-
Rhizobia are widely known for their ability fix nitrogen in symbiotic association with plants especially legumes. Nitrogen fixation occurs symbiotically through the formation of root nodules in leguminous plants which is induced by the production of nod factors[1]. In symbiotic nodules, the fixed nitrogen is converted to ammonia for uptake by the host plants[2]. This is a natural process which reduces the need and use of synthetic fertilizers resulting in reduced costs of production[3].
Apart from their well-known ability of fixing nitrogen, rhizobia have a good ability in suppressing soil-borne pathogens which affects plants[4,5]. Some rhizobia species such as R. japonicum, S. meliloti, B. japonicum, and R. leguminosarum have been observed to protect the plants from infection by below root-rot disease causing fungal pathogens such as Phytophthora clandestine, Pythium ultimum, Fusarium solani, F. oxysporum, Pythium sp., Rhizoctonia bataticola, and Macrophomina phaseolina[1,6,7]. As for other legumes, soybean plants are highly prone to soil-borne pathogens which cause root-rot diseases which limits growth and development resulting in low yields of plants[8,9]. Macrophomina phaseolina, F. solani, F. oxyporum, and R. solani are among the most significant soybean root pathogens[1,6]. Some of the soybean nodulating rhizobial species, Bradyrhizobium sp., S. meliloti, and Rhizobium sp, have been witnessed to suppress F. solani, M. phaseolina, F. oxysporum, and R. solani in soybean rhizosphere[1].
Several mechanisms, such as production of hydrocyanic acid, antibiotics, and mycolytic enzymes have been observed to limit the growth of soil pathogens in the rhizosphere of leguminous plants, including soybean[10]. The particular species of rhizobia have been observed to be effective in suppression of F. solani, M. phaseolina, F. oxysporum, and R. solani in the rhizosphere of soybean plants[1,10]. Furthermore, Trichoderma harzianum has been witnessed to be effective in the biocontrol of root rot fungal pathogens[11] and hence used as a control organism in different in vitro and greenhouse experiments. However, rhizobia are affected by some agro-ecological factors including management practices[12,13] therefore, multi-location on farm trials need to be conducted to test the biocontrol effectiveness of particular rhizobia strains.
Root rot diseases are among the major factors which contribute to the low yield of soybean, although the impact is neglected as they are difficult to evaluate. The pathogens causing the diseases in the roots are the inhabitants of soil. In this case, the plant may be infected by more than one disease causing pathogen, causing huge plant and yield losses. Various studies have demonstrated the effectiveness of rhizobia as biocontrol agents in suppressing root rot fungi; nonetheless, their usage receives limited attention, particularly when dealing with complicated effects of root rot diseases caused by more than one fungal pathogen. Therefore, exploration of the new rhizobia species capable of suppressing the root rot fungi is essential in reducing the loss of yield due to their infection in plants. Furthermore, the soil can be colonized by more than one pathogenic fungi. It is also worth testing the ability of rhizobia in suppressing the combined effect of fungal pathogen. This study aimed to test the ability of Bradyrhizobium sp. TZSR41A and Rhizobium sp. TZSR12C and TZSR25B from the soils of Tanzania to supress the growth of F. solani, M. phaseolina, F. oxysporum, and R. solani. The findings of this study will contribute to enriching the knowledge by which researchers can tap into, for the benefit of further studies the use of rhizobia inoculant in suppressing the growth of root rot fungi and their effectiveness in controlling the complex infection caused by more than one pathogen.
-
The fungal pathogens, Rhizoctonia solani, Fusarium oxyporum, and Fusarium solani were isolated from the roots of infected soybean plants during the growing season[14]. Macrophomina phaseolina was isolated from the infected soybean seeds. The identification and sampling of infected soybean plants was done in the farmers' fields at Mbeya city council. The plants were sampled based on the visible symptoms on the shoots of plants. Specific symptoms such as blighted leaves by Rhizoctonia solani, stunted growth, yellowing and death of older plants by Fusarium solani and Fusarium oxyporum and premature death with leaves attached associated with the gray or silver on lower stem and tap root for Microphomina phaseolina were observed during sampling. The sampling was done at seedling (Fig. 1a), flowering (Fig. 1b), and maturity (Fig. 1c) stages before the drying of plants. Microphomina phaseolina symptoms are mostly visible at maturity and it is spread to seeds.
Isolation of fungi from infected roots and seeds
-
After sampling, the plants were brought to the Laboratory of Seed Health Center at Sokoine University of Agriculture for isolation of fungal pathogens. The isolation of fungi from roots was done as per the protocol explained previously[1,10]. First, the roots of plant samples were cut at the crown and then washed with tap water to remove the soil debris, and seeds from the soybean plants infected with M. phaseolina were prepared. The roots and seeds were dipped in 1% sodium hypochlorite (NaOCl) for 2 min. Then the samples were dipped in sterile distilled water for 5 min to allow the removal of disinfectant. After 5 min, the samples were placed on sterile filter papers to allow drying.
After drying, the roots were cut into small pieces of about 2−3 mm while concentrating on the infected portions with the disease symptoms such as rusty-brown and dry sunken lesions for R. solani and dark brown discoloration and rot of tap roots for F. solani and F. oxyporum. Afterward, the roots and seeds were placed by punching in a sterile PDA in plates. The plates were kept in the incubator at 28 °C for 3−5 d. The growth of fungi was observed daily. The purification of cultures was done by cutting and picking a small piece from the mycelial edge for each fungus and then inoculating on a fresh PDA. Further purification was done by picking a single spore isolation under microscope (Olympus CX23 at ×20 magnification) placed in an aseptic condition under a biosafety cabinet system (BSC-IIA2-2F). The observation of the features of R. solani, F. oxyporum, F. solania, and M. phaseolina was recorded under a microscope. The plates were then incubated at 28 °C for 7 d to allow the growth of fungal pure colonies for the test of antifungal activity of rhizobia under controlled conditions.
Preparation of rhizobia cell-free filtrates
-
Fresh cultures of rhizobia isolates were prepared by recovering them through spot inoculation on YEMA plates and then incubated at 28 °C for 72 h to allow the growth of colonies. Then the cultures were transferred in 4 ml of sterile YEMB in 15 ml falcon tubes, followed by incubation at 28 °C in orbital incubator at the speed of 180 rpm for 72 h to allow the multiplication of cells. After incubation, 1 ml of the broth was transferred in 1.5 ml Eppendorf tubes, followed by two times centrifugation at 3,000 rpm for 20 min to obtain the culture filtrates. The supernatants with rhizobial cells were transferred in new Eppendorf tubes for use in testing in vitro antifungal activity[1] and the pellets were discarded.
Preparation of Czapek's Dox Agar and thick filter paper discs
-
Czapek's Dox Agar medium containing 30.0 g of sucrose (C12H22O11), 2.0 g of sodium nitrate (NaNO3), 1.0 g of dipotassium phosphate (K2HPO4), 0.50 g of Magnesium sulphate (MgSO4), 0.5 g of potassium chloride (KCl), 0.010 g of ferrous sulphate (FeSO4) and 15.0 g of Agar was dissolved in 1,000 ml of distilled water. The final pH of the medium was adjusted to 7.2 at room temperature. The 5 mm discs of the thick filter paper were prepared by punching with a paper punching machine and then placed in a beaker covered with aluminum foil. Both the medium and the discs were sterilized by autoclaving at 15 pound-force per square inch (lbs) and 121 °C for 15 min.
In vitro antifungal activity of cell-free cultures of rhizobial filtrates
-
The inhibition of root-infecting fungi was tested by growing rhizobial strains and 5 mm discs of fungi on Czapek's Dox Agar. The previously sterilized filter paper discs were placed by using a neo syringe, one at the center of the medium and three on the sides at equal dimensions in petri dish plates. The fungal mycelial were inoculated on the disc and were loaded with 10 μl of the rhizobial cell filtrates[15]. Each test was in three replicates and the Petri dishes were incubated at 28 °C for 7 d. The distance between the fungal colony and the disc will be considered an inhibition zone which was measured in mm to obtain the average for three replicates[1,16].
Testing the activity of rhizobia against fungal pathogens under greenhouse conditions
Preparation of fungal inoculum for inoculation in experimental soil
-
The fungal inocula were prepared on some sterilized wheat groats. About 2 kg of wheat groats were divided into four portions of 0.5 kg and were soaked in water for 60 minutes. After soaking, the portions were placed in the separate clean sulphate bags and then sterilized in an autoclave at 15 pound-force per square inch (lbs) and 121 °C for 15 min and then allowed to cool at room temperature under aseptic conditions. Each portion of the sterile wheat groats was then inoculated with a respective fresh and actively growing pure cultures of fungi, F. oxyporum, F. solani, R. solani and M. phomina and then incubated at the room temperature 25−29 °C for 14 d to allow the growth of cultures on wheat groats before inoculating the experimental soils.
Rhizobium antagonism against fungal pathogens under greenhouse conditions
-
For experimentation under greenhouse conditions, three soil samples were collected at a depth of 0−30 cm uncultivated area in the vicinity of NM-AIST banana farm and CREATES green houses. A composite sample was prepared through mixing and removing the roots and crumps. The sample was air dried, ground then sieved with 2 mm mesh. After preparation, fertility status of the sample was evaluated for its for its suitability in supporting rhizobia activities and growth of soybean plants for optimum yield. According to Nelson & Sommers[17], the wet digestion (oxidation) method of Walkley-Black was used to characterize soil organic carbon. As per the description by Thomas, the pH of the soil was measured electrochemically using a 1:2.5 (w/v) water suspension in a potentiometric manner[18]. According to Motsara & Roy[19], total nitrogen was determined using the micro-Kjeldahl digestion-distillation method. As the soil sample's pH was less than seven, the extraction of P was determined using the Bray 1 technique[20]. A 1 M solution of ammonium acetate (NH4OAc) was used to determine the cation exchange capacity at pH 7. By using a flame photometer, the exchangeable cations potassium (K+) and sodium (Na+) were identified. Atomic absorption spectrophotometer measurements were made for the exchangeable bases calcium (Ca2+) and magnesium (Mg2+), as well as the micronutrients iron (Fe), copper (Cu), zinc (Zn), and manganese (Mn)[18,19]. Exchangeable Na by CEC (×100) was divided to calculate the exchangeable sodium percentage (ESP)[21]. The rankings of physico-chemical parameters were according to[19, 21].
Prior to sowing of rhizobia inoculated seeds, the soil for pot experiment was sterilized by oven drying and at 70 °C for 48 h. After cooling to room temperature under aseptic conditions, the soils were filled in 2 L plastic pots which were previously sterilized with 70% ethanol for 1 minute and rinsed with 0.25% NaClO solution[22]. The sterile soils in pots were watered and then inoculated/contaminated with 4 g of wheat groats containing actively growing fungal cultures and left to acclimatize for 14 d before sowing the soybean seeds. During the 14 d of acclimatization, the contaminated soils were sprayed with sterile distilled water to provide the optimum moisture for the growth of fungi. Before sowing, the seeds were sterilized by soaking in 70% ethanol for 1 min and 0.25% NaClO solution for 3 min followed by washing with distilled water five times to remove the disinfectants[23] before sowing. The solid Biofertilizers formulations containing the two Rhizobium sp., TZSR12C, TZSR25B and one Bradyrhizobium sp. TZSR41A were used. The seeds were inoculated with each respective rhizobia inoculant through damp inoculation by adding about 0.5 ml of sugar solution as a sticking agent, followed by addition of inoculum and then thorough mixing again to ensure all seeds are coated. After inoculation, seeds were left to air dry in the shade for about 30−60 min. The seeds were then sown at the depth of 2.5 cm in the respective plots as designed.
The experiment was set in RCBD layout and the plan for layout was generated in Genstat software 15th Edition. The treatments included the fungal cultures alone (FC), rhizobia as biocontrol (BC) + fungi, rhizobia (BC) alone, no contamination and biocontrol (NOB) and, control fungi alone, control fungi + fungal cultures, rhizobia (BC) and combination of fungal cultures and control fungi and combination of fungal pathogens alone as well as effective rhizobia with the combinations of biocontrol with effective rhizobia and fungal cultures (Table 1). For each treatment, 10 seeds were sown. To estimate the efficiency of rhizobia biocontrols, T. harzianum (commercial biocontrol) was used as a positive control.
Table 1. In vitro growth inhibition of root-rot fungal pathogens by cell free culture filtrates of rhizobial strains in agar disc-diffusion assay.
Treatment Diameters of inhibition zones (mm) Colony diameters (mm) FS MP RS FO FS MP RS FO Control 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 Rhizobium sp. TZSR12C 5.0 1.3 2.3 *** 8.7 6.3 23.7 22.7 Rhizobium sp. TZSR25B 8.3 1.7 *** *** 5.3 4.7 24.0 25.0 Bradyrhizobium sp. TZSR41A 5.7 2.3 *** *** 7.7 4.0 23.0 23.7 *** Growth inhibited but no zone of inhibition was formed. FS = F. solani, Mp = M. phaseolina, RS = R. solani and FO = F. oxyporum. The data for germination was collected 7 d after emergence and for disease incidence (DI) and disease severity (DS) (Fig. 2) on seedlings the data was collected at 14 d after emergence. The disease incidence was determined by uprooting six seedlings and then counting the seedlings with symptoms of diseases out of six. The disease severity was determined by using a six point scoring scale of 0−5 (where 0 = no symptoms and 5 = plant wilted or dead) as previously described by Win & Jiang[24].
Statistical data analysis
-
GenStat 15th Edition was used to statistically evaluate the germination data, and Excel 2016 running on Windows 10 was used to create the graphs. Using Jamovi version 2.3.2.0, the mean and standard errors within the treatments for fungal diseases and biocontrols were computed. The mean separation between the treatments was calculated using a one-way analysis of variance (ANOVA) that was applied to the factor effect model as displayed in Eqn 1. The 15th Edition of GenStat was used to apply the Tukey's-HSD multiple comparison test at a threshold of 5% to distinguish between mean values among replications of fungal pathogens and biocontrols. As a result, sample replications were treated as a random effect, and only one factor the treatment with various biocontrols and fungal pathogens was considered the fixed main effect.
$ Y_{i}=\mu+\alpha_{i}+\varepsilon_i $ (1) Where Yi is the observed response variable in the ith factor; µ is the overall (grand) mean; αi is the main effect of the treatment; εi is the random error associated with the observation of response variable in the ith factor.
-
Out of 25 rhizobia strains, only three of them, Rhizobium sp. TZSR12C, TZSR25B and Bradyrhizobium sp. TZSR41A, were capable of suppressing the growth of four fungal pathogens which are F. solani, M. phaseolin, R. solni, and F. oxyporum under in vitro conditions (Fig. 3; Table 1). Interestingly, each of the three strains of rhizobia were able to suppress the growth of all four fungal pathogens, though, in different capacities. Also, modes of suppression included the formation or no formation of inhibition zone, depending on the fungal isolate. The largest diameter (8.3 mm) of inhibition zone was formed by Rhizobium sp. TZSR25B against F. oxyporum which is followed by (5.7 mm) in Bradyrhizobium sp. TZSR41A whereby the smallest (1.3 mm) was in Rhizobium sp. TZSR12C against M. phaseolina. Rhizobium sp. TZSR12C was capable of forming clear zones in three fungi, F. solani, M. phaseolina, and R. solani while Rhizobium sp. TZSR25B and Bardyrhizobium sp. TZSR41A were able to form the inhibition zones on only two fungi which are F. solani and M. phaseolina. On the other hand, the largest colony of 25.0 mm against F. oxyporum, and closely followed by 25 mm against R. solani while the smallest colony of 4.7 mm against M. phaseolina were formed by Rhizobum sp. TZSR25B. Interestingly, the largest colonies are found in fungal growth inhibition with smallest of without the formation of inhibition zones.
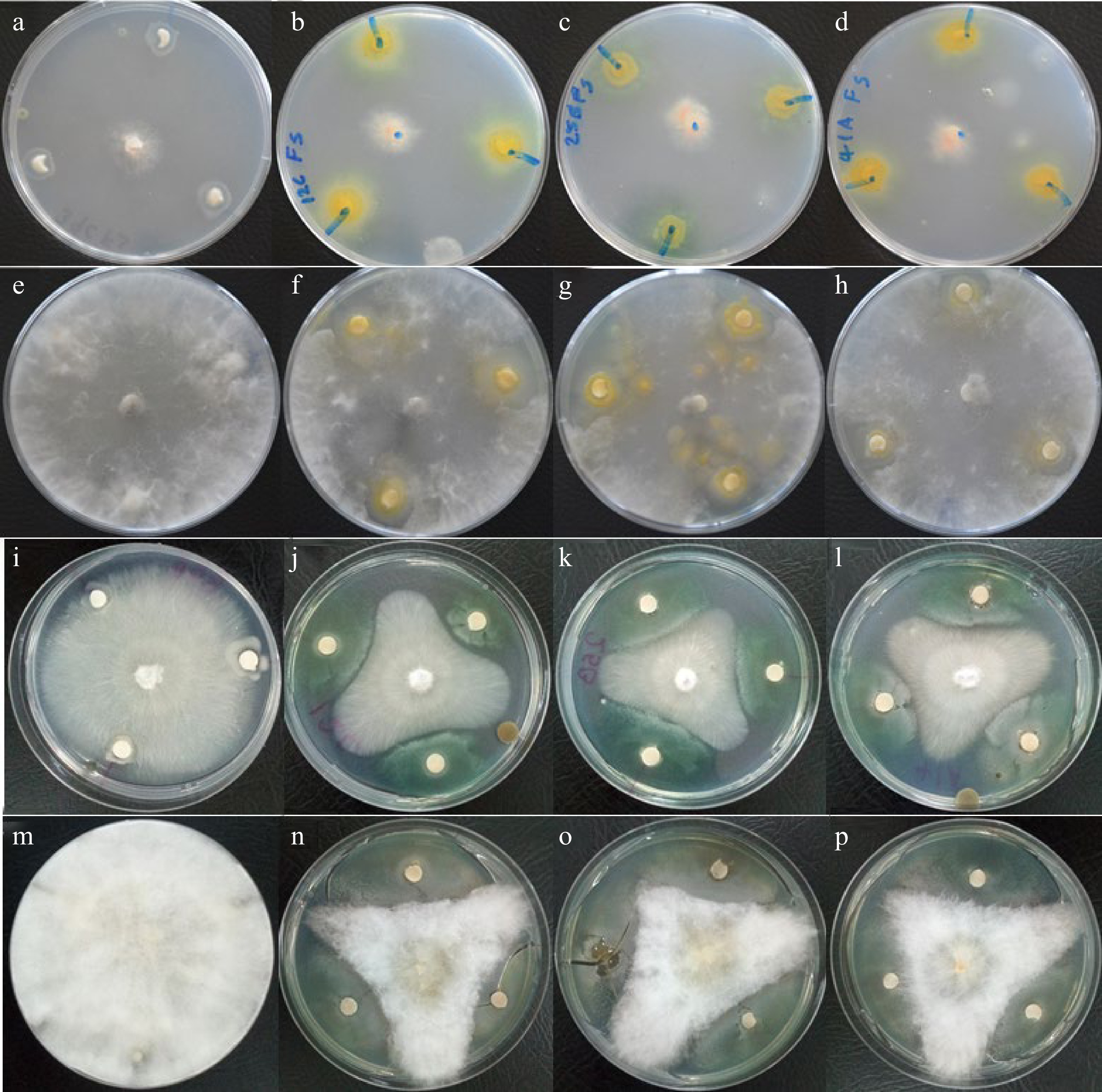
Figure 3.
Fungal growth inhibition by cell free culture filtrates of rhizobial strains in agar disc-diffusion assay. No inhibition of (a) F. solani, (e) M. phaseolina, (i) R. solani, and (m) F. phaseolina by B. japonicum TZSR39C, inhibition of (b) F. solani, (f) M. phaseolina, (j) R. solani, and (n) F. oxyporum by Rhizobium sp. TZSR12C, inhibition of (c) F. solani, (g) M. phaseolina, (k) R. solani, and (o) F. oxyporum by Rhizobium sp. TZSR25B, and inhibition of (d) F. solani, (h) M. phaseolina, (l) R. solani, and (p) F. oxyporum by Bradyrhizobium sp. TZSR41A.
Physico-chemical properties of the soil used for potting experiments
-
The soil used in the pot experiment had its physical and chemical characteristics evaluated, and the results are shown in Table 2. The soil's texture was sandy clay loam, and its pH ranged from neutral (pH = 6.93 in water). The proportion of organic carbon (OC) was 3.42 (high), and the total nitrogen content was 0.39 (medium). The soil had very high levels of copper (2.34 mg·kg−1), zinc (8.17 mg·kg−1), manganese (11.77 mg·kg−1), and iron (9.98 mg·kg−1) as well as very high levels of extractable phosphorus (75.64 mg·kg−1). The soil showed very high concentrations of the exchangeable bases calcium (13.95 cmol(+) kg−1) and magnesium (2.99 cmol(+) kg−1), medium concentrations of potassium (0.43 cmol(+) kg−1) and very low concentrations of sodium (0.07 cmol(+) kg−1). Furthermore, cation exchange capacity-CEC of an experimental soil was 18.22 cmol (+) kg−1 which is rated as medium.
Table 2. Physicochemical parameters of an experimental soil.
Parameter Mean Ratings SD Soil pH (H2O) 6.60 N 0.01 Electrical conductivity (dS/m) 0.22 NYR 0.04 Organic carbon (%) 3.20 VH 0.07 Total nitrogen (%) 0.36 M 0.1 Extractable phosphorus (mg·kg−1) 83.95 VH 0.63 Copper (mg·kg−1) 2.28 H 0.05 Zinc (mg·kg−1) 8.09 VH 0.01 Manganese (mg·kg−1) 10.96 VH 0.36 Iron (mg·kg−1) 10.53 VH 0.09 Exchangeable calcium (cmol(+) kg−1) 14.05 VH 0.01 Exchangeable magnesium (cmol(+) kg−1) 3.12 VH 0.09 Exchangeable sodium (cmol(+) kg−1) 0.09 VL 0.01 Exchangeable potassium (cmol(+) kg−1) 0.40 M 0.03 Cation exchange capacity (cmol(+) kg−1) 19.12 M 0.56 Exchangeable sodium percentage (%) 0.56 M 0.05 Texture Sandy Clay Loam VH = very high, H = high, M = medium, VL = very low, N = neutral and NYR = No yield reduction. S.D. = Standard Deviation[25]. Influence of rhizobia isolates and Trichoderma harzianum on germination of soybean seeds in the soils contaminated with fungal pathogens under greenhouse conditions
-
The ability and efficiency of rhizobia isolates and T. harzianum in enhancing the germination of seeds in the soils contaminated with fungal pathogens was tested under greenhouse conditions. The test was biocontrols (rhizobia or T. harzianum) without fungal pathogens (Fig. 4c), with one (Fig. 4d), two (Fig. 4a), three (Fig. 4e) and four (Fig. 4b) pathogens. Rhizobia isolates and T. harzianum were observed to possess different capacities in enhancing the germination of soybean seeds, in the soils without and those with contamination of different fungal pathogens under greenhouse conditions.
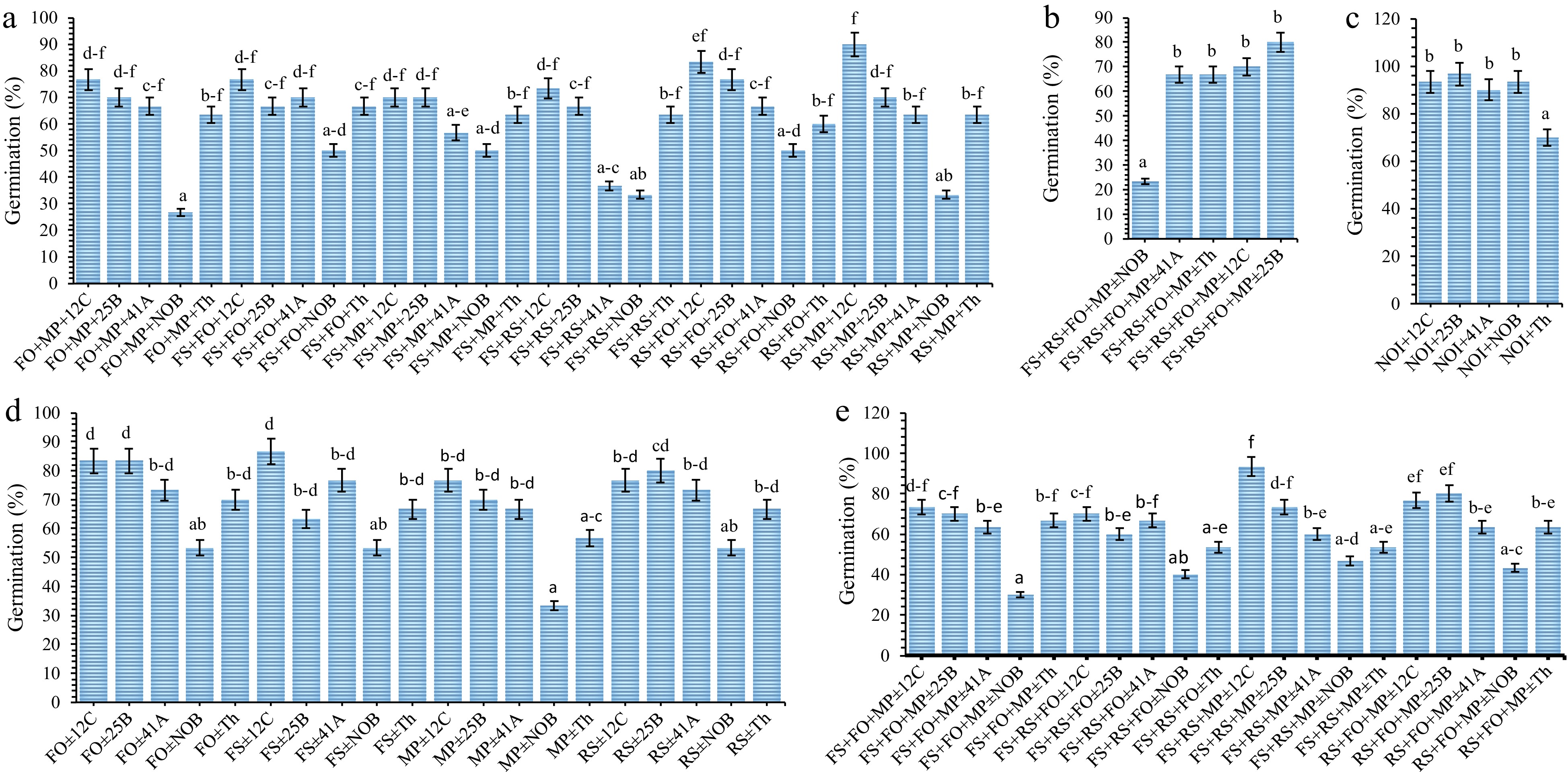
Figure 4.
The effect of rhozibia species and Trichoderma harzianum on germination of soybean seeds in the soils having root rot fungi contaminats. In front of each ID numbers, 12C, 25B and 41A there is common ID TZSR. FS = F. solani, MP = M. phaseolina, RS = R. solani and FO = F. oxyporum. The X-axis is biocontrol agents, in front of each ID number, 12C, 25B and 41A there is common ID TZSR. The labels (a)−(e) are biocontrols with (a) combination of two fungi, (b) four fungi, (c) without fungi, (d) one pathogen, and (e) three pathogens.
In the soils without fungal pathogens, Rhizobium sp. TZSR25B yielded the highest germination of 96.67% which was closely followed by 93.33% in Rhizobium sp. TZSR12C and soils without biocontrol and then 90% in Bradyrhizobium sp. TZSR41A, whilst the lowest germination of 70% was observed in T. harzianum. In the test of rhizobia against one fungal pathogen, the highest (86.67%) germination was observed in F. solani, which is closely followed by 83.33% in F. oxyporum both by Rhizobium sp. TZSR12C, and then 83.33% by Rhizobium sp. TZSR25B. In this group, the lowest germination 33.33% was observed in M. phaseolina, followed by 53.33% germination in F. oxyporum, F. solani, and R. solani, all without biocontrol. In the combination of two fungal pathogens, Rhizobium sp. TZSR12C exhibited the highest germination of 83.33% in combination of R. solani with F. oxyporum, this was followed by 76.67% and two combinations of F. solani with F. oxyporum, and F. oxyporum with M. phaseolina in Rhizobium sp. TZSR25B. For the combination of two fungi, the lowest germination of 26.67% was observed in F. oxyporum and M. phaseolina, followed by 33.33% in both F. solani with R. solani, and R. solani with M. phaseolina, all without any biocontrol.
The highest germination (93.33%) was observed in Rhizobium sp. TZSR12C against the combination of three fungal pathogens, F. solani, R. solani, and M. phaseolina which was distantly followed by 80% in Rhizobium sp. TZSR25B and 76.67% in Rhizobium sp. TZSR12C against R. solani, F. oxyporum and M. phaseolina. On the other hand, the three lowest germination were 30% in F. solani, F. oxyporum and M. phaseolina, 40% in F. solani, R. solani, and F. oxyporum and 43.33% in R. solani, F. oxyporum and M. phaseolina, all without biocontrol. In the combination of four fungal pathogens, the highest germination of 80% was attained by Rhizobium sp. TZSR25B, followed by 70% in Rhizobium sp. TZSR12C and 67% in Bradyrhizobium sp. TZSR41A and T. harziunum, all against F. solani, R. solan, F. oxyporum and M. phaseolina. The lowest germination of 23% was observed in the soil contaminated with all four fungi without any biocontrol agent. In all treatments, Rhizobium sp. TZSR12C and TZSR25B were observed to performed well.
Ability of rhizobia in supressing the root rot fungal pathogens under greenhouse conditions
Antagonistic activity of rhizobia and T. harzianum against individual fungal pathogens
-
The strains of rhizobia and T. harzianum exhibited the better (0.00%) performance in suppressing the individual fungal pathogen in the rhizosphere of soybean plants under greenhouse conditions (Table 3). All the biocontrol rhizobia were observed to be effective in suppressing F. oxyporum and F. solani with 0.00% infection in both the roots and foliage. Conversely, in the treatments of rhizobia isolates and T. harzianum, the highest infection of 16.67% with severity of 5.33% in roots was observed in Rhizobium sp. TZSR12C against F. solani. For the case of treatments without biocontrols, R. solani caused the highest infection of 66.67% with severity of 38.67% in roots, this resulted to the infection of 39.00% with severity of 7.67% in plant foliage. Despite the minor infections and severity of F. solani in Rhizobum sp. TZSR12C, R. solani in Bradyrhizobium sp. TZSR41A and, F. oxyporum, F. solani, and R. solani in T. harzianum, no infection was spread in the foliage parts of the plants. Furthermore, Rhizobium sp. TZSR25B was observed to perform better by completely suppressing F. oxyporum, R. solani, and M. phaseolina with very minimum infection of 5.67% and severity of 1.00% by F. solani in the roots.
Table 3. The effect of rhizobia species and Trichoderma harzianum as biocontrol inoculants on single root rot fungi of soybean plants.
Treatment RDI (%) RDS (%) FDI (%) FDS (%) FO + Rhizobum sp. TZSR12C 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a FO + Rhizobum sp. TZSR25B 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a FO + Bradyrhizobium sp. TZSR41A 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a FO + NOB 61.33 ± 5.67b 20.00 ± 4.04ab 50.00 ± 9.81b 10.00 ± 1.73b FO + T. harzianum 11.33 ± 5.67a 2.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a FS + Rhizobum sp. TZSR12C 16.67 ± 9.53a 5.33 ± 3.93a 0.00 ± 0.00a 0.00 ± 0.00a FS + Rhizobum sp. TZSR25B 5.67 ± 5.67a 1.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a FS + Bradyrhizobium sp. TZSR41A 0.00 ± 0.00a 0.00 ± 0.00a 5.67 ± 5.67a 0.00 ± 0.00a FS + NOB 61.33 ± 5.67b 29.00 ± 5.86ab 38.67 ± 5.67b 8.00 ± 1.00b FS + T. harzianum 5.67 ± 5.67a 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a MP + Rhizobum sp. TZSR12C 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a MP + Rhizobum sp. TZSR25B 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a MP + Bradyrhizobium sp. TZSR41A 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a MP + NOB 5.67 ± 0.00a 1.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a MP + T. harzianum 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a RS + Rhizobum sp. TZSR12C 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a RS + Rhizobum sp. TZSR25B 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a RS + Bradyrhizobium sp. TZSR41A 11.00 ± 11.00a 2.33 ± 2.33a 0.00 ± 0.00a 0.00 ± 0.00a RS + NOB 66.67 ± 9.53b 38.67 ± 22.26b 39.00 ± 11.00b 7.67 ± 2.33b RS + T. harzianum 16.67 ± 9.53a 3.33 ± 2.03a 0.00 ± 0.00a 0.00 ± 0.00a p-Value <0.001 <0.001 <0.001 <0.001 Antagonistic activity of rhizobia and T. harzianum against combination of two fungal pathogens
-
Rhizobia and T. harzianum acted differently in suppressing the growth of combined two fungal pathogens under greenhouse conditions (Table 4). The highest (100%) infection with 33.33% severity in roots, and 77.67% infection with 25.67% severity in foliage was observed in the combination of R. solani with F. oxyporum in contaminated soils without biocontrol rhizobia and T. harzianum. In the plants treated with biocontrols, Rhizobum sp. TZSR12C cleared the infection up to 0.00%, in both roots and foliage of plants, in the soils contaminated with combinations of F. oxyporum and M. phaseolina, R. solani and F. oxyporum and R. solani and M. phaseolina. On the other hand, the highest infection (22.33%) and severity (6.33%) in roots, with no infection on the foliage parts for Bradyrhizobium sp. TZSR41A against F. solani and R. solani was noticed. Rhizobia strains and T. harzianum in most treatments were able to reduce infection and severity in roots up to either 1.0% or 0.00% infection in foliage parts.
Table 4. The effect of rhizobia species and T. harzianum as biocontrol inoculants on combination of two root rot fungi of soybean plants.
Treatment RDI (%) RDS (%) FDI (%) FDS (%) FO + MP + Rhizobum sp. TZSR12C 5.67 ± 5.67a 1.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a FO + MP + Rhizobum sp. TZSR25B 5.67 ± 5.67a 1.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a FO + MP + Bradyrhizobium sp. TZSR41A 5.67 ± 5.67a 1.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a FO + MP + NOB 72.33 ± 14.68b 32.33 ± 3.93b 39 ± 14.74b 7.67 ± 2.91a FO + MP + T. harzianum 22.33 ± 5.33a 6.33 ± 3.33a 5.67 ± 5.67a 1.00 ± 1.00a FS + FO + Rhizobum sp. TZSR12C 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a FS + FO + Rhizobum sp. TZSR25B 11.33 ± 5.67a 2.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a FS + FO + Bradyrhizobium sp. TZSR41A 5.67 ± 5.67a 1.00 ± 1.00a 5.67 ± 5.67a 1.00 ± 1.00a FS + FO + NOB 83.33 ± 16.67b 43.33 ± 8.82b 61 ± 14.74bc 22.33 ± 7.86bc FS + FO + T. harzianum 16.67 ± 9.53a 3.33 ± 2.03a 5.67 ± 5.67a 1.00 ± 1.00a FS + MP + Rhizobum sp. TZSR12C 11.33 ± 5.67a 2.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a FS + MP + Rhizobum sp. TZSR25B 5.67 ± 5.67a 1.00 ± 1.00a 5.67 ± 5.67a 1.00 ± 1.00a FS + MP + Bradyrhizobium sp. TZSR41A 17.00 ± 0.00a 3.00 ± 0.00a 5.67 ± 5.67a 1.00 ± 1.00a FS + MP + NOB 88.67 ± 5.67b 30.00 ± 6.81b 72.33 ± 5.33c 24.33 ± 5.93bc FS + MP + T. harzianum 11.33 ± 5.67a 2.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a FS + RS + Rhizobum sp. TZSR12C 5.67 ± 5.67a 1.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a FS + RS + Rhizobum sp. TZSR25B 5.67 ± 5.67a 1.00 ± 1.00a 5.67 ± 5.67a 1.00 ± 1.00a FS + RS + Bradyrhizobium sp. TZSR41A 22.33 ± 5.33a 6.33 ± 3.33a 0.00 ± 0.00a 0.00 ± 0.00a FS + RS + NOB 83.33 ± 9.53b 33.33 ± 3.76b 61.33 ± 5.67bc 33.33 ± 6.67c FS + RS + T. harzianum 11.33 ± 5.67a 2.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a RS + FO + Rhizobum sp. TZSR12C 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a RS + FO + Rhizobum sp. TZSR25B 5.67 ± 5.67a 1.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a RS + FO + Bradyrhizobium sp. TZSR41A 5.67 ± 5.67a 1.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a RS + FO + NOB 100 ± 0.00b 33.33 ± 6.67b 77.67 ± 5.33c 25.67 ± 4.67bc RS + FO + T. harzianum 11.33 ± 5.67a 2.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a RS + MP + Rhizobum sp. TZSR12C 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a RS + MP + Rhizobum sp. TZSR25B 5.67 ± 5.67a 1.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a RS + MP + Bradyrhizobium sp. TZSR41A 11.33 ± 5.67a 2.00 ± 1.00a 5.67 ± 5.67a 1.00 ± 1.00a RS + MP + NOB 88.67 ± 5.67b 28.67 ± 4.33b 67.00 ± 0.00bc 13.00 ± 0.00ab RS + MP + T. harzianum 11.33 ± 5.67a 2.00 ± 1.00a 5.67 ± 5.67a 1.00 ± 1.00a p-Value <0.001 <0.001 <0.001 <0.001 FS = F. solani, MP = M. phaseolina, RS = R. solani and FO = F. oxyporum, NOB = no biocontrol and TZSR = Tanzania soybean rhizobia, RDS = root disease severity, FDI = foliar disease incidence, and FDS = foliar disease severity. Antagonistic activity of rhizobia and T. harzianum against combination of three fungal pathogens
-
Rhizobia strains and T. harzianum possessed different abilities in suppressing the growth of combined three fungal pathogens in the rhizosphere of plants (Table 5). For the contaminated soils without biocontrol, the highest (100.00%) infection with severity of 40.00% in roots, and 83.00% infection with severity of 27.67% in foliage was determined in combination of R. solani, F. oxyporum and M. phaseolina. For the plants treated with biocontrols (rhizobia and T. harzianum), the highest (33.33%) infection with severity of 10.00% in roots and 0.00% infection in foliage was observed in T. harzianum against the combination of R. solani, F. oxyporum and M. phaseolina. Except for Rhizobum sp. TZSR25B against the combination of F. solani, R. solani and F. oxyporum having the infection of 5.67% and severity of 1.00%, the two Rhizobium sp. TZSR12C and TZSR25B had cleared infection in the foliage for all treatments.
Table 5. The effect of rhizobia species and T. harzianum as biocontrol inoculants on combination of three root rot fungi of soybean plants.
Treatment RDI (%) RDS (%) FDI (%) FDS (%) FS + FO + MP + Rhizobum sp. TZSR12C 22.33 ± 5.33a 4.33 ± 1.33a 0.00 ± 0.00a 0.00 ± 0.00a FS + FO + MP + Rhizobum sp. TZSR25B 17.00 ± 0.00a 3.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a FS + FO + MP + Bradyrhizobium sp. TZSR41A 22.33 ± 5.33a 4.33 ± 1.33a 11.33 ± 5.67a 2.00 ± 1.00a FS + FO + MP + NOB 89.00 ± 11.00b 46.67 ± 6.67b 44.33 ± 5.67bc 17.67 ± 2.33b FS + FO + MP + T. harzianum 22.33 ± 5.33a 4.33 ± 1.33a 5.67 ± 5.67a 1.00 ± 1.00a FS + RS + FO + Rhizobum sp. TZSR12C 17.00 ± 0.00a 3.00 ± 0.00a 0.00 ± 0.00a 0.00 ± 0.00a FS + RS + FO + Rhizobum sp. TZSR25B 17.00 ± 0.00a 3.00 ± 0.00a 5.67 ± 5.67a 1.00 ± 1.00a FS + RS + FO + Bradyrhizobium sp. TZSR41A 5.67 ± 5.67a 1.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a FS + RS + FO + NOB 100.00 ± 0.00b 40.00 ± 11.55b 72.33 ± 5.33cd 29.00 ± 2.00c FS + RS + FO + T. harzianum 22.33 ± 14.68a 7.67 ± 6.23a 16.67 ± 16.67ab 3.33 ± 3.33a FS + RS + MP + Rhizobum sp. TZSR12C 5.67 ± 5.67a 1.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a FS + RS + MP + Rhizobum sp. TZSR25B 11.33 ± 5.67a 2.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a FS + RS + MP + Bradyrhizobium sp. TZSR41A 22.33 ± 5.33a 6.33 ± 3.33a 5.67 ± 5.67a 1.00 ± 1.00a FS + RS + MP + NOB 100.00 ± 0.00b 40 ± 11.55b 83.00 ± 0.00d 27.67 ± 5.33c FS + RS + MP + T. harzianum 22.33 ± 14.68a 4.33 ± 2.96a 5.67 ± 5.67a 1.00 ± 1.00a RS + FO + MP + Rhizobum sp. TZSR12C 5.67 ± 5.67a 1.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a RS + FO + MP + Rhizobum sp. TZSR25B 11.33 ± 5.67a 2.00 ± 1.00a 0.00 ± 0.00a 0.00 ± 0.00a RS + FO + MP + Bradyrhizobium sp. TZSR41A 11.33 ± 5.67a 2.00 ± 1.00a 17.00 ± 0.00ab 3.00 ± 0.00a RS + FO + MP + NOB 94.33 ± 5.67b 43.33 ± 3.33b 72.33 ± 5.33cd 29.00 ± 2.00c RS + FO + MP + T. harzianum 33.33 ± 16.67a 10.00 ± 5.77a 0.00 ± 0.00a 0.00 ± 0.00a p-Value <0.001 <0.001 <0.001 <0.001 * FS = F. solani, MP = M. phaseolina, RS = R. solani and FO = F. oxyporum, NOB = no biocontrol and TZSR = Tanzania soybean rhizobia, RDS = root disease severity, FDI = foliar disease incidence and FDS = foliar disease severity. Antagonistic activity of rhizobia and T. harzianum against combination of four fungal pathogens
-
The ability of rhizobia strains and T. harzianum against the combination of four fungal pathogens is presented in Table 6. The highest infection of 77.78% with severity of 46.67% in roots, and infection of 72.22% with severity 40.00% in foliage was observed in the treatment without any biological control agents. This was distantly followed by 38.89% of infection with 7.78% severity in roots, and 5.67% infection with 1.00% severity in T. harzianum against the combination of four fungal pathogens. The lowest infection (27.78%) with 5.56% severity in roots and without any spread of infection in foliage was observed in Rhizobium sp. TZSR12C against the combination of four fungal pathogens.
Table 6. The effect of rhizobia species and Trichoderma harzianum as biocontrol inoculants on combination of four root rot fungi of soybean plants.
Treatment RDI (%) RDS (%) FDI (%) FDS (%) FS + RS + FO + MP + Rhizobium sp. TZSR12C 27.78 ± 5.56a 5.56 ± 1.11a 0.00 ± 0.00a 0.00 ± 0.00a FS + RS + FO + MP + Rhizobium sp. TZSR25B 27.78 ± 5.56a 7.78 ± 2.94a 5.56 ± 5.56a 1.111 ± 1.11a FS + RS + FO + MP + Bradyrhizobium sp. TZSR41A 27.78 ± 5.56a 10.00 ± 3.33a 11.11 ± 5.56a 2.222 ± 1.11a FS + RS + FO + MP + NOB 77.78 ± 14.7b 46.67 ± 8.82b 72.22 ± 11.11b 40.00 ± 10.00b FS + RS + FO + MP + T. harzianum 38.89 ± 5.56ab 7.78 ± 1.11a 5.67 ± 5.67a 1.00 ± 1.00a p-Value 0.02 <0.001 <0.001 <0.001 FS = F. solani, MP = M. phaseolina, RS = R. solani and FO = F. oxyporum, NOB = no biocontrol and TZSR = Tanzania soybean rhizobia, RDS = root disease severity, FDI = foliar disease incidence, and FDS = foliar disease severity. -
In this study, Bradyrhizobium sp. TZSR41A and two Rhizobium sp. TZSR12C and TZSR25B have been observed to be capable of suppressing the selected root rot fungi (F. solani, F. oxyporum, R. solani and M. phaseolina) both under in vitro conditions. Similarly, Parveen et al.[1] identified the ability of Rhizobium sp. and Bradyrhizobium sp. in inhibiting the growth of particular fungal pathogens under in vitro conditions. The strains of rhizobia were observed to produce different colours in inhibiting the growth of different fungal pathogens. The changes in the colour of colonies in an antagonistic activity against fungi indicates the production of antibiotics, HCN or mycolytic enzymes[10,26].
Experimental soil's ability to support microbial activities and growth of soybeans
-
An experimental soil was examined for a variety of physico-chemical traits prior to validation of the ability of rhizobia isolates to prevent the infection of seedlings by fungal pathogens under greenhouse conditions. According to Al-Saedi et al.[27], the soil's neutral pH and medium Sandy Clay Loam (SCL) texture indicate that it is suitable for a different rhizobia populations and their activities. According to previous studies[28,29], soil pH is the best indicator for determining the availability of macro- and micronutrients that support plant growth and development for optimum yield. The very high percentage of soil organic carbon and the moderate total nitrogen levels found in this study suggest that the soil has been enriched with organic matter[28,30,31] . Higher carbon levels are crucial for supplying energy for rhizobial activities, while medium to low nitrogen levels are enough for the biological nitrogen fixation process[30,31]. A very high concentration of available P in an experimental soil notifies its sufficiency for energy acquisition, storage, and utilization in microbial activities[32,33]. Much higher levels of micronutrients Zn, Mn, and Fe, and a higher level of Cu suggest the availability of particular nutrients for uptake by plants and serving various functions for the growth of plants and microbial interactions[29−32].
Exchangeable bases, Ca, Mg, and K serves different essential roles in soils to support the activities rhizobia and the growth and development of plants[34−36]. The higher level of calcium in this study will promote the abundance of rhizobia, development of roots, hence enhance the access of rhizobia to root hairs, root infection by rhizobia and hence nodulation[37−41]. Magnesium, is important in stabilization of cell membrane, nucleic acids, and ribosomes of the microbes[42]. Additionally, the higher concentrations of potassium will help in the regulation of water by the plant and influence the growth of roots. Furthermore, the CEC of an experimental soil was medium, indicating the balance of the soil's exchangeable bases and acid forming cations, Al3+ and H+[43]. The very low exchangeable Na is however, required for microbial activities to avoid salinity stress. Exchangeable potassium can replaced by the presence of potassium[37,44]. In general, physico-chemical properties of soil used in this study was suitable in supporting rhizobia activities.
Influence of rhizobia strains in enhancing the germination of inoculated seeds
-
The strains of rhizobia in this study were observed to enhance the germination of soybean seeds in the soils inoculated with sole or a combination of different fungal pathogens. The ability of rhizobia in enhancing the germination of legumes including soybean have been demonstrated by different studies[6,26]. In this study, Rhizobium sp. TZSR12C improved the germination up to 93.33% in soils inoculated with F. solani, R. solani and M. phaseolina and up to 96.67% by Rhizobium sp. TZSR25B in non-contaminated soils however, the seeds inoculated with T. harzianum had the least germination of 70%. The improvement percentages germination by rhizobia strains in this study are distinctly higher than the 83.3% obtained in previous studies[6,45], suggesting the higher efficiency of the tested rhizobia species in influencing the germination of seeds.
Antagonistic activities of rhizobia strains under greenhouse conditions
-
The ability of rhizobia in suppressing root rot disease-causing pathogens such as Fusarium solani F. oxysporum, Macrophomina phaseolina, and F. solani have been observed in different studies[1,4,6,46]. Different strains that form symbiotic nodules and fix nitrogen in soybeans have been studied on their abilities to perform the dual purpose of fixing nitrogen as well as suppressing root rot diseases[1,6,47,48]. In this study, Bradyrhizobium sp. TZSR41A and two Rhizobium sp. TZSR12C and TZSR25B as well as T. harzianum have been observed to be capable of suppressing root rot fungi (F. solani, F. oxyporum, R. solani and M. phaseolina) in soybean seedlings under greenhouse conditions. Similarly, some studies[1,15,49,50] identified the ability of Bradyrhizobium sp. and Rhizobium sp. to suppress F. oxyporum, F. solani, R. solani, and M. phaseolina in the roots of soybean plants.
On the other hand, previous studies[6,51], demonstrated the ability and efficiency of R. japonicum in inhibiting the infection of soybean roots and foliage parts. In this study, the rhizobia were capable completely inhibiting the infection and severity to 0.00% which is equivalent to 100% inhibition for the occurrence of single pathogen. The ability of Rhizobium sp. TZSR12C in clearing the infection to 0.00% in F. solani with F. oxyporum, R. solani with F. oxyporum and R. solani with M. phaseolina indicates the highest efficiency of the strain against the particular pathogens[6,52]. However, there are limited reports on the ability of rhizobia in suppressing the combination of three or four fungal pathogens in the rhizosphere of plants. Being capable of suppressing the combination of different fungal pathogens is an indication of the highest efficiency of inhibiting the growth and protecting the plant roots from infection by pathogens. However, this needs the confirmation of their abilities under field conditions.
-
This study identified three stains of rhizobia, Rhizobium sp. TZSR12C, Rhizobium sp. TZSR12C and Bradyrhizobium sp. TZSR41A, which are capable of suppressing the common species of root rot fungal pathogens affecting the soybean, both under in vitro and greenhouse conditions. Under in vitro conditions, Rhizobium sp. TZSR25B had the highest performance by forming the largest colony diameter and inhibition zone against F. oxyporum. Under greenhouse conditions, Rhizobium sp. TZSR12C and TZSR25B had better performance in enhancing germination of seeds in the soils contaminated with fungal pathogens. Rhizobium sp. TZSR25B possessed the highest performance in suppressing the infection against individual fungi and in enhancing the germination in most of the treatments while Rhizobum sp. TZSR12C had the highest performance in combination of two, three, and four pathogens. The results of this study give an insight into the suitability of the tested rhizobia species as effective biocontrol agents for the selected root rot fungi. However, the test was conducted only under in vitro and greenhouse conditions hence, future research should focus on on-farm trials to validate the biocontrol potential of the identified rhizobia strains in real-world agricultural settings. Additionally, further investigations into the mechanisms of interaction between the rhizobia strains and specific fungal pathogens would contribute to a more comprehensive understanding of their biocontrol capabilities.
-
The authors confirm contribution to the paper as follows: conceptualization and methodology: Nakei MD, Ndakidemi PA, Venkataramana PB; original draft preparation: Nakei MD; review and editing: Ndakidemi PA, Venkataramana PB. All authors have read and agreed to the published version of the manuscript.
-
The datasets generated or analyzed during the current study are available from the corresponding author on reasonable request.
-
The authors thank all staff and technical experts from Nelson Mandela African Institution of Science and Technology (NMAIST), Arusha-Tanzania for their guidance and support during sampling, laboratory and greenhouse experiments.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Nakei MD, Venkataramana PB, Ndakidemi PA. 2024. Evaluating Rhizobium and Bradyrhizobium species as potential biocontrol agents for root rot fungi in soybean seedlings. Technology in Agronomy 4: e010 doi: 10.48130/tia-0024-0007
Evaluating Rhizobium and Bradyrhizobium species as potential biocontrol agents for root rot fungi in soybean seedlings
- Received: 03 October 2023
- Revised: 27 March 2024
- Accepted: 07 April 2024
- Published online: 20 May 2024
Abstract: Soybean (Glycine max) is among the legumes that are highly prone to soil-borne pathogens that causes root-rot diseases limiting growth and development and resulting in low yields of plants. This study was conducted to test the ability of three rhizobia strains, Rhizobium sp. TZSR12C, Rhizobium sp. TZSR25B and Bradyrhizobium sp. TZSR41A, in comparison with the commercial biocontrol (Trichoderma harzianum in suppressing the growth of root rot fungal pathogens (Fusarium solani, Rhizoctonia solani, Fusarium oxyporum, and Macrophominaphaseolina) in vitro and greenhouse conditions. The rhizobium cell filtrates were used in testing their activities against fungal pathogens under in vitro while the solid biofertilizer formulations containing the respective rhizobia inoculants were used to inoculate the soybean seeds sown in pathogen-contaminated soil under greenhouse conditions. Results showed that all rhizobia isolates and T. harzianum were capable of suppressing the fungal pathogens both under in vitro and greenhouse conditions with the highest inhibition zone (8.3 mm) and colony diameter (25.0 mm) being in Rhizobium sp. TZSR25B against F. oxyporum under in vitro conditions. In greenhouse experiments, Rhizobium sp. TZSR12C had the highest performance in inhibiting the infection of plant up to 27.78% with severity of 5.56% in roots and 0.00% infection in foliage against the combination of F. solani, R. solani, F. oxyporum, and M. phaseolina. It was found that, on their performance, the tested rhizobia strains can potentially be utilized as biocontrol agents against the fungal pathogens in the rhizosphere of soybean plants.
-
Key words:
- Fungal pathogens /
- Rhizobia strains /
- Root rot disease /
- Suppression














