-
Nigrospora Zimm. (Ascomycota, Sordariomycetes, Xylariales) are ubiquitous fungi revealed on plants[1−4], lichens[5], insects[6,7], marine organisms[8], humans[9], and in the soil[10]. These fungi are associated with different plants as endophytes, saprobes, parasites, and pathogens[2,4,11−13].
Nowadays, with the emerging possibilities of accurate species identification of fungi, studying the patterns of their occurrence and abundance, as well as various characteristics is an urgent task. In recent years the revision of Nigrospora genus was carried out and new species were described on the basis of the molecular and morphological evidence[1,4,10,13−17]. The identified genetic diversity of Nigrospora indicates that many species of this genus have not yet been discovered. Moreover, the information on the distribution and host plants of known Nigrospora species remains often fragmentary. At least 30 Nigrospora species were detected in China on various substrates[10,11,13,14,16,17], among them, 14 Nigrospora species were associated with Poaceae plants[11,13,14].
In Russia, the N. gorlenkoana Novobr. occurrence on wheat, barley, oats, maize, and rapeseed were previously confirmed by molecular genetic methods[18], while there is no information on the distribution of other Nigrospora species. In our previous study, we suggested that one strain of Nigrospora sp. MFG 70052, isolated from Phragmites australis stem can be described as a potential new species based on morphology and phylogenetic analysis data, which represented the distinct genetic line, sister to N. chinensis Mei Wang & L. Cai[18].
The aim of study was to identify the Nigrospora sp. strain isolated from Phragmites australis collected in North Asia.
-
The Nigrospora sp. strain was isolated from the Phragmites australis collected in Primorsky Krai of Russia in 2010. The fragments of stems with necrosis were carefully washed with tap water, and were surface sterilized by soaking in 5% NaOCl (3 min), and then rinsing with sterile water. Then, the plant tissues were placed on potato sucrose agar (PSA) containing 1 mL/L mixture of antibiotics (HyClone, Austria), and 0.4 μL/L Triton X-100 (Panreac, Spain). Following incubation at 25 °C, the pure Nigrospora culture was sub-cultures onto fresh PSA. Suspension 20 μL consisting of 20−50 conidia and sterilized distilled water was transferred to a Petri dish on PSA containing Triton X-100 and spread with a sterile spatula over the surface of the medium prepared the day before. Incubation continued for 48 h at 25 °C, after one germinated spore was selected and transferred to a new dish with PSA. The fungal growth was recorded after 3 d of incubation. The single-spored Nigrospora strain MFG 70052 was deposited in the culture collection of the All-Russian Institute of Plant Protection (VIZR, St. Petersburg, Russia).
Molecular genetic analysis
-
Genomic DNA was extracted from fungal mycelium grown on PSA (7 d, 25 °C, in the dark) using the CTAB protocol[19]. The fragments of the internal transcribed spacer (ITS), β-tubulin gene (tub), and translation elongation factor EF-1a (tef) were amplified with ITS1/ITS4[20], T1/T2[19], and EF1-728F/EF1-986R[21] primer pairs and according to the authors' protocols, respectively. The sequencing of the fragments was carried out on an ABI Prism 3500 sequencer (Applied Biosystems, Japan) using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, USA).
The obtained combined sequences of three loci (ITS, tub, and tef) as well as the sequences of representative 88 Nigrospora spp. strains (Table 1) were aligned using the MEGA X 10.1 program. The bestfitting substitution model was determined using the IQ-TREE 2 v.2.1.3 program. The T92+G model was chosen for the multilocus analysis. The phylogenetic analysis was performed using maximum likelihood (ML) also in IQ-TREE 2 v.2.1.3 program. Bootstrap supports values were calculated with 1,000 replicates. To further infer phylogenetic relationships among taxa, a Bayesian analysis was conducted with MrBayes 3.2.1 using 2,000,000 generations of Markov chain Monte Carlo (MCMC), and trees were sampled at every 1000th generation.
Table 1. The information on the strains used for phylogenetic analysis in this study.
Species Strain ID Substrate Origin GenBank accession numbers ITS tub tef Nigrospora anhuiensis QY-2 Rice China OP677969 PP103614 PP103590 N. aurantiaca CGMCC 3.18130 T Nelumbo sp., leaves China KX986064 KY019465 KY019295 N. aurantiaca LC 7034 Musa paradisiaca China KX986093 KY019598 KY019394 N. aurantiaca LC12063 Saccharum officinarum, leaves China MN215769 MN329933 MN264008 N. bambusae CGMCC 3.18327 T Bamboo, leaves China KY385307 KY385319 KY385313 N. bambusae LC 7244 Bamboo, leaves China KY385306 KY385320 KY385314 N. brasiliensis CMM1214 T Nopalea cochenillifera Brazil KY569629 MK720816 MK753271 N. brasiliensis CMM 1217 Nopalea cochenillifera Brazil KY569630 MK720817 MK753272 N. camelliae-sinensis CGMCC 3.18125 T Camellia sinensis China KX985986 KY019460 KY019293 N. camelliae-sinensis LC12070 Saccharum officinarum, leaves China MN215773 MN329937 MN264012 N. chinensis CGMCC 3.18127 T Machilus breviflora China KX986023 KY019462 KY019422 N. chinensis LC4433 Castanopsis sp. China KX986013 KY019536 KY019436 N. chinensis LC 4660 Quercus sp. China KX986026 KY019548 KY019445 N. chinensis QY Rice China OP677966 PP103612 PP103588 N. coryli W18 Corylus heterophylla China PP218065 PP320372 PP461302 N. cooperae SFC20230324-M03 Ishige sp. South Korea OQ726361 OQ735179 OQ735196 N. cooperae BRIP 72408b Themeda arguens Australia OP035047 OP039537 OP039538 N. covidalis CGMCC 3.20538 T Lithocarpus sp. China OK335209 OK431479 OK431485 N. covidalis LC15837 Lithocarpus sp. China OK335210 OK431480 OK431486 N. endophytica URM8462 T Manihot esculenta Brazil OM265233 OP572420 OP572416 N. endophytica ARM687 Manihot esculenta Brazil OM265226 OP572418 OP572415 N. falsivesicularis CGMCC 3.19678 T Saccharum officinarum, leaves China MN215778 MN329942 MN264017 N. falsivesicularis LC13553 Saccharum officinarum, roots China MN215779 MN329943 MN264018 N. ficuum ZHKUCC 22-0143 Ficus sp. China OR164911 OR166318 na N. ficuum ZHKUCC 22-0125 Ficus sp. China OR164910 OR166317 na N. globosa CGMCC3.19633 T Soil China MK329121 MK336134 MK336056 N. globosa LC 12441 Soil China MK329122 MK336135 MK336057 N. globospora CGMCC 3.20539 T Petasites hybridus China OK335211 OK431481 OK431487 N. globospora LC 15839 Petasites hybridus China OK335212 OK431482 OK431488 N. gorlenkoana CBS 480.73 Vitis vinifera Kazakhstan KX986048 KY019456 KY019420 N. gorlenkoana MFG 70030 Wheat, grain Russia OK563236 OK626358 OK626374 N. gorlenkoana MFG 70036 Wheat, grain Russia OK563241 OK626363 OK626379 N. gorlenkoana MFG 70038 Barley, grain Russia OK563242 OK626364 OK626380 N. gorlenkoana MFG 70051 Oats, grain Russia OK563250 OK626372 OK626388 N. guangdongensis CFCC 53917 T Cunninghamia lanceolata China MT017509 MT024495 MT024493 N. guangdongensis Tly068 Cunninghamia lanceolata China MT017510 MT024496 MT024494 N. guilinensis CGMCC 3.18124 T Camellia sinensis China KX985983 KY019459 KY019292 N. guilinensis LC 7301 Nelumbo sp., stem China KX986063 KY019608 KY019404 N. hainanensis CGMCC 3.18129 T Musa paradisiaca, leaves China KX986091 KY019464 KY019415 N. hainanensis LC13514 Saccharum officinarum, leaves China MN215780 MN329944 MN264019 N. humicola CFCC 56884 T Soil China ON555686 ON557392 ON557394 N. humicola CFCC 56885 Soil China ON555687 ON557393 ON557395 N. humicola MFG 70052 Phragmites australis Russia OK563251 OK626373 OK626389 N. lacticolonia CGMCC 3.18123 T Camellia sinensis China KX985978 KY019458 KY019291 N. lacticolonia LC 7009 Musa paradisiaca, leaves China KX986087 KY019594 KY019454 N. lacticolonia LC12059 Saccharum officinarum, leaves China MN215783 MN329947 MN264022 N. macarangae MFLUCC 19-0141 T Macaranga tanarius, dead leaves Taiwan, China MW114318 na na N. macarangae NCYUCC 19-0177 Macaranga tanarius, dead leaves Taiwan, China MW114319 na na N. macarangae NCYUCC 19-0312 Macaranga tanarius, dead leaves Taiwan, China MW114320 na na N. magnoliae MFLUCC 19-0112 T Magnolia liliifera, leaves China MW285092 MW438334 na N. magnoliae LC 6704 Camellia sinensis China KX986047 KY019571 KY019373 N. manihoticola URM8461 T Manihot esculenta Brazil OM265224 OM869479 OM914791 N. marylouisemclawsiae BRIP 74865b T Spinifex sericeus Australia PP125567 PP209362 PP209361 N. mercuriadeae BRIP 75764a T Chromolaena odorata Australia PP707904 PP712794 PP712793 N. musae CBS 319.34 T Musa paradisiaca, fruit Australia KX986076 KY019455 KY019419 N. musae LC 6385 Camellia sinensis China KX986042 KY019567 KY019371 N. oryzae LC 6759 Oryza sativa China KX986054 KY019572 KY019374 N. oryzae LC 2707 Rhododendron simiarum China KX985954 KY019481 KY019307 N. osmanthi CGMCC 3.18126 T Osmanthus sp. China KX986010 KY019461 KY019421 N. osmanthi LC 4487 Hedera nepalensis China KX986017 KY019540 KY019438 N. pernambucoensis SCUA-Saf-N16 Arthrocnemum macrostachyum Iran PP256498 PP263820 PP263806 N. pernambucoensis URM8463 T Manihot esculenta Brazil OM265234 OM869481 OM914793 N. philosophiae-doctoris CGMCC 3.20540 T Disporum sessile China OK335213 OK431483 OK431489 N. philosophiae-doctoris LC 15838 Disporum sessile China OK335214 OK431484 OK431490 N. pyriformis CGMCC 3.18122 T Citrus sinensis China KX985940 KY019457 KY019290 N. pyriformis LC 2694 Rubus reflexus China KX985945 KY019472 KY019300 N. rubi CGMCC 3.18326 T Rubus sp. China KX985948 KY019475 KY019302 N. saccharicola CGMCC3.19362 T Saccharum officinarum, leaves China MN215788 MN329951 MN264027 N. saccharicola LC 12057 Saccharum officinarum, leaves China MN215789 MN329952 MN264028 N. sacchari-officinarum CGMCC3.19335 T Saccharum officinarum, roots China MN215791 MN329954 MN264030 N. sacchari-officinarum LC 13531 Saccharum officinarum, roots China MN215792 MN329955 MN264031 N. singularis CGMCC3.19334 T Saccharum officinarum, roots China MN215793 MN329956 MN264032 N. singularis LC 12068 Saccharum officinarum, roots China MN215794 MN329957 MN264033 N. shadeganensis IRAN 4958C T Halocnemum strobilaceum Iran PP256499 PP263821 PP263812 N. shadeganensis SCUA-Saf-N28 Aeluropus lagopoides Iran PP256500 PP263822 PP263813 N. sphaerica LC 7298 Nelumbo sp., leaves China KX985937 KY019606 KY019401 N. sphaerica LC13530 Saccharum officinarum, roots China MN215795 MN329958 MN264034 N. stoneae BRIP 75022a Cyperus aromaticus Australia OR608744 OR604067 OR604065 N. stoneae BRIP 75019a T Cyperus aromaticus Australia OR608743 OR604066 OR604064 N. vesicularifera CGMCC3.19333 T Saccharum officinarum, leaves China MN215812 MN329975 MN264051 N. vesicularifera LC 12055 Saccharum officinarum, leaves China MN215814 MN329977 MN264053 N. vesicularis CGMCC 3.18128 T Musa paradisiaca, leaves China KX986088 KY019463 KY019294 N. vesicularis LC 0322 unknown host plant Thailand KX985939 KY019467 KY019296 N. yunnanensis GUCC24-0008 T Juglans regia China PP915796 PP947937 PP947933 N. yunnanensis GUCC24-0009 Juglans regia China PP915797 PP947938 PP947934 N. zimmermanii CBS 290.62 Saccharum officinarum Ecuador KY385309 KY385317 KY385311 N. zimmermanii LC 13534 Saccharum officinarum Pakistan MN215824 MN329987 MN264063 Nigrospora sp. LC 2725 Symplocos zizyphoides China KX985960 KY019487 KY019313 Nigrospora sp. LC 4566 Lithocarpus sp. China KX986022 KY019545 KY019354 Apiospora arundinis CBS 114316 Hordeum vulgare, leaves Iran KF144884 KF144974 KF145016 The analyzed Nigrospora strain is indicated in bold. T – the ex-type strain; na – the sequence is unavailable. Morphological study
-
The morphological characteristics of Nigrospora sp. strain MFG 70052 were observed during its growth on PSA in the dark at 25 °C for 7–30 d, while the diameter of the colony was measured on the third day of cultivation in two perpendicular directions and the average diameter of strain colony was calculated. The micromorphological structures were investigated and documented using Olympus ВХ53 microscope (Olympus America, USA) connected to a SUBRA camera (Jenoptik, Germany).
-
Phylogenetic analysis of the adjusted and aligned sequences of Nigrospora sp. MFG 70052, as well as reference strains, included the following lengths: ITS − 459 bp, tub − 588 bp, tef − 566 bp. The number of parsimony-informative sites per genome locus was 58 (12.6%), 184 (31.3%), and 305 (53.9%), respectively. The topology of the phylogenetic tree was concordant with that reconstructed previously[1,2,10]. The analyzed strain MFG 70052 formed a clade with the reference strains N. humicola Qin Yang & Ning Jiang CFCC 56884 and CFCC 56885 with high bootstrap support ML/BP 100/1.0, which confirms the belonging of the strain MFG 70052 to the N. humicola species (Fig. 1).
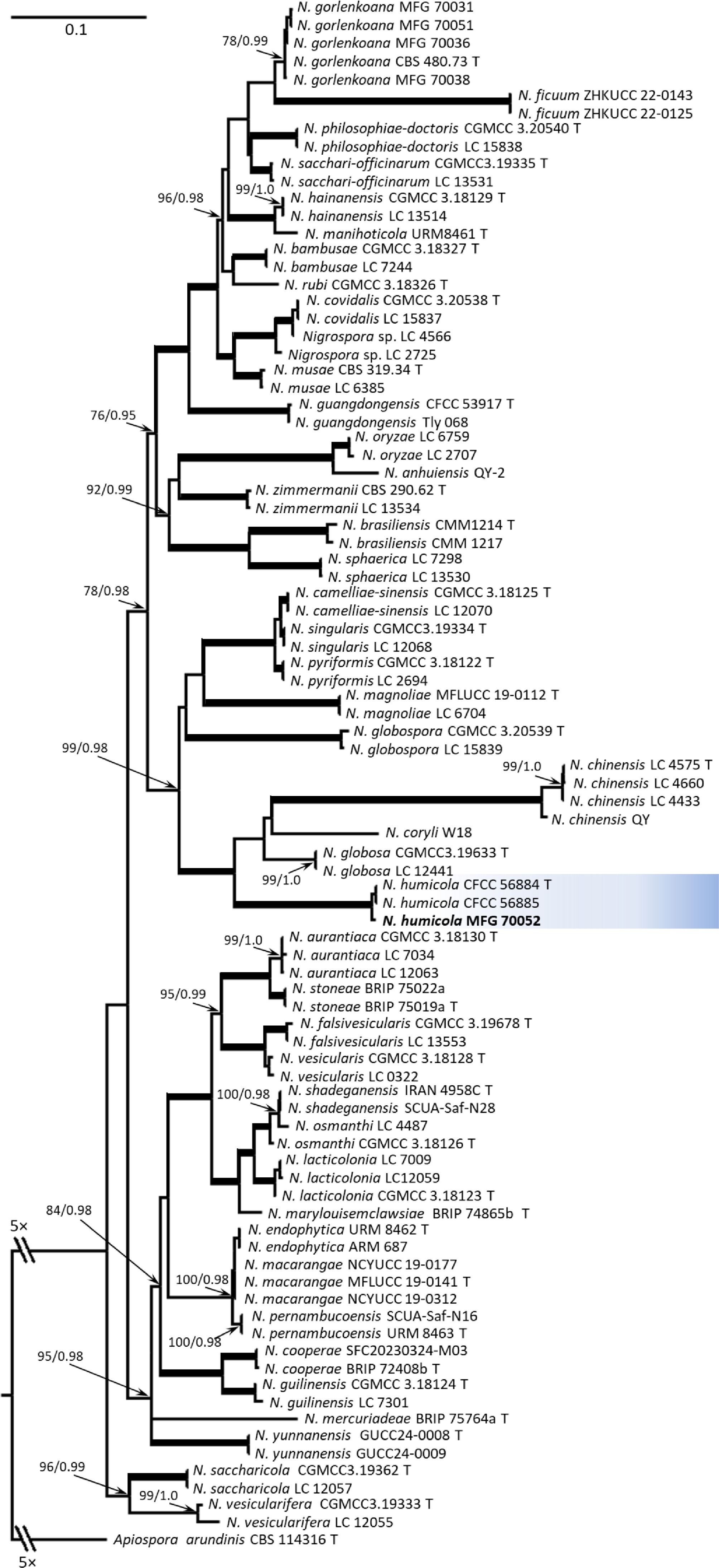
Figure 1.
Maximum likelihood (ML) phylogenetic tree based on DNA sequence data from three loci (ITS, tub, and tef) of Nigrospora fungi. ML bootstrap support values > 70 %, followed by Bayesian posterior probability (BP) scores > 0.95 are shown at the nodes. Thickened lines indicate ML/BP of 100/1.0. The tree was rooted on sequences of Apiospora arundinis CBS 114316. The analyzed strain is indicated in bold. T, ex-type strain.
Description morphology
-
The colony of Nigrospora humicola MFG 70052 reaching 80 mm on the third day of cultivation on PSA at 25 °C. Mycelium is flat, floccose, edge entire, initially white, later becoming gray when sporulation is abundant, and the edge is smooth/filamentous (Fig. 2). The reverse is smoke-gray, with age a noticeable pattern of dark hyphae and sporulation is noted.
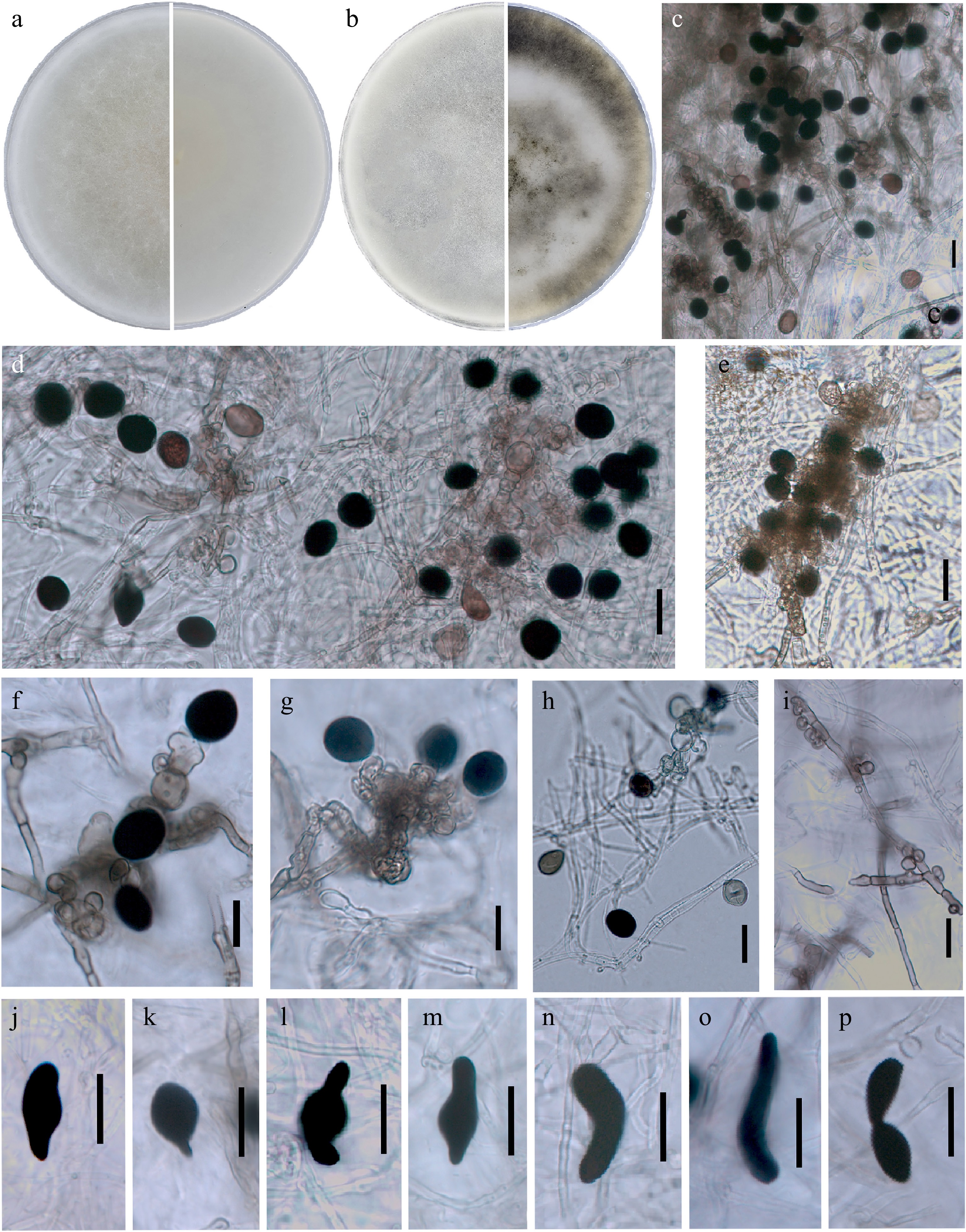
Figure 2.
Nigrospora humicola MFG 70052 (PSA, 25 °C in the dark). Upper surface and reverse overview after growth for (a) 7 d, and (b) 30 days. (c)−(h) Conidiophores and conidiogenous cells giving rise to brown-black conidia. (i) Septate hyphae and spherical vesicles aggregating in conidiophores. (j)−(p) Sterile cells. Scale bars = 20 μm.
Hyphae 2.5–6 μm, smooth, branched, septate, hyaline, some becoming brownish with age. Setae not observed. Conidiophores are mostly reduced to conidiogenous cells, macronematous, aggregated in clusters on hyphae. Conidiogenous cells 4–10 × 4–8 μm, hyaline to pale brown, their coloration is especially noticeable when they are aggregated, ampulliform, spherical, or subspherical.
Conidia 10.8–16.7 × 11.2–18.7 μm (13.7 × 15.6 μm), solitary, acrogenous, spherical or broadly ellipsoidal, egg-shaped, aseptate, pale brown at the beginning of formation, later black, shiny, smooth, aseptate. In some light-colored young conidia of N. humicola the presence of equatorial slits is noticeable. Sterile cells 27.0–40.8 × 11.2–21.4 μm (33.6 × 16.7 μm), terminally formed on hyphae, dark brown to black, elongated-ellipsoid or club-shaped, sometimes curved or angular.
-
At present, investigations of fungal diversity rely almost invariably on sequencing several informative genome loci and the development of multilocus phylogeny, for this approach provides greater resolution than the sum of morphological features. In this study, using the phylogenetic analysis of the ITS, tub, and tef loci Nigrospora strain MFG 70052 isolated from reed in 2019 from the Primorsky region of Russia bordered with China, was accurately identified as N. humicola. This is the first finding of this species on the plant (Poaceae) and the first detection in Russia. Nigrospora humicola was described recently when two strains isolated from soil in 2021 in the Hebei Province of China were studied in detail[10].
Some Nigrospora species are associated with different plants, e.g. N. oryzae (Berk. & Broome) Petch is the most ubiquitous species of this genus and has been reported from 40 different plant host genera including Poaceae family plants[11], and largely distributed in the territory of Russia, N. gorlenkoana was found in mycobiota of cereal crops and rapeseed plants[18]. Overall, Nigrospora species lacks host specificity[14], so the host range of N. humicola may also be wide.
Morphologically, MFG 70052 strain similar to N. humicola strains described by Zhang et al.[10] in the cultural features, sizes and shapes of conidiogenous, and conidial cells. However, there are some morphological differences that we consider important for identification of this species. One of them was a formation of sterile cells that looks like conidia due to dense dark pigmentation, but they are much larger and have an irregular shape. The sterile cells are formed directly on hyphae, not on conidiogenous cells and frequently observed in some Nigrospora species[11]. These morphological structures were described in the phylogenetically closest to N. humicola species N. chinensis, representing the sister clade[11]. In our opinion, they are an additional diagnostic feature. Moreover, in the culture of N. humicola, stained clusters of conidiophores and conidiogenous cells are formed, the same as described in N. oryzae and N. lacticolonia Mei Wang & L. Cai[11]. Equatorial slits are present in some young conidia of N. humicola, as in N. gorlenkoana[11], but not mentioned in the description of the species[10].
The results obtained in this study indicate that analysis of morphology is likely to be useful in refining our understanding of the Nigrospora species limits. The accurate identification of Nigrospora species is possible only by combined analysis of genetic, morphological, ecological, and physiological data[2,11,22]. The diversity of Nigrospora species in nature as well as their characteristics require further investigation.
This work was supported by the Russian Science Foundation (Project No. 19-76-30005).
-
The authors confirm contribution to the paper as follows: study conception and design, data collection: Orina AS, Gagkaeva TYu, Gavrilova OP; analysis and interpretation of results: Orina AS, Gagkaeva TYu; draft manuscript preparation: Orina AS, Gagkaeva TYu. manuscript review and editing: Gavrilova OP. All authors reviewed the results and approved the final version of the manuscript.
-
The data generated and analyzed during this study are available in this article. DNA sequence data are available in the GenBank database, and the accession numbers are provided in Table 1. The Nigrospora humicola MFG 70052 strain has been deposited in the fungal culture collection of the Laboratory of Mycology and Phytopathology (VIZR, St. Petersburg, Russia).
-
The authors declare that they have no conflict of interest.
- Copyright: © 2025 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Orina AS, Gavrilova OP, Gagkaeva TYu. 2025. First report of Nigrospora humicola (Apiosporaceae, Xylariales) on Phragmites plant from North Asia. Studies in Fungi 10: e005 doi: 10.48130/sif-0025-0006
First report of Nigrospora humicola (Apiosporaceae, Xylariales) on Phragmites plant from North Asia
- Received: 22 January 2025
- Revised: 03 March 2025
- Accepted: 07 March 2025
- Published online: 28 April 2025
Abstract: In this study, one Nigrospora sp. strain was isolated from a stem of Phragmites australis collected in North Asia and identified as N. humicola based on the results of multilocus phylogeny of the internal transcribed spacer (ITS), β-tubulin (tub), translation elongation factor EF-1a (tef) genes, and a detailed analysis of the cultural and morphological characters of the fungus. This is the first detection of N. humicola in Russia and its first occurrence on a plant, so the substrate range of this species may be wider than currently known. Morphological analysis of the strain revealed the additional characteristics that are considered important for the identification of N. humicola: sterile cells; formation of stained clusters of conidiophores and conidiogenous cells; equatorial slits in some young conidia. Thus, further analysis of morphology would be useful in refining our understanding of the Nigrospora species limits.
-
Key words:
- Poaceae /
- Reed /
- Fungi /
- Morphology /
- Phylogeny












