-
In addition to removing carbon and nutrients, biological wastewater treatment plants (WWTPs) are essential for biodegrading organic materials, turning harmful substances into harmless byproducts, and biodegrading dangerous chemicals. As a result, they are crucial for applications in biotechnology and essential for promoting sustainable development. Therefore, it is essential to have a better grasp of how to keep wastewater treatment plants (WWTPs) functioning steadily despite the dynamic environmental conditions caused by continuously shifting wastewater influent[1,2]. The majority of nutrient removal occurs in activated sludge (AS) microbial communities, and the diversity, shape, and composition of these communities dictate the efficiency of wastewater treatment plants (WWTPs). The fundamental mechanics, nevertheless, are still viewed as a mystery. Understanding the processes by which microbial communities are assembled offers a viable approach for identifying the critical factors affecting the operation of WWTPs[3]. Microbial assembly is thought to be simultaneously impacted by stochastic processes such as dispersal limitation, birth-death events, or drift as well as predictable processes such as selection enforced by environmental filtration or biotic interactions[2]. The relative effects of deterministic and stochastic processes on the emergence of microbial communities have been the subject of intense debate to date in both naturally occurring and artificial ecosystems. For instance, researchers have indicated that stochastic mechanisms, such as random immigration, are the main drivers, while others have indicated that predictable factors have a greater impact on community assembly in WWTPs. The degree to which stochastic and deterministic mechanisms have an impact on the AS microbial community under various environmental circumstances in WWTP studies may be the cause of this discrepancy[4]. Numerous elements, such as influent characteristics, operational parameters, process design, and ambient conditions have an impact on the microbial communities in WWTPs. It has been established that influence features have the most significant role in determining the structure and makeup of microbial communities[5]. Furthermore, because each had unique influent components and organic loading, which led to varied bacterial development and metabolism, the microbial communities from industrial and municipal WWTPs were dissimilar[6]. Biodegradability can be used to reflect the level of available nutrients and the toxicity of influent because not all organic materials can be broken down by microorganisms through biodegradation, and it is much easier to quantify than wastewater composition. A variety of techniques have been developed to gauge the biodegradability of wastewater, but one of the most widely used surrogates is the proportion of biological oxygen demand (BOD) to chemical oxygen demand (COD)[5,7]. Living organisms can breakdown or change harmful organic pollutants into less toxic substances through the bioremediation process[4]. Since the majority of bioremediation research has been mostly on the use of bacteria, bioremediation utilizing fungi has received little attention over the previous two decades. Nevertheless, fungi have recently attracted much attention due to their potential for bioremediation, which is attributed to the enzymes they generate. The ability of fungal hyphae to penetrate polluted soil and reach pollutants is another advantage that fungi have over bacteria[8].
-
The inability of traditional wastewater treatment to successfully remove some pharmaceutically active chemicals (PhACs) is a result of their recalcitrance. As a result, their presence in surface water and potential environmental effects have sparked alarm worldwide. The use of white-rot fungi (WRF) and their oxido reductase enzymes has been suggested as a low-cost and environmentally benign method for the biological transformation of these toxins for water treatment. The ability of a fungal culture to remove PhACs depends on several variables, including the species of the fungus, the enzymes produced, the molecular makeup of the target compounds, the content of the culture media, etc. Numerous researchers have worked over the past 20 years to understand the removal mechanisms and how crucial operational parameters such as temperature and pH affect the enzymatic treatment of PhACs. In short, increased hydrophobicity and the presence of electron-donating groups such as amines and hydroxyls in the chemical structure make PhACs more effectively degraded by fungi[8]. Redox mediators such as syringe aldehyde are used to break down resistant chemicals, which increase the efficiency of the process but run the risk of harmful effects on the effluent and enzyme deactivation. The stability and reusability of enzymes can be improved by immobilizing the enzyme on supports. To reduce costs and allow for regeneration, the immobilization technique should be carefully chosen. Nevertheless, more research is required to understand the mechanisms underlying total phosphorus (TP) toxicity levels and enzymatic degradation, as well as to improve the entire treatment approach for both economic and technical reasons[8]. One of the most important factors that controls sorption onto biomass and can facilitate the removal of particular chemicals is the hydrophobicity of PhACs, which can be quantified as log kow. Researchers have investigated how extracellular enzyme biodegradation and biosorption affect the clearance of hydrophobic substances (log kow > 4) and found that both processes have a significant impact. Additionally, they discovered that the biosorption of hydrophobic chemicals promoted their biodegradation. However, because biosorption plays a limited role in the removal of hydrophilic chemicals (log kow < 3), it has been noted that for some compounds, its impact is insignificant compared to biodegradation. There were notable variations in the effects of treatment with whole-cell WRF and extracted enzymes because whole-cell fungal treatment involves extracellular, intracellular, and mycelium-bound enzymes. The complete elimination of some substances in whole-cell fungal reactors highlights the significant function of intracellular and mycelium-bound enzymes as well as their beneficial interaction with external enzymes[9]. The catalytic core of each monomer of laccase, as shown in Fig. 1a, contains four copper atoms that are separated into three types (1, 2, and 3). The type 1 atom (T1) catalyzes the oxidation of the substrate and gives the enzyme its color. The supplied electron from the substrate is then internally transported from T1 to the copper sites at T2, and T3, where the reduction of oxygen to water occurs. A radical is produced by one-electron oxidation at site T1 and two molecules of water are produced by four-electron reduction at sites T2, and T3. The first free radical is somewhat unstable and can be converted to a quinone either spontaneously or through an additional enzyme-catalyzed step. Additional nonenzymatic radical reactions are also possible for phenolic polymers, including humic acids, which may lead to their partial breakdown[10]. Because lignin peroxidase (LiP) has a high redox potential, it can oxidize substances that other enzymes are unable to. It can attack both phenolic and nonphenolic structures, causing reactions such as demethoxylation, hydroxylation, carbon‒carbon cleavage, aromatic ring fission, phenolic oxidation, methylation, and dimerization. LiP uses a peroxidase catalytic mechanism in which hydrogen peroxide oxidizes the native enzyme to produce LiP-I with two electron deficiencies. LiP-I is reduced to one electron-deficient LiP-II after oxidation of the target compound. LiP-II transforms back into the original LiP when it oxidizes a different target molecule. Poor-molecular-weight redox mediators play a significant role due to their low mobility and accessibility of enzyme active sites for target molecules[11].
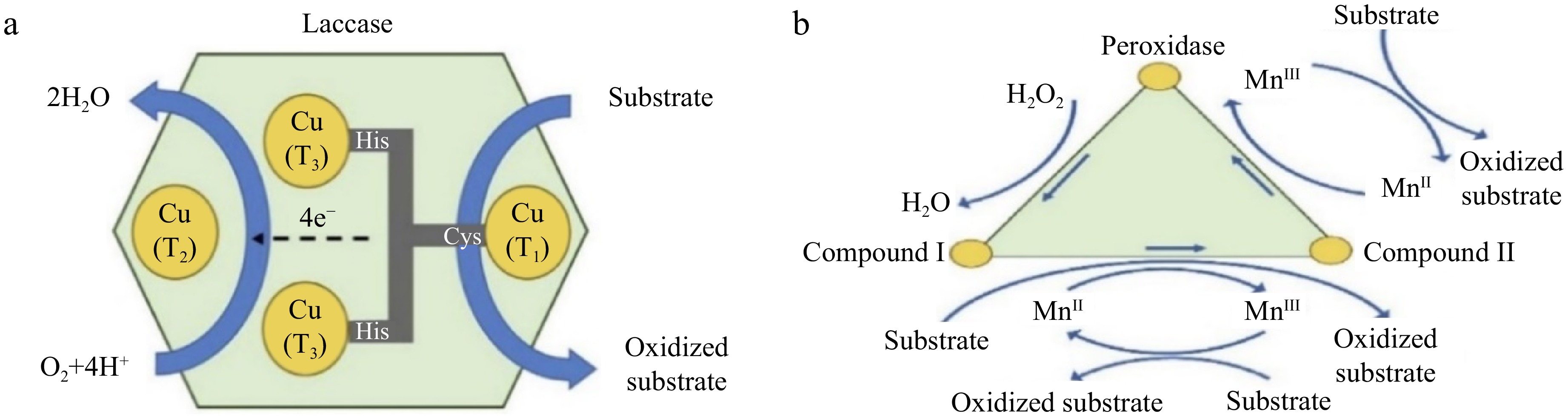
Figure 1.
Oxidation mechanism of compounds: (a) using laccase enzyme, and (b) using peroxidase enzyme[8].
As shown in Fig. 1b, manganese peroxidase (MnP) serves as a redox mediator to attack organic molecules and oxidize them through the abstraction of one hydrogen and one electron, converting Mn2+ to Mn3+, which can be stabilized by chelators such as organic acids. Like LiP, MnP initially combines with hydrogen peroxide to produce MnP-I, which has two more electrons than MnP. Target molecules are oxidized by MnP-I, which then transforms into MnP-II, which is gradually reduced to native MnP and requires Mn2+ to complete the catalytic cycle. All four extracellular enzymes cannot be produced by a single fungal species, and different WRF species have different combinations of ligninolytic enzymes. Even the profile of enzymes secreted varies among WRF species. Additionally, the content of nutrients, such as carbon and nitrogen, as well as the characteristics of the growth medium, such as temperature and pH can affect the release of enzymes. In addition to the above-described enzymes, cytochrome P450 (CYP450), an internal enzyme system in WRF, was shown to play a significant role in the breakdown of certain contaminants. Therefore, there are three methods for using these fungi to remove PhACs: (i) whole-cell culture, (ii) crude culture extract or pure enzyme, and (iii) immobilized enzymes[12]. WRF is effective at removing a variety of organic compounds from fungal species that are resistant to bacterial degradation. Specifically, the intracellular system and extracellular ligninolytic enzymes, i.e., MnP, laccase (Lac), and LiP. In comparison to the use of a single enzyme, whole-cell fungal therapy can remove a larger spectrum of PhACs, including antibiotics, anti-inflammatories, and antiepileptics. This is due to the combined effects of intracellular/extracellular enzymes and the sorption of PhACs on the biomass. WRF are appealing for use in the removal of PhACs due to several characteristics, including (1) the nonspecificity of their produced enzyme, which enables the degradation of a wide range of micropollutants, and (2) quick colonization through hyphal growth, which allows WRF to access pollutants. It is important to highlight that the degradation of persistent pollutants by WRF is a cometabolic process, meaning that it occurs in the presence of a substrate that is easily degradable. The downside of the requirement of cosubstrate addition is that glucose typically increases the cost but also increases the degradation efficiency. An example of the fungal method for pollutant removal is shown in Fig. 2. As a result, the target pollutants may be adsorbed onto the fungus or into cells, where they will then be broken down by extracellular, and intracellular enzymes[13,14].
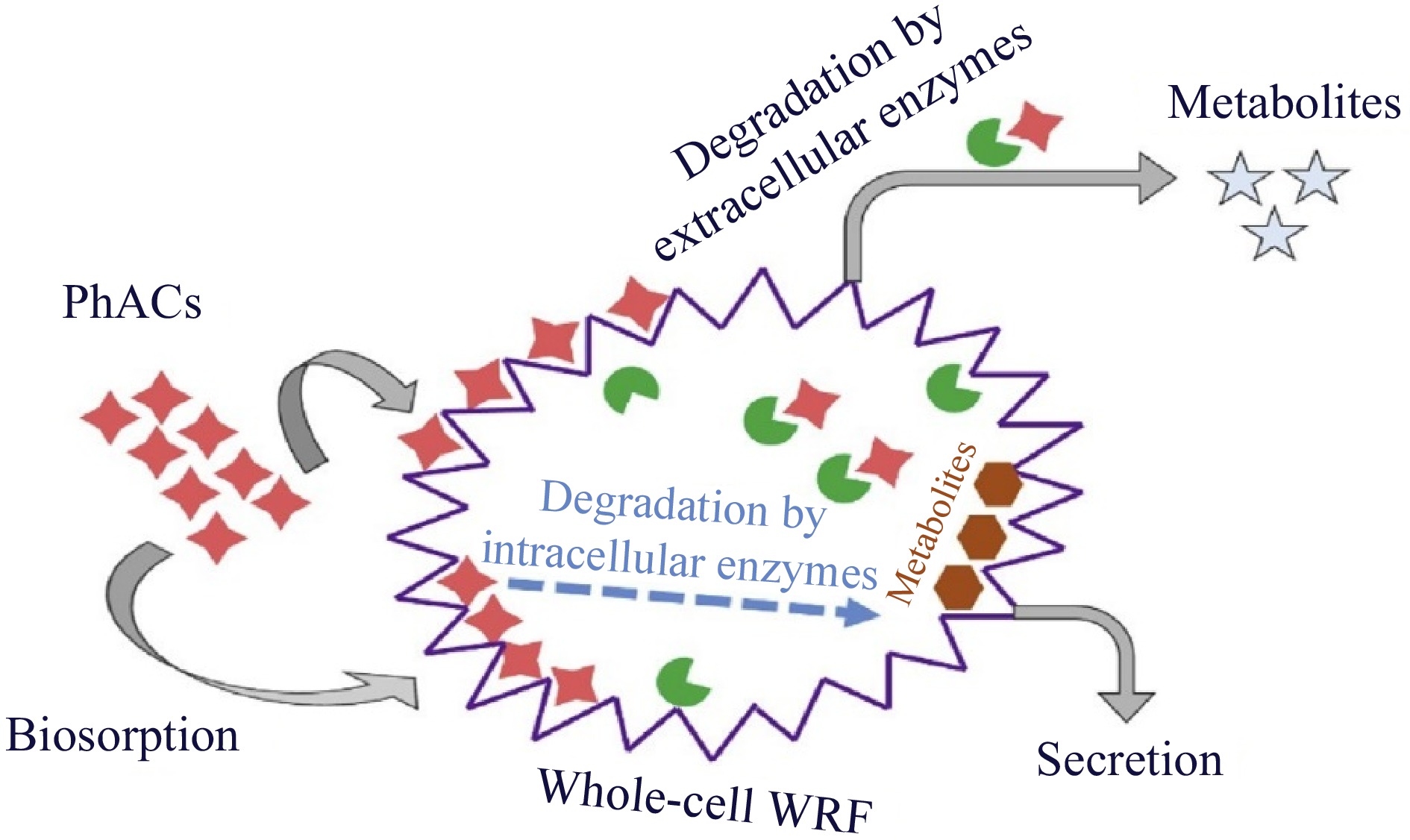
Figure 2.
Schematic diagram of pollutant removal using white-rot fungi[8].
-
The most well-known of these dyes, which pose a major threat to the environment, include azo dye, methyl orange dye, and methylene blue. Numerous fungal strains or consortia contribute significantly to the breakdown and mineralization of various dyes. Major sources of dye pollutants in wastewater include the textile, leather, paper, and printing industries. These industries release substantial amounts of dyes, such as azo dyes, anthraquinone dyes, and triphenylmethane dyes, into the environment. Effective fungal bioremediation can help mitigate the environmental impact of these pollutants by degrading and mineralizing complex dye structures. The capacity to accelerate fungal metabolism to create ideal environmental conditions is the main benefit of utilizing fungi to treat dye-containing wastewater[15]. Enzymes that are both intracellular and extracellular, such as Lac, Lip, and MnP, can increase their metabolic activity and aid in the treatment of wastewater containing dye[16]. In the modern world, water pollution, which primarily results from manufacturing industries, has become a severe environmental problem. The large amount of dye-contaminated wastewater created by dyeing procedures is a major cause of water and soil pollution in developing and impoverished nations. Due to its detrimental effects on human health and contamination of aquatic ecosystems[13].
Biodegradation mechanisms of Azo dyes
-
Azo dyes are man-made, persistent, recalcitrant macromolecules that are not biodegradable. For azo dye mitigation, several physicochemical technologies have been suggested. However, there are still significant drawbacks, including high operational costs, high energy consumption, convoluted processes, partial mineralization, and secondary waste creation. As an alternative, azo dye detoxification from wastewater is thought to be a clean, efficient, and secure technology. Fungi, yeast, bacteria, and algae can be used in biological treatment; these organisms have drawn attention for their eco-friendliness[17]. Fungi can be considered biodegraders or biosorbents for dyes. They are advantageous bioadsorbents due to their abundance of fungal biomass, cost-effectiveness, strong mechanical properties, and chemical stability in both acidic and alkaline environments. The physical qualities of the stiff structure of macrofungi increase their biosorption ability. As the temperature and dye reaction kinetics increased, the fungus had a greater adsorption capacity. A decrease in the number of active sites[17] or deactivation of the adsorbent surface prevented decolorization with increasing temperature. Compared to activated carbon, Cunningham Ella elegans and Trametes versicolor were found to be more effective at removing dyes. Additionally, several fungal species can participate in decolorization and degradation processes. Phenol oxidase and peroxidase, which may degrade azo dyes, are produced by filamentous fungi[18]. Since fungal mycelia can dissolve insoluble substrates through enzymatic action, they have advantages over single-celled organisms. Aspergillus ochraceous, Trametes versicolor, Pleurotus, Bjerkan-dera adusta, and Phlebia are other fungi that have attracted significant attention for their ability to degrade dyes. Due to the complete mineralization of dye at low cost and safety, the use of fungus for dye decolorization is a preferred method. However, its use in decolorization systems is limited by the long duration of hydraulic retention and the preservation of fungi in bioreactors for complete decolorization[17−19]. It is possible to decompose an aqueous solution of azo dyes using enzymatic, electrochemical, photocatalytic, microbiological, or hyphenated processes[20]. In virtually all currently known degradation processes, the azo dye decolorization process starts with chemical reductive cleavage of the -N−N bond. The different processes used to completely degrade azo dyes in wastewater include electrochemical (anodic and cathodic processes), photocatalytic (metal and nonmetal photocatalysts), enzyme-mediated (activated sludge treatment, anoxic degradation, and direct enzymatic reduction), and anaerobic (chemical and redox-mediated reduction) processes. To degrade dyes, fungi, bacteria, algae, or groups of these organisms are used[21]. It can act as a substrate-specific immobilizer quickly, improving the degradation effectiveness, especially for that kind of substrate. Enzymes naturally self-degrade; therefore, they have no negative environmental effects. A medium supplement for the growth of microbial cultures can be made from the byproducts of enzyme-mediated breakdown[22]. Because certain enzymes are substrate-specific and some are not, distinct processes are used for their degradation. Cofactors and redox-active coenzyme intermediates such as flavin adenine dinucleotide hydrogen (FADH), nicotinamide adenine dinucleotide phosphate hydrogen (NADPH), and nicotinamide adenine dinucleotide hydrogen support the direct enzyme degradation of dye (NADH). These intermediaries can assist with the transfer of electrons from an enzyme (such as AZR) to an azo dye. The azo-bonds (-N−N-) in the dye molecules became weaker as a result of electron transfer, and the bonds eventually broke to produce nontoxic, tasteless, and colorless aromatic amine intermediates. Aerobic conditions cause further degradation of aromatic amines, resulting in the production of low-molecular-weight compounds such as water, carbon dioxide, and ammonia[23]. In essence, the majority of azo dye degradation begins with the anaerobic process of decreasing double bonds, which is followed by the aerobic phase of full mineralization. During the breakdown of azo dyes, a few auto oxidized byproducts of harmful and carcinogenic aromatic compounds are also produced as intermediates of aromatic amines. Further disintegration of aromatic amines under aerobic conditions produces small molecular weight compounds such as water, carbon dioxide, and ammonia[23]. In brief, most azo dye degradation starts with reducing double bonds under anaerobic conditions, followed by aerobic processes for complete mineralization. Some autooxidized byproducts of toxic and carcinogenic aromatic compounds also formed during azo dye degradation (Fig. 3).
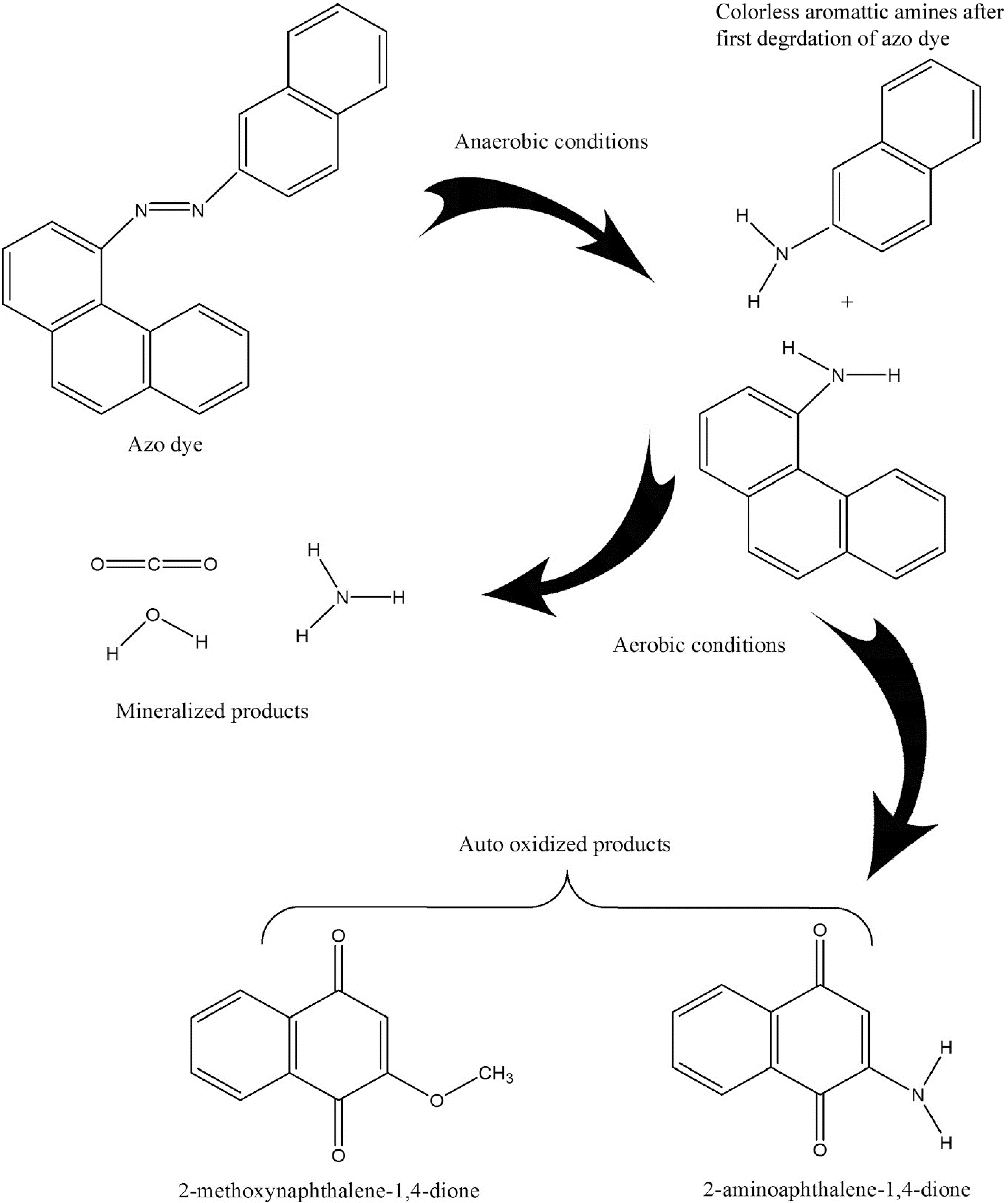
Figure 3.
Azo dye degradation mechanism in the presence of isolated enzymes[17].
The most widely employed enzymes for the breakdown of azo dyes are azoreductase and laccase. Both the cytoplasm and the membrane contain the azo-reductase enzyme. Due to its complicated chemical makeup and the cytoplasmic azo-subpar reductase's capacity to diffuse across the cell membrane sulfonated azo dye cannot be degraded by this enzyme. Nonsulfonated azo dye molecules are typically degraded by water-soluble cytoplasmic azoreductase enzymes. Different pathways are activated by redox-active intermediate-dependent cytoplasmic and membrane-bound azoreductase enzymes to accomplish azo dye reduction[24].
-
Heavy metal pollution of the environment caused by industrial emissions is becoming a global issue. Technologies for treating wastewater containing dyes and heavy metals that are practical, affordable, and environmentally acceptable are crucial. The remediation of heavy metals by various microorganisms, such as algae, bacteria, fungi, and enzymes, is an exciting biologically based method for treating effluents since it offers significant advantages over conventional treatment methods. Due to the world's increasing population, strict agriculture, and rapid industrialization, increased urban wastewater outputs have been revealed to be humanity's most significant environmental issue[25]. There may be a connection between the lack of clean water in many developing nations and the improper treatment of wastewater or the discharge of wastewater below a permissible level in compliance with neighboring water bodies. The metals that accumulate in living organisms include arsenic, cadmium, chromium, copper, lead, mercury, nickel, and zinc. These substances are toxic and persistent, meaning they do not break down easily in the environment. As a result, even small amounts of these metals in water can accumulate in the tissues of living organisms, leading to detrimental health, and environmental effects. Prolonged exposure can cause serious health issues, including organ damage, neurological problems, and an increased risk of cancer, highlighting the need for effective monitoring and remediation of these contaminants in water sources[26].
Fungi-based biological remediation of heavy metals
-
Fungi have a cell wall that can make up 30% of their dry weight and is mostly made up of polysaccharides, which make up approximately 80% of the dry weight. Fungi can serve as efficient biosorbents due to their abundance of cell wall components with strong metal binding capacities[27]. In addition to the small amount of glycoprotein, the cell walls of fungi include significant amounts of chitin, chitosan, glucan, and mannan. These polymers frequently contain metal-binding ligands, including carboxyl (R−COOH or R−CO2H), hydroxyl (−OH), amine (NH2), and phosphate (PO4−, PO3−) functional groups. The biological adsorption of Zn2+ HM ions in fungal mycelia, the vegetative component of fungi made up of thread-like hyphae, has also been documented. Fungi are used in biological adsorption processes for a variety of reasons, but one benefit is their ease of large-scale growth due to their quick multiplication cycle and high biomass yield. It is simple to culture and utilizes straightforward fermentation techniques and an inexpensive growing medium. In particular, the primary fungi used as biological adsorbents are nonpathogenic and safe to handle. Examples of species used for biological adsorption processes include Aspergillus niger, Penicillium chrysogenum, and Rhizopus arrhizus. These fungi have been shown to effectively adsorb and remove various contaminants from wastewater, making them valuable tools in bioremediation efforts[26].
The enzyme degradation mechanism of white rot fungi and heavy metal remediation
-
White-rot fungi (WRF) are widely known for their broad capacities for the breakdown of organic compounds and as a resource with potential environmental applications. These materials can transform or breakdown various organic contaminants in the environment due to the distinctive oxidative and extracellular ligninolytic systems of WRF, which exhibit low substrate specificity. The use of WRF for environmental applications has grown in recent years as a result of knowledge expansion and improvements in the removal of heavy metal pollution. There have been reports of the ability of certain WRF strains to remove heavy metals such as Pb, Cd, Cr, Cu, Ni, Zn, and Hg. These strains include Phanerochaete chrysosporium, Pleurotus ostreatus, Ganoderma lucidum, Trametes versicolor, IPEX lacteus, Lentinus edodes, Coriolus versicolor, and Schizophy[28]. The lignin-degrading enzyme system, which consists of LiP, MnP, and Lac, is primarily responsible for the degradation of pollutants by white rot fungi. These enzymes can either attach to the cell wall or be secreted outside to create a synergistic degradation system[29].
Catalytic mechanism of LiP and MnP
-
LiP has a high redox potential and, with the help of free radical catalysis, can directly or indirectly oxidize a wide range of compounds. Figure 4 illustrates the mechanism by which lignin peroxidase is directly oxidized: LiP(Fe3+) is excited by H2O2 and loses electrons to form compound I ([Fe4+]+•); compound I then gains electrons to become compound II (Fe4+), generating a free radical (R•) at the same time; compound II then continues to lose electrons to form Fe3+ and produces a free radical (R•). Free radicals are prone to instability and reduction reactions. Substances that are easily oxidized to produce free radicals can aid in the indirect oxidation of lignin peroxidase[30].
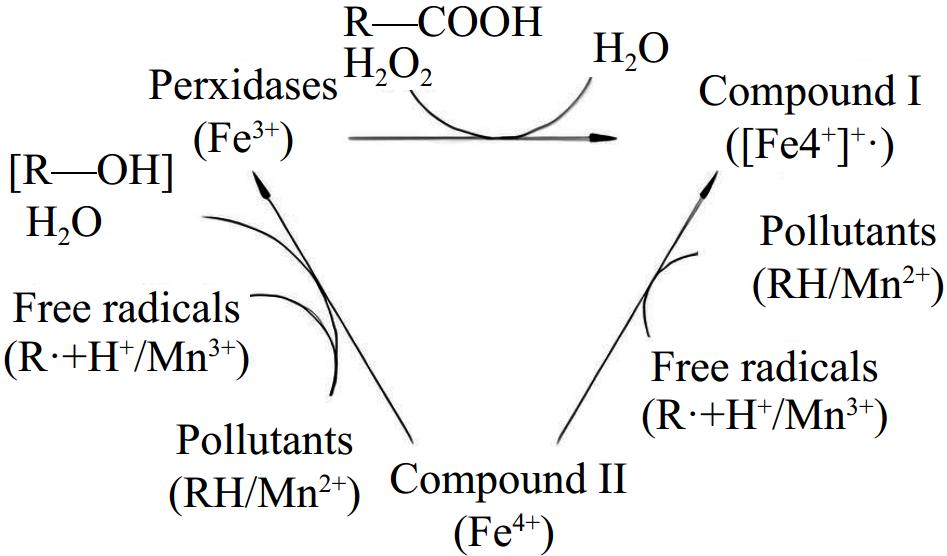
Figure 4.
Generic scheme of the catalytic cycle of peroxidases[30].
-
Laccase is an oxidase protease containing copper ions. The synergistic action of four copper ions within laccase molecules and the generation of free radicals are the main components of its catalytic mechanism. The oxidative degradation mechanism of laccase is depicted in Fig. 5. The white-rot fungal laccase type I Cu+ reacts with O2 to form the intermediate oxide type II Cu+. It combines with H2O to make type III Cu2+, which can then produce inactivated laccase type III Cu2+. Inactivated laccase type III Cu2+ loses four electrons to transform into laccase type I Cu+, which simultaneously produces water and free radicals from the substrate molecule. Free radicals in the substrate are typically unstable and prone to redox[30].
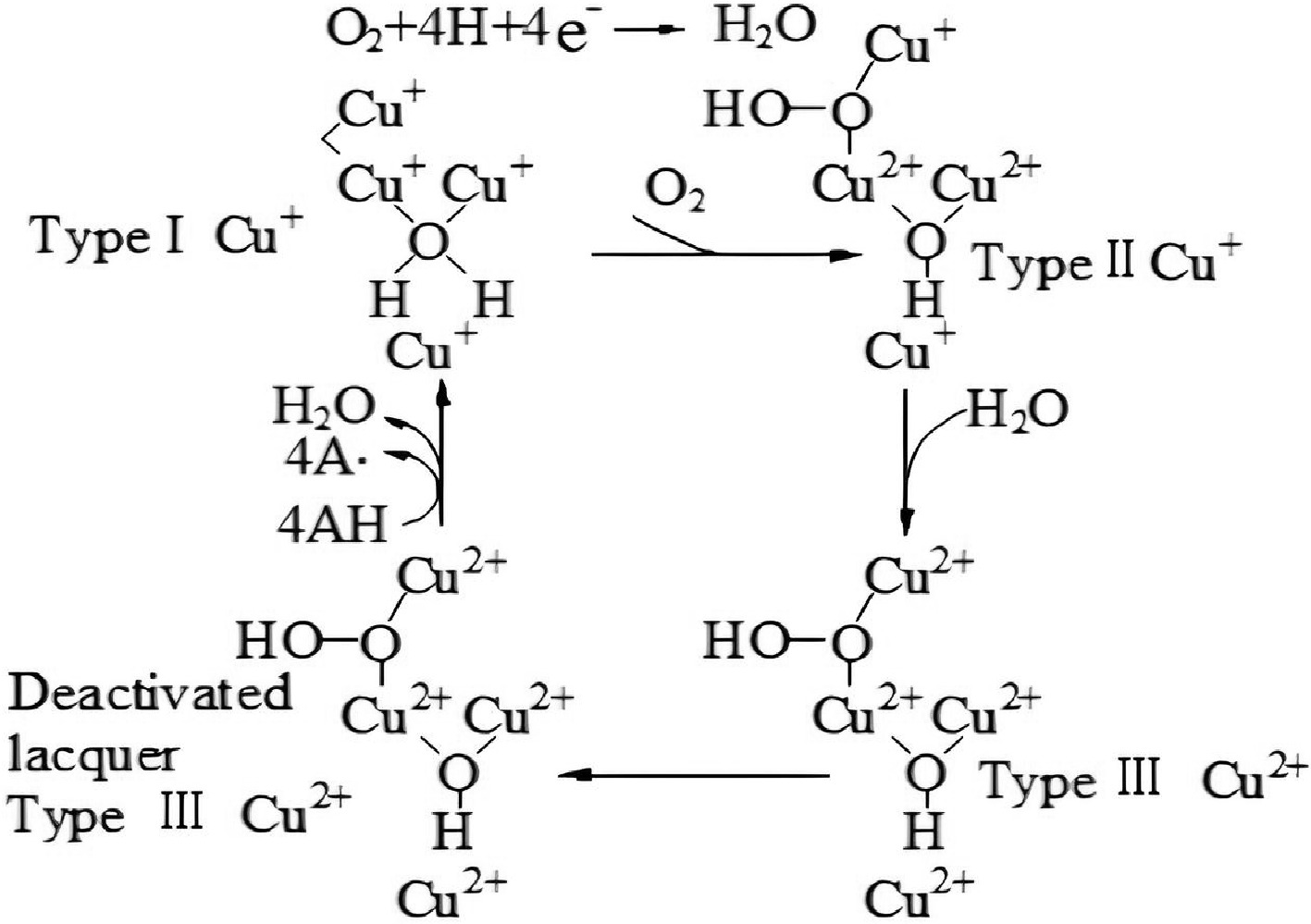
Figure 5.
The catalytic cycle of laccases[30].
The accumulation of heavy metals in organisms can result in many different types of damage through thiol binding, protein denaturation, essential metal displacement, and other side effects, which are extremely harmful to human health and the safety of the environment. The fungal cell wall has a high content of polysaccharides and proteins and contains a variety of essential metals. Unlike organic pollutants, heavy metal ions are nonbiodegradable and easily accumulate through the biological chain. Many researchers have become interested in the removal of heavy metals by biosorption in recent years[30].
-
The first line of defense against heavy metal stress is the sorption of heavy metals by the cell walls and membranes of WRF, which is necessary for bioremediation[31]. The biosorption process may be broken down into three steps: a fast surface adsorption stage, a sluggish transfer stage from external to internal, and a stage at which equilibrium is reached according to research on the heavy metal adsorption capabilities of WRF. For WRF to effectively remove heavy metals, cell wall adsorption is essential. The significance of cell wall adsorption in the remediation process was confirmed by early studies that separated cell wall components to reveal their contributions and discovered that cell wall adsorption accounted for up to 37.8%−76.8% of heavy metal remediation[32]. An essential mechanism for the adsorption of heavy metals is the interaction between functional groups found on the cell wall and the metals. In another study, Fourier transform infrared spectroscopy revealed that the biosorption of heavy metals into WRF involves the −OH, −NH, C−H, C−O, −COOH, and C−N groups. The effectiveness of Cd2+ and Ni2+ ion biosorption on the surface of P. chrysosporium by interactions with functional groups on fungal mycelia, which are responsible for the biosorption of heavy metals, was also demonstrated by Noormohamadi et al.[33]. At the same time, the interaction of heavy metals with functional groups on the surface of WRF may change the morphology of the mycelial surface. In the study of Cr ion adsorption, the interactions between Cr ions and amino, carboxyl, hydroxyl, and phosphate groups on the mycelial surface were found to form cage-like structures under scanning electron microscopy. This can be attributed to the production of intramolecular or intermolecular composites. During the removal of uranium ions using fungi, needle-like structures containing uranium appear along the cell wall and at the cell membrane. Through this binding, they prevent further entry of uranium ions into the cell with toxic effects[34]. It was noted that a few studies on the adsorption of heavy metals by white rot fungus found that heat-inactivated WRF mycelia have a comparatively greater capacity to do so. The heat treatment can create more binding sites by denaturing proteins on cell wall structures. Nevertheless, several investigations have suggested that living biomass is more efficient[35]. After carefully examining related articles, a difference between how dried biomass and heat-inactivated biomass were treated was discovered (dried at 30 °C for 24 h vs heat-inactivated at 90 °C for 15 min), which may account for the superior effect of live biomass over dried biomass. Sharma et al.[35] did not observe the adsorption of heavy metals, indicating that they accumulate inside cells as opposed to adhering to cell surfaces. This might be connected to how certain strains handle cleanup. The implementation of WRF for heavy metal remediation is complicated by variations in the preferences and methods of heavy metal remediation by various strains[35]. WRF has a limited capacity to absorb and enrich heavy metals. The biological activity of WRF can be inhibited by high concentrations of heavy metal ions because of the considerable differences in the adsorption capacities of the same strain for various heavy metal ions and different strains for the same heavy metal ion.
-
The contamination of soils, groundwater, sediments, surface water and air with harmful and toxic hydrocarbon chemicals is currently one of the main issues facing the industrialized world. Fungi appear to be especially promising because of their potent and unique biodegradative effects. This is especially true in the context of soil, which is the preferred environment for saprotrophic fungi. In particular, filamentous fungi have an apical growth mode in their hyphal network that allows them to penetrate soil more effectively than bacteria. The mechanical pushing of hyphae enables fungi to enter difficult-to-reach interstices where pollutants are present. Fungi can take advantage of substrates even with low moisture and low nutrient concentrations because of the range of degradation processes, and certain species can survive dry periods through cryptobiosis. In addition, fungi, which are frequently favored by acidic pH, help to accelerate acidification by secreting organic acids[36]. Numerous local fungi that thrive in polluted areas have evolved adaptable systems that allow them to use hydrocarbons as the sole carbon source[37]. Fossil fuels, such as natural gas, coal, and petroleum, are mostly composed of hydrocarbons. Crude oil, in particular, is made up of hundreds of different hydrocarbons, ranging from C1 (methane) to more than C30. The results of the Saturate, Aromatic, Resin, and Asphaltene (SARA) study shows that aliphatic hydrocarbons in petroleum are virtually entirely alkanes; in addition to gases, the main constituents are open-chain linear or branched alkanes and cycloalkanes (also known as naphthene). Usually, aromatic hydrocarbons are abundant in the heaviest fractions and serve as monomers for resins (which also include aliphatic unsaturated monomers). The so-called asphaltenes, which are crucial to the synthesis of asphalt, are composed of polycyclic (and heterocyclic) aromatic structures that are soluble in toluene but insoluble in n-alkanes[38]. Petroleum hydrocarbons are produced by both geological and biological processes, but when they enter the environment, they become serious pollutants. Dispersal and spills can occur during exploration, extraction, refinery, transport, and storage activities[39]. Utilizing the metabolism and cometabolism of microorganisms is the foundation of biological therapy. Since bacteria and fungi can utilize petroleum molecules as a source of carbon, metagenomic research has shown that the bacterial population from estuarine soil cometabolizes polycyclic aromatic hydrocarbons. Therefore, bioremediation is a 'green' technique that is easy to develop and economical from an energy and financial standpoint[40].
Fungal degradation mechanisms of aliphatic hydrocarbons involving enzymes
-
The biodegradation processes in which naturally occurring bacterial and fungal communities engage are among the primary mechanisms by which hydrocarbon pollutants can be eliminated from the environment. The degradation process in fungi typically follows an aerobic (oxygen present) mode; the anaerobic (oxygen absent) mode is primarily expressed by yeasts but is also observed in filamentous fungi[41,42]. As most organic pollutants, hydrocarbons are mostly broken down by fungi in aerobic environments; in reality, fungi act through enzymes that oxidize hydrocarbons to produce water and nontoxic or less toxic residues. According to how vulnerable they are to microscopic parasite attack, hydrocarbon compounds are often rated in the following decreasing order: n-alkanes are followed by branched alkanes, low-molecular-weight aromatics, cyclic alkanes, high-molecular-weight aromatics, and polycyclic aromatic compounds[43]. Another key feature determining degradability among aliphatics is the position of the double bond; for example, alkenes with a double bond in the terminal position are more easily degradable than alkenes with an internal double bond. Furthermore, because the main step is hampered by the lack of an exposed terminal group, cyclic aliphatics are the most resistant molecules to fungal attack[44]. The oxidation of the substrate in microsomes by cytochrome P450 (CYP) mono-oxygenases, which contain alkane-oxygenase enzymes, is necessary for the metabolic pathways observed in fungi[45]. The production of toxins, the metabolism of various types of endogenous and xenobiotic compounds, and other biological processes involving these enzymes, which are members of the heme-protein superfamily, all contribute to the fitness of organisms[46]. Depending on the way of life, different fungal species have different numbers of CYP genes. Compared to filamentous fungi, particularly plant pathogens, yeast, and yeast-like fungi have relatively few CYPs (three in Saccharomyces cerevisiae, six in Cryptococcus neoformans, and 10 in C. albicans). For instance, 121 genes are encoded by Cryphonectria parasitica, while 107 genes are encoded by Magnaporthe oryzae[47]. In fact, Shin et al. reported that CYP enzyme responses play a crucial role in the pathogenesis of fungi [47]. However, compared with those of plants and animals, few fungal CYPs have been functionally characterized[48]. The terminal oxidases in electron transfer chains connected to NADPH reductases are often CYP monooxygenases. When the other oxygen atom is converted to water, these enzymes provide the electrons necessary for the insertion of one oxygen atom into the aliphatic chain[49]. Higher eukaryotes often include a variety of CYP families with numerous distinct P450 forms that may work together as a group of isoforms to contribute to the metabolic conversion of a substrate[50]. The cytochrome P450 isoform catalyzes a variety of reactions, one of which is the monooxygenase reaction, which can result in the synthesis of epoxides, the oxidation of alcohols, or the hydroxylation of aliphatic or aromatic molecules. By using the various microsomal forms of cytochrome P450, some yeast species, including Yarrowia lipolytica, Candida albicans, C. tropicalis, C. maltose, C. apicola, and Debaryomyces hansenii, may breakdown hydrocarbons. Aliphatic breakdown starts with a process called hydroxylation, which involves the addition of an oxygen atom to the terminal methyl group to create an alcohol[51]. The alcohol is subsequently oxidized to the associated al-dehyde and then to the associated fatty acid. The resultant fatty acid undergoes p-ox-dation, which involves the first activation of the fatty acid to create an acyl-CoA ester and is then integrated within the major catabolic pathways. Together with the creation of electrons in electron transport chains, the latter are frequently further catabolized in the citric acid cycle[52,53]. The molecule is oxidized to a dicarboxylic acid and processed by oxidation via the diterminal route. Subterminal oxidation involves oxidizing an aliphatic hydrocarbon to a secondary alcohol, followed by the matching ketone and ester[52]. Furthermore, fungi can oxidize unsaturated and branched-chain aliphatic alkanes, as well as cyclic alkanes, through a variety of pathways, producing epoxides, alcohols, diols, and carboxylic acid units (Fig. 6). However, there is relatively little information in the literature about how these chemicals degrade. Figure 7 depicts the breakdown pathway of a generic PAH by fungi to compare the various degradation mechanisms of aliphatics and PAHs.
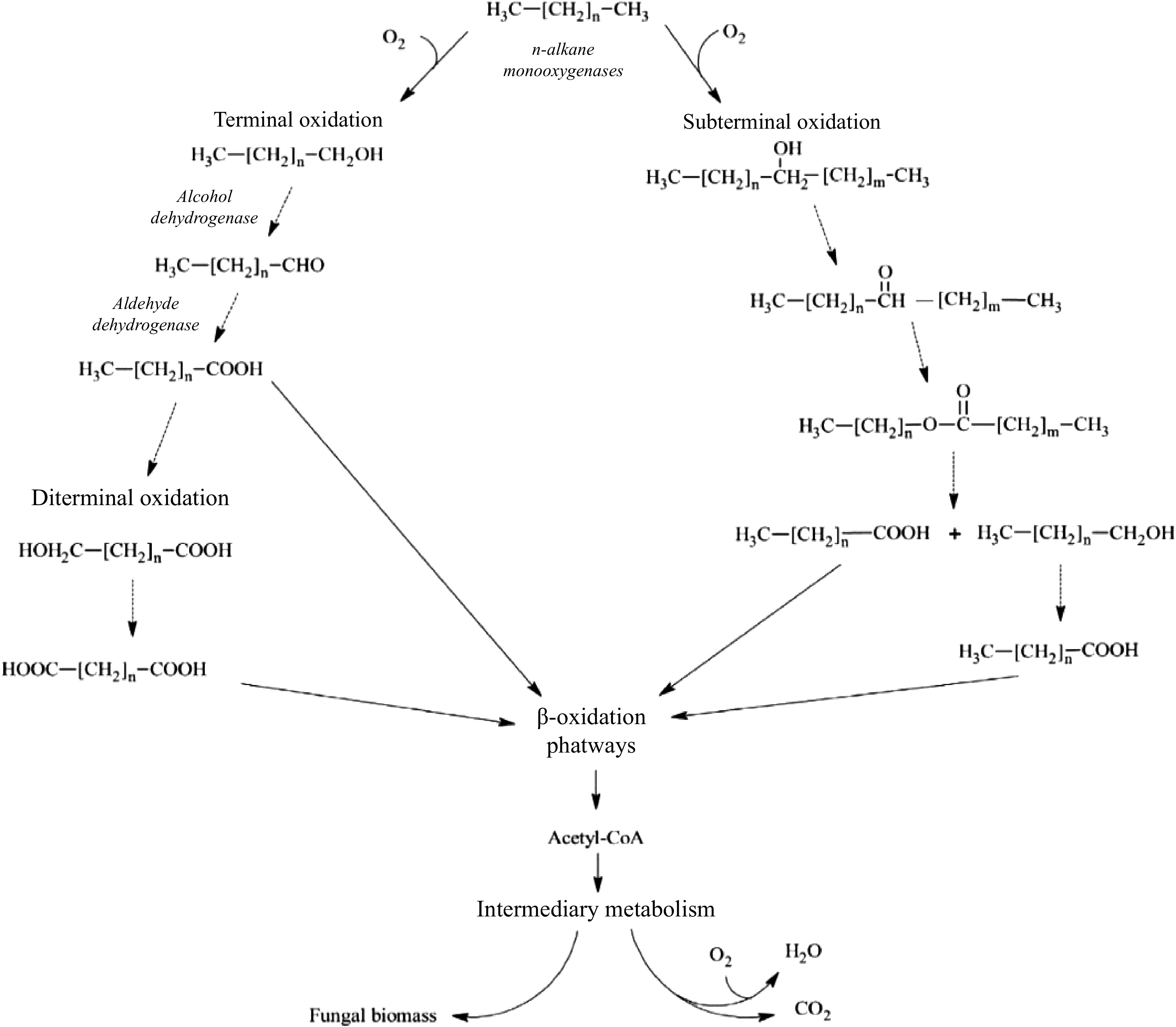
Figure 6.
Aerobic degradation of aliphatic hydrocarbons by fungi. The figure shows the terminal, diterminal and subterminal possible pathways for the degradation of n-alkanes (modified from Prenafeta-Boldú et al.[52]).
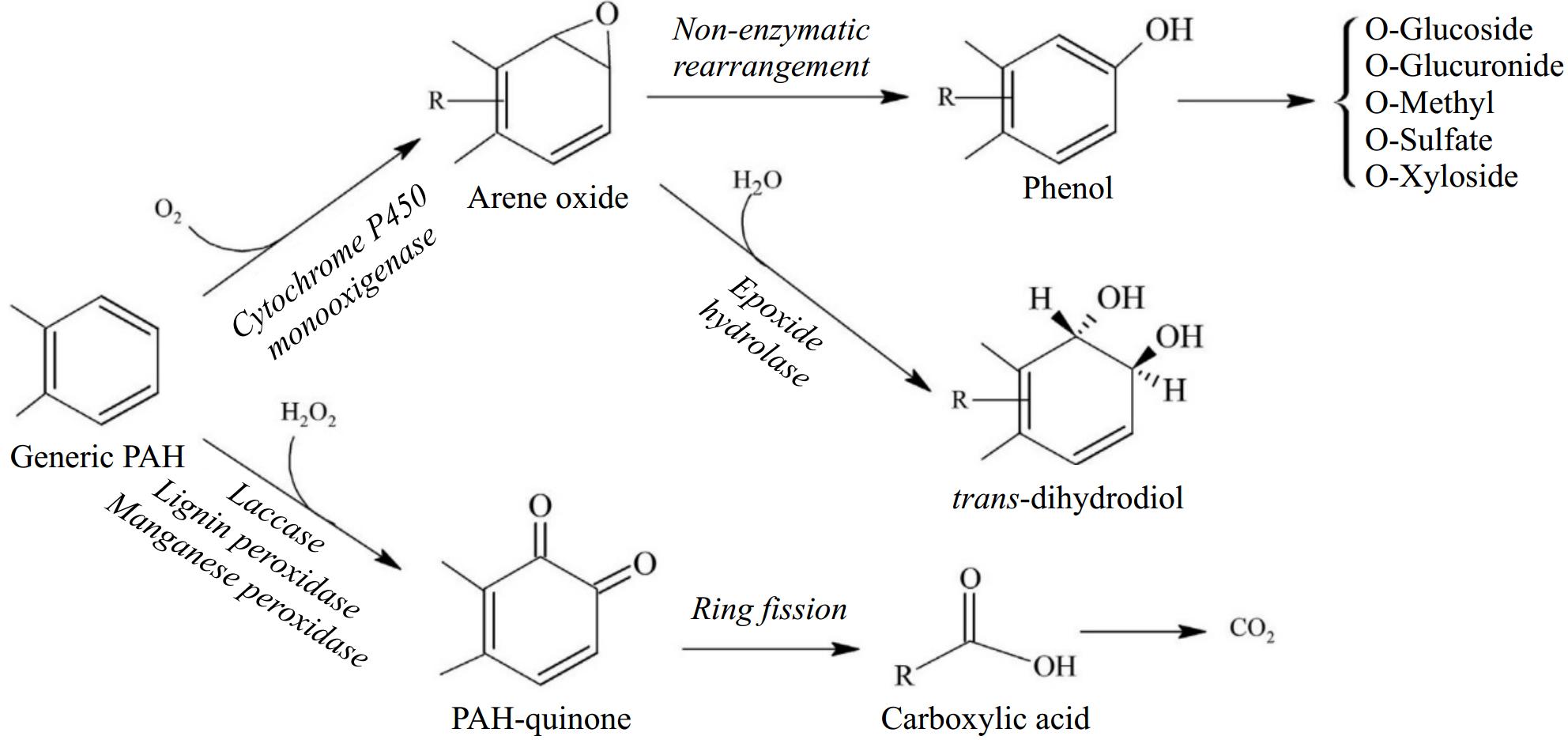
Figure 7.
Different pathways for the aerobic degradation of a generic PAH by fungi (modified from Kadri et al.[81]).
Although the aliphatic degradation pathways in fungi have been previously described, less is known about the enzymes involved in these pathways and, subsequently, about the genes that encode these enzymes. Nevertheless, the following enzymatic pathways have been suggested[36,52]:
(1) The bacterial enzymes that catalyze the breakdown of C1−C4 alkanes are similar to methane monooxygenases (from methane to butane).
(2) The enzymes known as nonheme iron oxidases catalyze the degradation of C5−C16 alkanes (from pentane to hexadecane).
(3) Enzyme systems that catabolize the degradation of C17 and longer alkanes are essentially unknown.
The identification of many genes involved in the metabolism of hydrocarbons has been made possible by the genome sequencing of organisms that breakdown hydrocarbons. However, the genomes of fewer fungal species have been sequenced than those of bacterial species, and most related research has concentrated on identifying genes that degrade PAHs[54]. Only saprotrophic or saprotroph-behaving necrotrophic taxa connected to lignocellulosic substrates, such as wood decay fungus, are capable of degrading aliphatic hydrocarbons. Additionally, P. strigosozonata was grown in oil-containing media, which led to the discovery of a significant upregulation of genes encoding three enzymes with potential use in xenobiotic biotransformation[55]. These genes have been linked to diene lactone hydrolase, carboxylesterase (type B), and B-class CYP monooxygenase. The degradation of aliphatic hydrocarbons does not appear to depend on the same enzymatic complex known to be involved in lignolysis, including laccases, Mn peroxidases (MnP), lignin peroxidases (LiP), dye-degrading peroxidases (DyP), versatile peroxidases (VP), and dye-degrading peroxidases (D). Although a process involving LiP in the aerobic degradation of trichloroethylene (TCE) by Phanerochaete chrysosporium was hypothesized, it was also indicated that the presence or absence of LiP and MnP had no effect on the total mineralization of TCE by the same species[56]. Researchers discovered that TCE promoted lac-case secretion, yet even in the absence of this enzyme, TCE was destroyed. In contrast, it was hypothesized that CYP was a crucial enzyme for the breakdown of TCE because the presence of 1-amino-reduction benzotriazole in the enzyme made the process extremely difficult. At the end of the initial examination, trichloroacetic acid was produced via the aerobic degradation of perchloroethylene (PCE), demonstrating that CYP-mediated degradation resulted in the generation of halogenated organic compounds rather than total mineralization to CO2 (Fig. 8)[57]. Despite these previous findings, halogenated aliphatic hydrocarbons can also be broken down by wood-decaying fungi via pathways other than oxidation via cytochrome P450.
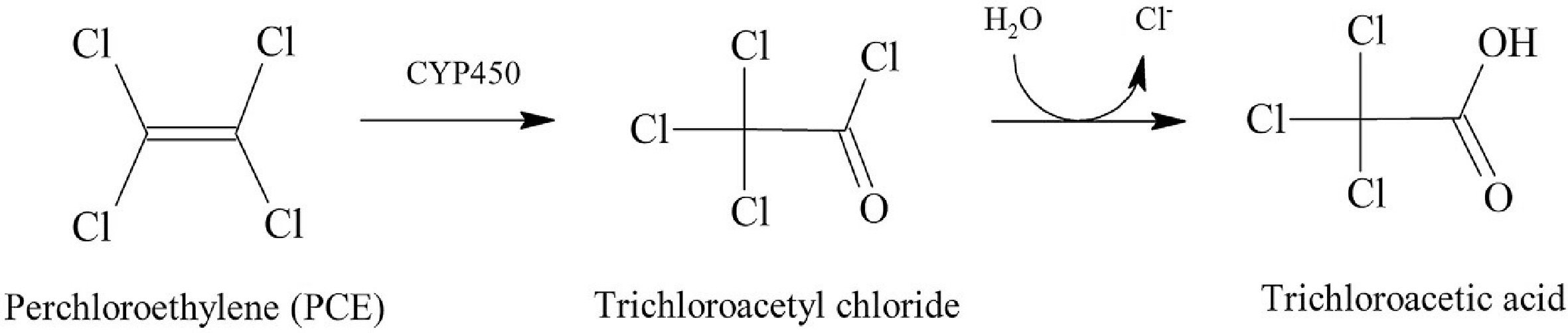
Figure 8.
Proposed pathway for partial enzymatic dehalogenation of PCE under aerobic conditions by T. versicolor[57].
-
In nature, quinones are organic compounds that are found in organisms and plants. They are created when fungi that cause wood decay (white rot) enzymatically depolymerize lignin. Because of their broad conjugation and similar stability, hydroquinones, and quinones are easily transformed into one another through reversible reduction. Therefore, even in an extracellular context, the quinone/hydroquinone pair offers a flexible redox system[57]. The superoxide anion radical (O−), HO, and hydroxyl radical (OH) are ROS-releasing processes that include the quinone redox cycle. It was originally noted that the quinone redox cycle was used by white-rot species to produce hydroxyl radicals[58]. Due to the high redox potential of ROS, hydroxyl radicals are regarded as the most active oxidants in fungal systems. Consequently, whether they are generated through enzymatic processes or the quinone redox cycle, ROS radicals can be implicated in the oxidation of both aliphatic and aromatic hydrocarbons[59].
The reactions that quinones undergo to produce hydroxyl radicals can be summarized as follows[60,61]:
(1) Reduction of quinones to hydroquinones by cell-bound dehydrogenases.
(2) Oxidation of hydroquinones to semiquinone radicals by laccases.
(3) Autooxidation of semiquinones catalyzed by Fe3+-EDTA, which is reduced to Fe2+ (this process results in the superoxide anion radical).
(4) The superoxide anion radical is dismutated to create H2O2 and O2.
(5) Decomposition of H2O2 and release of hydroxyl radicals by the comprehensive Fenton reaction;
$ {{\mathrm H}_2{\mathrm O}_2+{\mathrm{Fe}}^{2+}{\text -}{\mathrm{EDTA}}\to \cdot {\mathrm{OH}}+{\mathrm{OH}}^{-}+{\mathrm{Fe}}^{3+}{\text -}{\mathrm{EDTA}}} $ The results mentioned above support the notion that the pathways used to breakdown the aromatic and aliphatic parts of lignin typically work synergistically and with positive feedback. In this context, it has been demonstrated that selective white-rot species of wood-decaying fungi are efficient degraders of complicated hydrocarbon matrixes. Only a small percentage of fungal species have been documented to be obligately anaerobic; the majority are aerobic or facultatively anaerobic. Degradation occurs more quickly and effectively under aerobic conditions. The development of thin biofilm layers on the hydrocarbon matrix, which hinders the total separation of the organism from the aerobic environment, favors aerobic destruction. As the definition of 'aerobic' is limited to the presence of atmospheric oxygen, a number of oxygen-dependent activities are referred to under both aerobic and anaerobic conditions while utilizing different substrates than
$ {o}_{2} $ 1. Peroxidases, whose substrate is H2O2
2. The substrates of cytochrome P450nor are NO2− and NO3
Therefore, it can be difficult to distinguish between aerobic and anaerobic breakdown mechanisms in fungi. This is why it was thought that the degradative process could only begin under aerobic conditions, making oxygenases essential[64]. However, fungi have evolved methods that allow them to mineralize hydrocarbons in environments with severe oxygen limitation or absence, such as aquifers, muck, or ecosystems where mangroves predominate[65]. It is well known how bacteria degrade aliphatic compounds, such as alkenes, under anaerobic conditions. However, the anaerobic aliphatic breakdown routes of fungi are largely unexplored. However, it has been observed that some strains can use reductase enzymes to convert aromatic compounds to benzoyl-CoA[66].
-
Pesticides can aid in reducing crop losses caused by weeds, diseases, and pests, but their excessive use severely pollutes the environment. They persist in the environment, and they move up the food chain, posing a major threat to human health. The agriculture sector's reliance on chemicals to maintain productivity in arid settings has resulted in widespread environmental contamination with poisonous pesticides and nitrogen fertilizers that are altering the path of biogeochemical cycles. They include fungicides, insecticides, and herbicides and are among the factors contributing to water pollution. Some pesticides are also persistent organic pollutants that contaminate soil. Biodegradation is a very effective, less polluting, and more affordable method of removing harmful toxins than other methods[67]. Pesticide residues in water are a major problem since they endanger all kinds of biological communities, including human communities. Pesticides can enter water in a variety of ways, including accidental spills, industrial effluent, surface runoff, and transport from pesticide-treated soils, washing of spraying equipment after use, drift into ponds, lakes, streams, and river water, and aerial spray to control pests that inhibit the flow of water[68]. Typically, pesticides are transported from fields to other water reservoirs via runoff or drainage from irrigation or rain[69]. Similarly, a variety of events, such as spray drift, volatilization from the treated surfaces, and aerial application of pesticides, can contribute to the presence of pesticides in the air. The size of the droplets and the wind speed both affect how far the spray drifts. The rate of volatilization depends on the amount of time that has passed since the pesticide treatment, the surface it lands on, the temperature, humidity, wind speed, and the ingredient vapor pressure. The volatile or semivolatile nature of pesticide chemicals creates a significant risk of air pollution in large cities[70]. For instance, after agricultural spraying in California and Washington (USA), organophosphorus (OP) pesticides were found in air and surface environmental samples. Pesticides and their active metabolites contaminate ambient air and water bodies in Italian forests due to indiscriminate use, which may have an impact on the wellbeing of aquatic organisms, including fish, amphibians, and birds[71]. As a result, the advantages of using pesticides should be evaluated in light of the effects of their environmental persistence. While some pesticides can be eliminated through the degradation process, harmful degraded compounds still accumulate in the environment. Despite the vast amount of statistical data on pesticide degradation from regulatory bodies and testing organizations, it is still unclear how to specify the precise paths of pesticide degradation under a given set of ground conditions[72].
Biodegradation mechanisms of pesticides in the environment
-
Pesticide breakdown in soils is primarily mediated by microorganisms. It is a coincidental metabolic process in microorganisms. Pesticides are consumed with other sources of food and energy or are used as a source of carbon and energy by soil microorganisms such as bacteria, actinomycetes, fungi, algae, and protozoa. In 1 g of fertile soil, there are between 5,000 and 7,000 different species of fungi and unique populations of bacteria. These diverse soil microorganisms function as efficient and environmentally beneficial bioreactors to break down harmful chemical wastes[73]. Various structural and toxicological modifications to the parent pesticides are the result of such an enzyme biocatalytic reaction[72]. It is still unclear exactly how pesticides are biodegraded inside microbial cells. Recent studies have shown that microbes employ a certain genetic material (DNA-bound protein) to encrypt the required reactions to cope with a particular metabolic chemical. According to several additional studies, a particular microbial group may be responsible for the breakdown of a particular substrate. To be digested, the dissolved pesticides in the soil solution move across the cell membrane of particular microbial colonies[74]. Other microbial extracellular enzymes predigest pesticides that are poorly transported across the cell membrane and are released from the cell. Pesticides are digested by internal enzymes once they penetrate microbial cells. The rate of biodegradation of a specific pesticide is influenced by environmental factors that encourage biochemical processes and the availability of required enzymes. An effective degradation process requires a sufficient concentration of microorganisms, their variety, and a sufficient amount of extracellular enzyme-pesticide contact time. The kinetics of biodegradation are likely represented by a variety of mathematical models. Most models either support first-order kinetics or second-order kinetics. However, for first-order kinetics, the pace of degradation depends on the temperature, pH, and availability of limiting nutrients, but for second-order kinetics, the rate depends on the concentration of pesticides present as well as the size of the bacterial populations[75]. The pace of degradation in cases of biotic degradation, which has never occurred abiotically, is largely dependent on the activity of the encoded enzymes. For instance, the C−P bond in the herbicide glyphosate is unaffected by intense electromagnetic radiation, acid or base, or other environmental factors. C−P bonds can be broken or metabolized by a wide variety of environmental bacteria. A 14-gene operon encodes the enzyme that catalyzes the C−P lysis reaction[76]. Chemical processes are frequently used to break down pesticide compounds that do not contain appropriate reactive groups. High pH and low redox conditions, as well as the in situ synthesis of abiotic catalysts (such as poly-sulfides, surface-bound Fe(II), and MnO2), all affect the rate of these chemical reactions. Microorganisms can occasionally act as catalysts in these kinds of chemical transformations, which helps abiotic catalysts form more quickly.
-
Organophosphate pesticides (OPs) are a class of chemicals that have traditionally been utilized as chemical warfare agents and are used in agriculture, municipal hygiene, disease vector management, and the treatment of domestic pests worldwide[77]. The insecticidal properties of OPs, phosphorus-containing insecticides, were initially noted in Germany during World War II. Phosphotriesters, thiophosphotriesters, and phosphorothiolesters are the three main types. Three O-linked groups are present in the phosphate core of phosphotriesters, phosphoryl oxygens are replaced by sulfur in thiophosphotriesters, and one or more ester oxygens are replaced by sulfur in phosphorothiolesters (Fig. 9)[78].
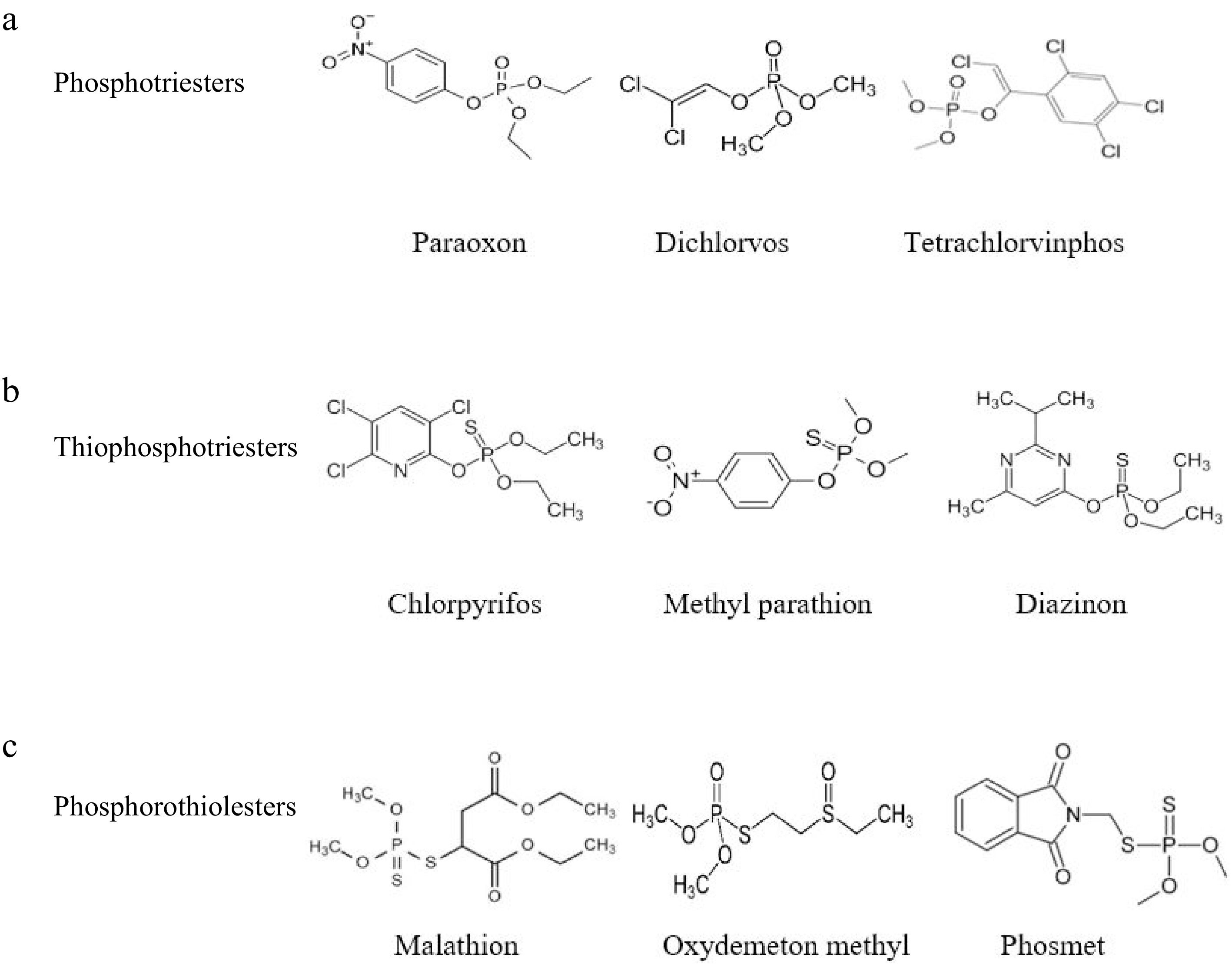
Figure 9.
Chemical structures of the main class of OPs. (a) Phosphotriesters; (b) Thiophosphotriesters; (c) Phosphorothio-lesters[18].
Organochlorine pesticides, which have been prohibited in the United States since the 1970s, have been replaced by this chemical class of pesticides. Due to their strong irreversible acetylcholinesterase (AChE) inhibitory properties, which have a dramatic impact on the neurological systems of exposed creatures, including humans, OPs are also extremely toxic[79].
The hydrolysis mechanism normally catalyzed by AChE relies on the attack of a serine residue at the active site on the carbonyl group in AChE, but in the presence of organophosphates, this residue is readily phosphorylated. A histidine residue at the active site captures a proton from the serine residue, increasing its nucleophilic character so that it readily attacks the electrophilic phosphorus atom, releasing the leaving group (X) (Fig. 10). The alkoxy substituent (R2) connected to the phosphorus atom can be dealt with by the phosphorylated enzyme, which reacts slowly with water in contrast to the acetylated enzyme. The organophosphate compounds thus inactivate acetylcholinesterase by phosphorylating serine at the enzyme active site. The result is the formation of a strong hydrogen bond between a protonated histidine residue of the catalytic site and the negatively charged oxygen atom of the inhibitor. Therefore, protonated histidine cannot function as a general base catalyst for the hydrolysis of the phosphorylated enzyme, which is a necessary step for the reactivation of AChE (Fig. 10)[80]. Pesticide waste in food analysis conducted by the Brazilian Food, Drug and Sanitary Surveillance Agency (ANVISA) revealed that OPs are the pesticides with the highest frequency in subpar samples. Chlorpyrifos, methamidophos, and acetate are the main active ingredients responsible for food contamination.
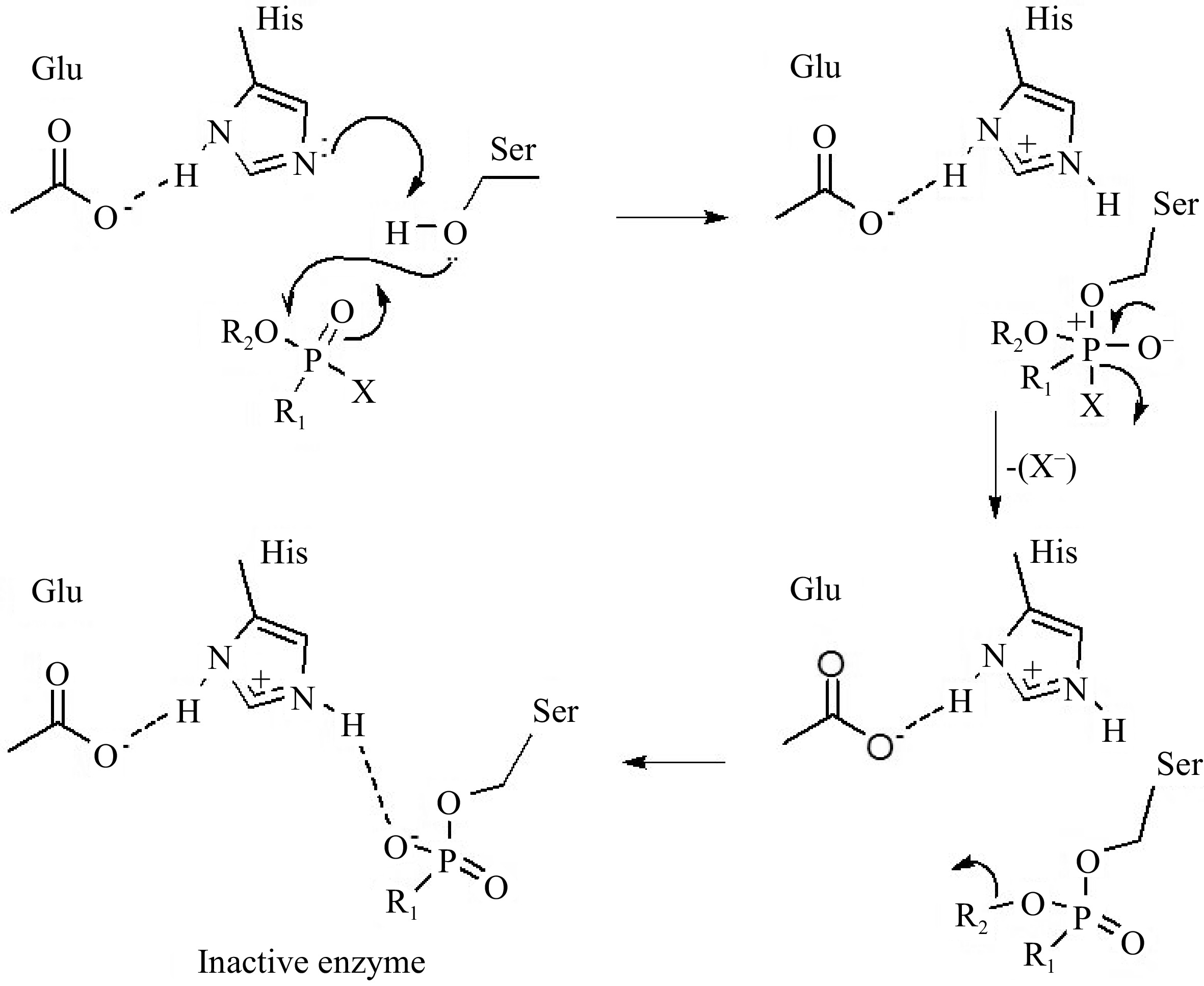
Figure 10.
Mechanisms of acetylcholinesterase inhibition by organophosphate pesticides[18].
-
Modern industrialized scientists have prioritized developing biocatalysts as potent tools for the degradation of pollutants because they frequently involve contaminant catalysts and are challenging to apply in large-scale contaminated field areas. Chemical techniques have several drawbacks in the degradation of pollutants. On the other hand, under the right safety, health, environmental, and financial conditions, biocatalytic processes can be conducted at room temperature and atmospheric pressure. In this context, it has been demonstrated that fungi possess a variety of catabolic processes that can be used to convert pollutants into less harmful molecules for the environment. More research is needed to better understand these capacities while considering natural variables and their application in large-scale contaminated fields, which is why the majority of the research on fungal bioremediation has been performed at the laboratory scale and under laboratory settings. Additionally, the breakdown of new contaminants from expanding industrial contamination requires the screening of new fungal strains with intriguing enzymatic activities. This microorganism screening will open the door for the future use of entire fungal cells and enzymes for bioremediation in conjunction with existing biotechnologies such as genetic engineering.
-
The authors confirm contribution to the paper as follows: draft manuscript preparation: Mohammed NN; study supervision and manuscript editing & revision: Zamel D; literature collection: Etman AE, Rabee MM; resources: Elmasry SA; revision and references validation: Khan AU. All authors reviewed the results and approved the final version of the manuscript.
-
All data generated or analyzed during this study are included in this published article
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Mohammed NN, Zamel D, Etman AE, Rabee MM, Elmasry SA, et al. 2024. Elucidation of the biodegradation mechanisms of fungi in efficient pollutant removal from wastewater. Studies in Fungi 9: e011 doi: 10.48130/sif-0024-0011
Elucidation of the biodegradation mechanisms of fungi in efficient pollutant removal from wastewater
- Received: 31 August 2023
- Revised: 15 July 2024
- Accepted: 21 July 2024
- Published online: 26 August 2024
Abstract: Microorganisms play a crucial role in various aspects of our lives and the environment, particularly in areas such as human health, and ecological balance. They are also versatile and powerful tools harnessed for various beneficial purposes, such as bioremediation, and biotechnology advancements. Continued research and innovation in the field of microbiology hold great promise for addressing environmental challenges, improving industrial processes, and enhancing human health and well-being. Bacteria, fungi, and algae are highly incorporated in several environmental applications; however, the fastidious nature of some bacteria limits their application in wastewater treatment. Therefore, in recent years, researchers have focused on the utilization of fungi in wastewater treatment applications owing to their easy adaptation and resistance to chemicals. This review article focuses on the mechanisms by which fungi biodegrade pollutants in wastewater, specifically highlighting their roles in removing contaminants such as industrial wastes, pharmaceuticals, and organic compounds.
-
Key words:
- Microorganisms /
- Wastewater /
- Purification /
- Treatment /
- Contaminants












