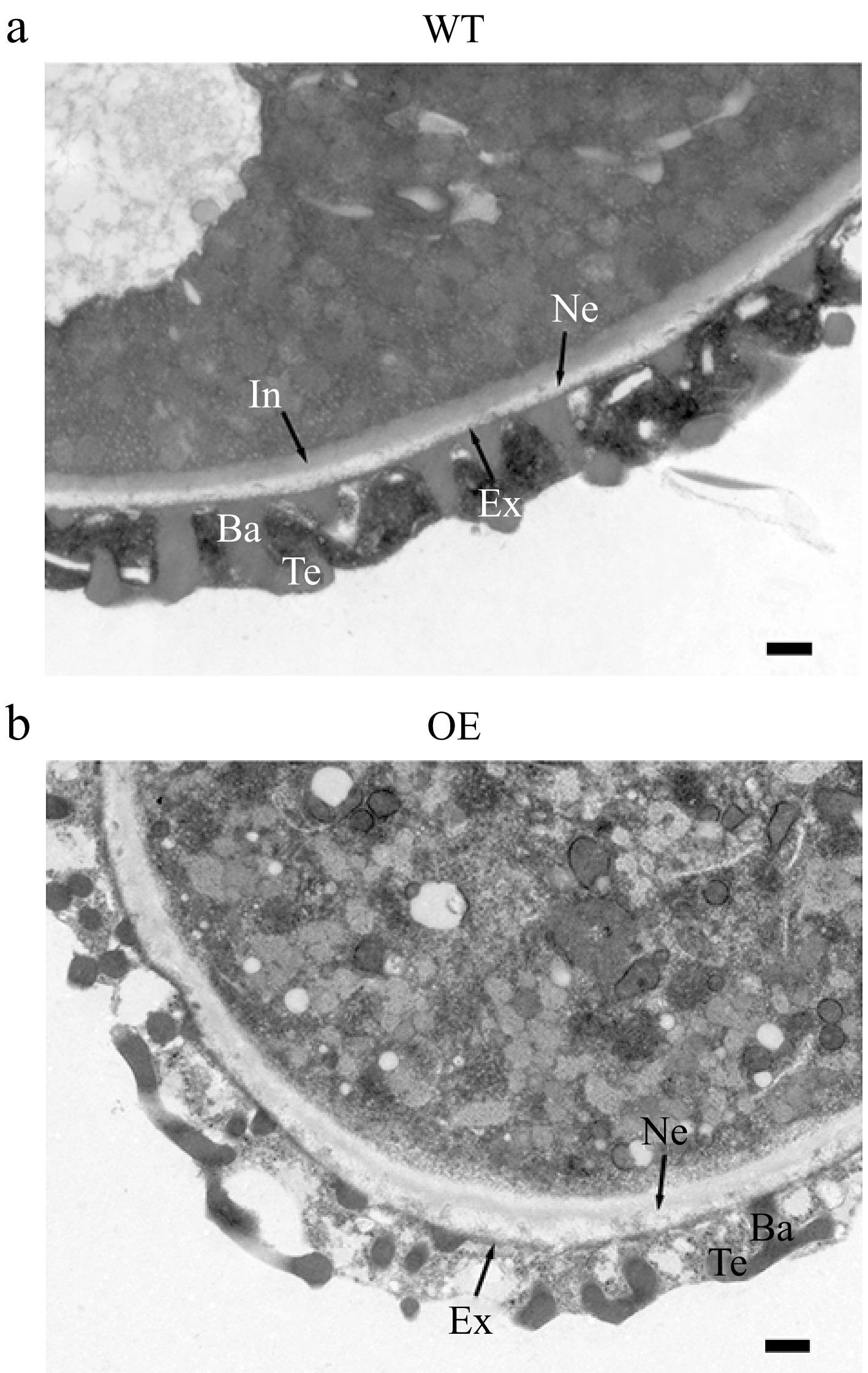
Figure 6.
Observation under transmission electron microscope of mature pollen cell wall of wild-type (WT) and overexpressed JsFLA3 (OE) A. thaliana. (a) WT. (b) OE. Ba, baculum; Ex, exine; Ne, nexine; Te, tectum; In, intine. Bar: 500 nm.
Overexpression of JsFLA3 changed cellulose contents and the expression of cellulose synthase genes in each tissue
-
The cellulose content was significantly higher in each tissue (stem, leaf, and flower) of WT compared to OE (Fig. 7). The cellulose content in OE flowers was 43.8% lower than that in WT flowers (Fig. 7). RNA-Seq data indicated that eight AtCesAs were down-regulated in OE lines (Supplementary Fig. S3). Compared to the WT, down-regulation of AtCesA1, AtCesA2, AtCesA3, AtCesA6, and AtCesA9 was observed in the OE lines (Fig. 8). Particularly in the OE flower, and almost 2-fold lower expression of AtCesA2 and AtCesA9 was observed (Fig. 8c).
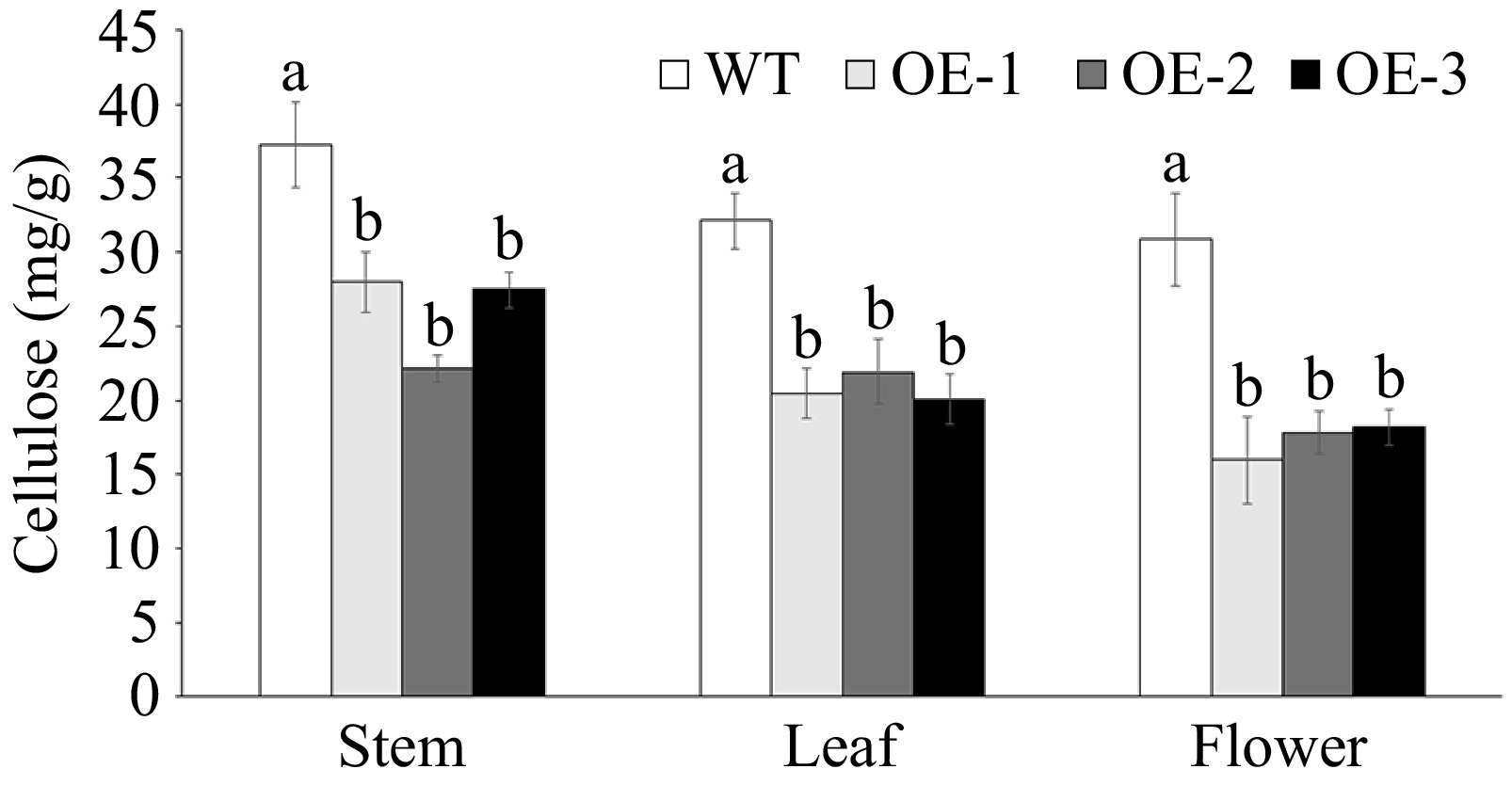
Figure 7.
Cellulose content assay in various tissues of wild-type (WT) and overexpressed JsFLA3 (OE) A. thaliana. Values are shown as mean ± SE (n = 4). Columns headed by a different letter indicate significantly different cellulose content (p < 0.05).
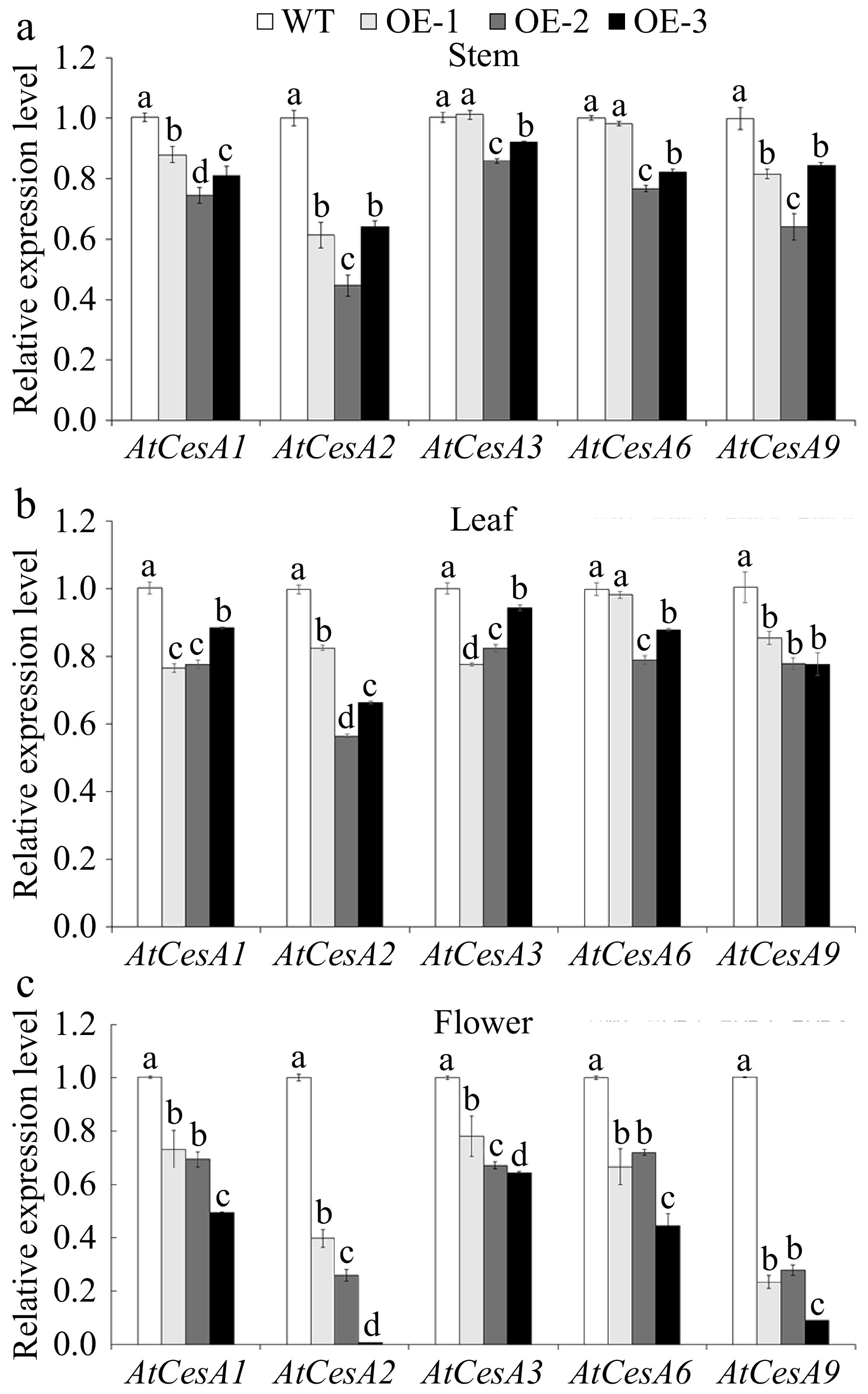
Figure 8.
The relative expression level of AtCesAs in various tissues of wild-type (WT) and overexpressed JsFLA3 (OE) Arabidopsis. (a) Stem. (b) Leaf. (c) Flower. Values are shown as mean ± SE (n =3). Columns headed by a different letter indicate significantly different AtCesAs transcript abundances (p < 0.05).
-
FLAs are hyperglycosylated proteins that participate in male and female gametophyte development, pollen tube growth and other processes[32,46,47]. The protein core accounts for approximately 10% of the entire AGP molecule weight and is composed of hydroxyproline, alanine, proline, threonine, and serine[17]. In this study, JsFLA3 was cloned from J. sambac 'Danbanmoli', and the JsFLA3 protein was rich in alanine, serine, proline, and threonine amino acids (Supplementary Table S3). This suggests that JsFLA3 is a characteristic AGP. The AGP molecule is equipped with a C-terminal GPI anchor, which allows it to be attached to the cell membrane[17]. GPI anchor affected the membrane location of AtFLA3[31]. In tomato, overexpression of AGP-1 reduced fruit yield, but the effect on seed development and fruit production was minimal when overexpressing the AGP-1ΔC allele without the GPI anchor domain[48]. In this study, the results showed that JsFLA3 had a GPI anchor at the C-terminus and located at the plasma membrane (Figs 1b & 2b), indirectly demonstrating the importance of the GPI anchor for membrane location of FLAs.
Previous studies have shown that some FLAs are abundant in pollen and play significant roles in pollen development, such as AtFLA3[31], AtFLA14[32], OsFLA1[46], and OsDEAP1[49]. In rice, deap1 mutant plants generated abnormal pollen grains with defective extine formation and delayed tapetum degradation[49]. The heterologous expression of BcMF18 in A. thaliana resulted in male fertility and the development of shorter siliques with a low seed set. This was observed because 46% of OE pollen grains were found to be aborted[50]. In the present study, JsFLA3 OE plants had short mature siliques with partial sterility (Fig. 3c−g). At the same time, the pollen viability and germination were much lower than WT plants (Figs 4a, b & 5n). In comparison with WT, a significant number of pollen grains in JsFLA3 OE plants exhibited abnormal morphology, characterized by shrunken and wrinkled appearance, and a higher frequency of pollen abortion (69.4%) was detected in JsFLA3 OE plants (Fig. 5a−m). The results were consistent with previous investigations, indicating that pollen abortion is the underlying cause of reduced male fertility and shorter siliques in the OE plants.
The formation of the intine, which is a component of pollen, is regulated by male gametophytic factors[11]. Recently, several members of FLAs have been identified to be involved in pollen intine formation. For instance, the overexpression of AtFLA14 resulted in the production of pollen grains with either a thickened intine or collapsed intine[32]. In Brassica campestris, putative AGP-encoding genes BcMF8 and BcMF18 were found to participate in pollen wall development. A disruption in BcMF8 led to the formation of abnormal pollen grains with aberrant intine development, while BcMF18 reduction resulted in deformed pollen grains lacking intine[51,52]. In addition, heterologous expression of BcMF18 in A. thaliana resulted in pollen abortion due to the abnormal and degraded intine layer[50]. In the present study, compared with WT pollen (Fig. 6a), the intine layer of OE pollen grains displayed abnormalities and ultimately degraded (Fig. 6b), which was consistent with previous studies. These results suggested that JsFLA3 plays a vital role in pollen intine formation.
Cellulose is a linear β-1,4-linked glucose polymer and constitutes a fundamental component of the plant cell wall, synthesized by CesA enzyme located in the plasma membrane in land plants[53]. The CesAs have been shown to play a significant role in cell wall formation. For example, the A. thaliana genome contains 10 CesA genes, and CesA4, CesA7, and CesA8 are essential for the deposition of secondary cell wall cellulose[54]. Mutations in AtCesA1 or AtCesA3 caused decreased cellulose crystallinity and pollen production[55]. Furthermore, CesA1, CesA2, and CesA3 have been proved to be key players in the formation of the secondary cell wall in loblolly pine and white spruce[56,57]. In Populus trichocarpa, the CesA4, CesA7, and CesA8 RNAi knockdown transgenic plants showed a remarkable reduction in cellulose content[58]. The loss of both AtCesA2 and AtCesA6 caused a decrease in cellulose content, and the cesa2 cesa6 cesa9 triple-mutant plants resulted in an abnormal inner wall[55]. In this study, the content of cellulose in the flower was significantly lower in OE than in WT (Fig. 7). In addition, both RNA-Seq data and qRT-PCR experiments showed that almost all AtCesAs were significantly down-regulated in OE plants (Fig. 8 & Supplementary Fig. S3). These results indicated that the reduced expression of CesAs genes caused a decrease in cellulose content, subsequently leading to aberrant intine formation and ultimate pollen abortion.
Several members of FLAs have been identified to play a significant role in cell wall cellulose deposition. For instance, GPI-anchored AGPs were secreted together with cellulose synthase to the cell surface, and they may be divorced from the GPI anchor and fused into the cell wall when AGPs bound to cellulose[59]. Both AtFLA11 and AtFLA12 influenced the deposition of cellulose and the cell wall matrix integrity[28]. BcMF18 reduction led to deformed pollen grains lacking intine because of abnormal cellulose distribution[51]. The unchanged expression level of genes associated with intine formation, like FLA3, USP, TEK, RGP1, and RGP2, and the genes related to cellulose synthesis, such as GALT2, FLA11, FLA12, SOS5, and FEI2 were observed in BcMF18 OE plants, indicating that BcMF18 might not influence the cellulose synthesis but be involved in the process of cellulose deposition[50]. Besides the above function, some FLAs have been reported to influence the cellulose synthesis. For example, compared with WT plants, the content of cellulose in the stem was decreased in the fla11/fla12 single and double mutants[28]. In cotton, GbFLA5 improved fiber strength by influencing the direction of the microfibril deposition and cellulose synthesis[60]. In P. alba, OreFLA11 altered cell wall composition by decreasing lignin content and increasing cellulose content[61]. Consequently, the down-regulation of CesA genes in OE plants suggested that JsFLA3 impaired intine formation by influencing cellulose synthesis. It is speculated that JsFLA3 interacts with CesA genes, thereby reducing CesA content. However, further investigation is needed to determine whether there is indeed an interaction between JsFLA3 and CesA genes. Since jasmine lacks both the VIGS system and a genetic transformation system, and only demonstrates the heterologous expression of JsFLA3 in Arabidopsis, this may differ from the actual situation in jasmine. Though the study did not fully elucidate the detailed molecular mechanism of JsFLA3 in regulating cellulose synthesis, it provides a reference for subsequent research.
-
In this study, fasciclin-like arabinogalactan protein 3 (JsFLA3) was cloned and characterized in jasmine. The heterologous expression of JsFLA3 in Arabidopsis resulted in male sterility. The results strongly suggest that JsFLA3 may play a significant role in cellulose biosynthesis, thereby affecting the pollen wall development. These results provide valuable insights into the role of FLAs in plant pollen development.
This work was financially supported by the National Natural Science Foundation of China (Grant Nos 32372748 and 31772338). We thank Chunmei Wang and Tong Wang from the Central Laboratory in Jiangsu Academy of Agricultural Sciences for their technical support on confocal laser scanning microscopy.
-
The authors confirm contribution to the paper as follows: conceptualization: Qi X, Deng Y; data curation: Qi X, Li X; formal analysis: Wang H, Chen H; funding acquisition: Deng Y; investigation: Qi X, Wang H, Li X; methodology: Qi X, Wang H; project administration: Deng Y; resources: Chen S; software: Wang H; supervision: Deng Y; validation: Ahmad MZ, Feng J; visualization: Feng J; writing – original draft: Qi X; writing – review & editing: Ahmad MZ, Deng Y. All authors reviewed the results and approved the final version of the manuscript.
-
The raw data for the transcriptome sequencing reads of A. thaliana in this research can be accessed on the NCBI under the BioProject accession number PRJNA1018719, along with the BioSample accession numbers SAMN37449609 and SAMN37449610.
-
The authors declare that they have no conflict of interest.
- Supplementary Table S1 The primer sequences of this study.
- Supplementary Table S2 The gene sequence of JsFLA3.
- Supplementary Table S3 The amino acid composition of jasmine JsFLA3.
- Supplementary Table S4 The expression of all genes in wild-type (WT) and overexpressed JsFLA3 (OE) Arabidopsis.
- Supplementary Table S5 The expression of DEGs in WT-VS-OE.
- Supplementary Table S6 The GO enrichment analysis of DEGs in WT-VS-OE.
- Supplementary Table S7 DEGs involved in reproductive development and cell wall formation processes.
- Supplementary Fig. S1 The number of genes and DEGs were detected in wild-type (WT) and overexpressed JsFLA3 (OE) A. thaliana. (a) The number of genes. (b) The number of DEGs.
- Supplementary Fig. S2 GO functional classification of DEGs. DEGs were identified in three categories: molecular function (MF, green), cellular component (CC, red) and biological process (BP, blue).
- Supplementary Fig. S3 Expression pattern analysis of AtCESAs between wild-type (WT) and overexpressed JsFLA3 (OE) Arabidopsis using FPKM data.
- Copyright: © 2025 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Qi X, Wang H, Li X, Ahmad MZ, Chen S, et al. 2025. The fasciclin-like arabinogalactan protein FLA3 of Jasminum sambac causes defects in pollen wall development. Ornamental Plant Research 5: e005 doi: 10.48130/opr-0025-0001
The fasciclin-like arabinogalactan protein FLA3 of Jasminum sambac causes defects in pollen wall development
- Received: 28 August 2024
- Revised: 18 December 2024
- Accepted: 24 December 2024
- Published online: 20 February 2025
Abstract: Jasmine [Jasminum sambac (L.) Aiton, Oleaceae] has serious reproductive barriers, preventing the generation of hybrids through crosses. Previous studies have identified several key genes involved in the interaction between pollen and pistil following jasmine pollination, including fasciclin-like arabinogalactan protein 3 (JsFLA3). In this study, the FLA3 was cloned and characterized. The results showed that JsFLA3 belongs to the arabinogalactan proteins (AGPs) family, which was specifically abundant in the stamen. JsFLA3 was found to be localized to the plasma membrane. Overexpression of JsFLA3 in Arabidopsis thaliana resulted in 64.9% of abnormal pollen grains displaying a wrinkled and shrunken appearance. Simultaneously, the intine of abnormal pollen disappeared due to degradation observed by transmission electron microscopy. In addition, transcriptome analysis revealed that some differentially expressed genes (DEGs) were associated with reproductive development and cell wall formation processes. Moreover, the cellulose content decreased in overexpressed plants, particularly in the flowers. RNA-Seq and qRT-PCR results revealed the down-regulation of the transcription levels of AtCesAs in transgenic A. thaliana. Taken together, these findings suggested that JsFLA3 plays a significant role in cellulose biosynthesis, thereby affecting the development of the pollen wall. This study provides valuable insights into the role of FLAs in plant pollen development.













