-
The demand for food has sharply increased because of the growing population[1,2]. Improving the LUE of crops represents the primary option for increasing crop yield potential[3]. LUE refers to the efficiency by which a crop produces biomass from absorbed light energy, which is measured by the accumulation rate of photosynthetically active radiation in relation to biomass per unit of intercepted or absorbed light[4]. The improvement of LUE depends on the rate of photosynthesis and the efficiency of converting light energy into fixed carbon[5]. The principal methods of improving LUE include (i) extending photosynthetic time, (ii) increasing the area of photosynthesis, and (iii) increasing the rate of photosynthesis. Therefore, enhancing photosynthetic efficiency is crucial for increasing yield[6]. Photosynthesis is the process by which plants and photosynthetic bacteria use light energy and CO2 to produce carbohydrates and O2[7]. During photosynthesis, plants convert light energy into chemical energy, which is used for plant growth. Chlorophyll plays an important role in photosynthesis in higher plants and has multiple functions in this process[8]. Related proteins bind with chlorophyll and form complexes that capture, convert, and redirect light energy[8]. Chl a and Chl b are the primary substances of the core protein complex and light-harvesting antenna protein complex of photosystems, respectively[9].
Role of LPOR in chlorophyll synthesis and chloroplast development
-
Chlorophyll biosynthesis is a complex process completed by multiple enzymes (Fig. 1). Accurate and stable chlorophyll synthesis is critical for plant growth and development because free chlorophyll and its precursors, the tetrapyrrole compounds can be photosensitive and phototoxic to cells. This effect is most pronounced in the photosystem II reaction center, which is highly exposed to oxidative damage[10−12]. Chlorophyll synthesis consists of two parts. The first part is the common pathway of tetrapyrrole substance synthesis, starting from the synthesis of glutamyl tRNA and ending with Pro. This pathway involves the enzymes glutamine tRNA reductase, delta-amino ketones pentanoic acid dehydratase, uroporphyrinogen III synthase, coproporphyrinogen III oxidase, and protoporphyrinogen oxidase. The second aspect involves Pro chelation with Mg2+ in the chlorophyll synthesis pathway, requiring involved enzymes Mg-chelatase, Mg-protoporphyrin IX methyltransferase, POR, chlorophyllide a oxygenase, chlorophyll synthase, and chlorophyllase. The rate-limiting enzyme POR performs a crucial catalytic role in chlorophyll synthesis by transforming Pchlide into Chlide[13]. LPOR also participates in chloroplast development and serves as the primary protein component in PLBs of etioplasts[14,15]. Two types of POR exist in nature: DPOR (EC 1.3.7.7) and LPOR (EC 1.3.1.33)[16]. From evolutionary terms, DPOR is the older enzyme, a multisubunit enzyme containing three separate subunits, and very similar to nitrogen-fixing enzymes. DPOR catalyzes Pchlide reduction in an ATP-dependent manner and is oxygen-sensitive, and is present in non-flowering land plants, algae, and cyanobacteria[17]; LPORs are thought to have evolved in cyanobacteria about 2 billion years ago as a result of increased atmospheric oxygen levels and are not sensitive to oxygen[18]. LPOR is present in a wide range of organisms, including plants, algae, cyanobacteria, and anaerobic photosynthetic bacteria[19−21]. However, only LPOR is present in angiosperms, indicating that induction with light is necessary for angiosperms to produce chlorophyll[22,23]. These characteristics make LPOR essential in chlorophyll biosynthesis and chloroplast development in angiosperms.
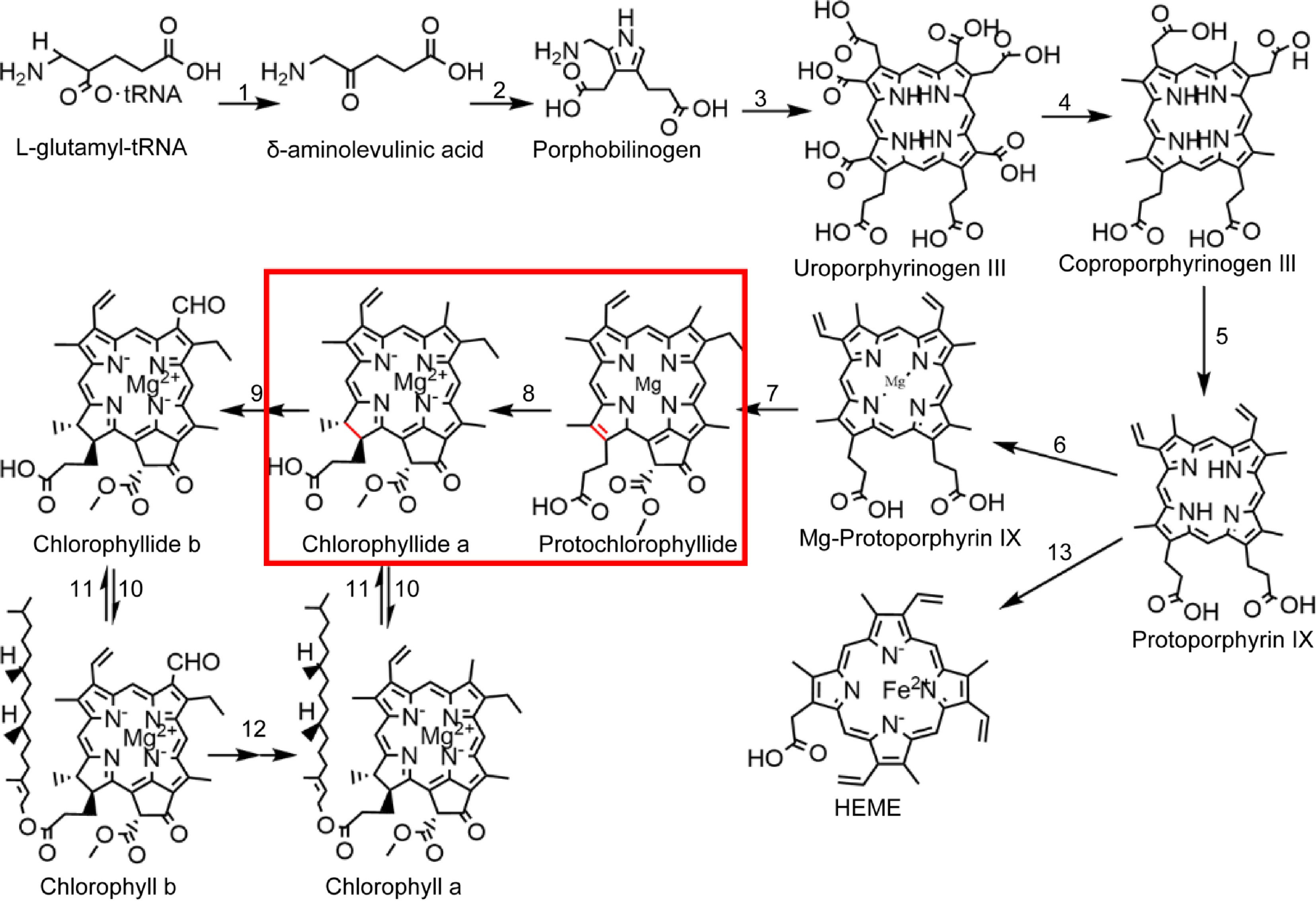
Figure 1.
Chlorophyll biosynthesis pathway. 1: glutamine-tRNA reductase, 2: delta-amino ketones pentanoic acid dehydratase, 3: urinary porphyrins original III synthetase, 4: coproporphyrin III oxidase, 5: protoporphyrin oxidase, 6: Mg-chelating enzyme, 7: Mg-protoporphyrin IX methyltransferase, 8: the original chlorophyll acid ester oxidoreductase, 9: chlorophyll acid ester a oxygenase, 10: chlorophyll synthase, 11: chlorophyllase, 12: chlorophyllide a oxygenase, 13: ferrous chelase.
A multitude of investigations have demonstrated that LPOR plays a pivotal role in the response to abiotic stress, influencing the biosynthesis of chlorophyll, and exhibiting a certain level of stress resistance. A synthesis was conducted of the research conducted domestically and internationally on LPOR, encompassing its structural characteristics, catalytic reaction mechanism, optical signal, temperature, moisture, and the factors influencing its activity.
-
LPOR is a single polypeptidase that is encoded within the nucleus. It has a transporter protein segment at its N-terminus. This transporter protein segment transports LPOR into the plastid after it is transcribed. LPOR is highly similar to the SDR family and needs photoactivation to reduce Pchlide and perform biological functions[24]. Previous studies investigated LPOR in several species, including Arabidopsis thaliana[25], Brassica oleracea[26], Oryza sativa[27], Hordeum vulgare[28], Zea mays[29], Nicotiana tabacum[30], Cucumis sativus[31], and Pisum sativum[32]. Pchlide, the precursor of chlorophyll synthesis, failed to undergo the requisite reduction in time, thereby allowing the production of a substantial amount of ROS upon exposure to light, which resulted in the oxidative damage and chlorophyll bleaching of the seedling[33]. In addition, the accumulation of ROS in chloroplasts also impairs the de novo synthesis of protein D1 (also known as photosystem b-a or PsbA), which is essential for PSII repair[34]. The enzyme LPOR catalyzes the conversion of pchlide into chlide, a process which enables the developing seedling to gain the capacity to perform photosynthesis[35]. The production of chlorophyll is thus facilitated, thus allowing the seedling to grow in an autotrophic manner[15].
The different gene expression patterns of LPOR subtypes prevent direct photooxidative harm in yellow seedlings when exposed to light. In Arabidopsis, three LPOR subtypes, namely, LPORA, LPORB, and LPORC were identified[36,37]. At the early stage of plant development, AtPORA and AtPORB are expressed during etiolation in Arabidopsis seedlings[35]. AtPORB and AtPORC are synthesized abundantly in slightly mature seedlings and, subsequently, in mature plants[35]. Upon light exposure, the expression levels of AtPORB and AtPORC are directly correlated to Chl a content and the stacking of thylakoids in seedlings[38,39]. The cpSRP43 as a chaperone, stabilizes the enzyme and provides the optimal quantity of PORB during leaf greening and heat shock[40]. In contrast, cpSRP54 enhances its binding to the thylakoid membrane, thus ensuring a sufficient level of metabolic flux during late chlorophyll biosynthesis[40]. Two LPOR subtypes, namely, OsPORA and OsPORB, were identified in rice[27]. During early leaf development, OsPORA is expressed in darkness, whereas OsPORB is expressed throughout the entire leaf development process regardless of light conditions[41]. Previous studies demonstrated that although OsPORA and OsPORB have overlapping biochemical functions, the response of OsPORB to constant light or physiological functions during reproductive growth cannot be substituted with OsPORA[27]. Barley also has two LPOR subtypes[16]. In vitro measurement revealed that HvPORA is expressed in etiolated seedlings, and its expression is downregulated after light exposure[42]. On the contrary, HvPORB is expressed during morphogenesis in leaf development regardless of light conditions[42]. In addition, barley etioplast contains a distinctive light-harvesting complex called LHPP, of which HvPORA is an essential component[42]. The LHPP complex is prepared in advance for the LPOR-catalyzed transformation of Pchlide under light to prevent the free Pchlide phototoxicity after light exposure, which causes photobleaching of seedlings[43].
Structural characteristics and functions of LPOR
-
A comparison of the amino acid sequences of LPOR in barley and Synechocystis reveals that the conserved Cys residue sequence is closely linked to the binding and catalysis of substrate Pchlide conversion. Point mutation experiments on various Cys residues revealed that Cys276 is the active site for Pchlide binding. Additionally, Cys303 functions as a pigment-binding site with low affinity. Both Cys residues participate in the assembly and stabilization of PORB in the etioplast[44,45].
LPOR’s structure, along with other SDR family members[24], contains the Rossmann-fold structure that binds dinucleotides[46]. The structure has three flexible regions positioned at amino acid residues 146–160, 228–255, and 284–291. Binding to the NADPH binding site of LPOR is closely associated with amino acid residues 146–160 and 228–255, whereas amino acid residues 284–291 plays a crucial role in regulating substrate Pchlide binding. In contrast to the other SDR family members, the SDR proteins stand out for utilizing Asn-Ser-Tyr-Lys to facilitate proton transfer from tetrads, leading to the production of stable reaction intermediates[46]. However, LPOR employs Thr residues, particularly Thr145, to replace the Ser residues. The structural modeling of the ternary enzyme-substrate complexes constructed from crystal and electron microscopy data also confirmed the differences in the orientation of LPOR to the substrate Pchlide and the structure of the LPOR active site[47−49]. Further results indicate that the conserved Tyr and Gln residues in LPOR are essential for Pchlide binding, while the active site Cys residue is crucial for both hydride and proton transfer reactions in LPOR[48].
LPOR participates in both chlorophyll biosynthesis and chloroplast development[50]. Chloroplasts originate from proplastids, and etioplasts are transitional forms of chloroplast development. Etioplasts are characterized by the lack of chlorophyll and possession of a distinctive membrane structure called PLBs[15]. LPOR is the main protein constituent of PLBs, comprising more than 90% of the total protein content of PLBs[51]. Carotenoids act in parallel with DET1 to regulate the transcriptional formation of LPOR and plastids PLBs, thereby controlling chloroplast development[52]. PLBs have diverse LHPP complexes with distinct absorption spectra, namely, LPOR-Pchlide633, LPOR-Pchlide640, and LPOR-Pchlide655, with peak absorptions at 633, 640, and 655 nm, respectively[26,53]. The photoactive binary complex of LPOR-Pchlide655 binds to NADPH after light detection and catalyzes Pchlide reduction, leading to light conversion and gradual formation of Chl[53]. Moreover, the decomposition of the PLB lattice structure initiates grana stacking, leading to the complete development of chloroplasts[50]. Nonetheless, LPOR-Pchlide633, which is not photoactive, can degrade after exposure to light, thereby triggering an outburst of ROS in chloroplasts. Severe instances of such degradations may result in cell death[32].
Reaction mechanism of Pchlide reduction to Chlide catalyzed by LPOR
-
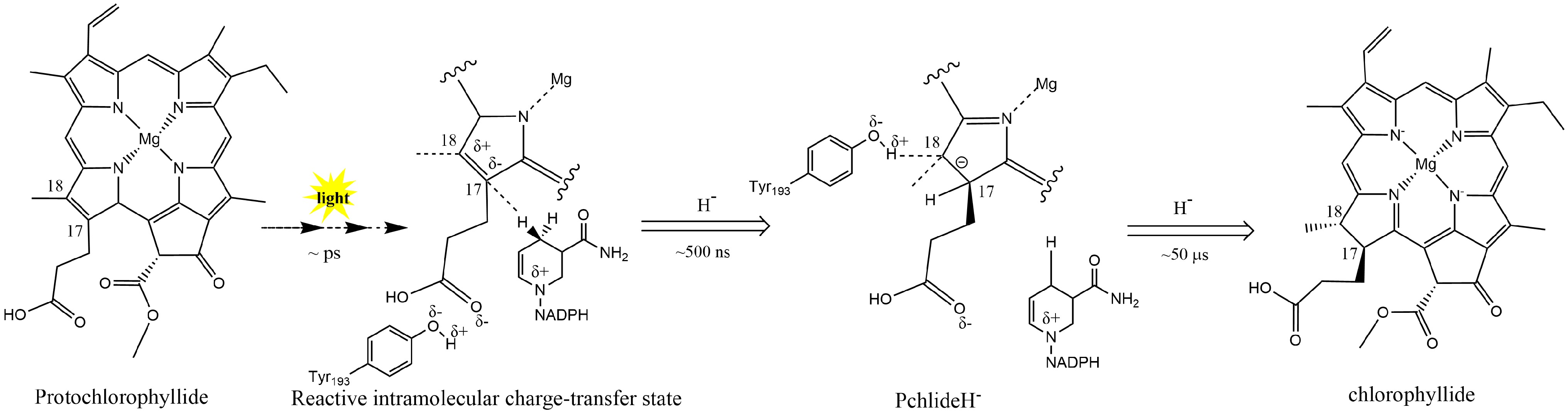
In plants, Pchlide reduction is an important rate-limiting step within the chlorophyll synthesis pathway. In angiosperms lacking DPOR, chlorotic seedlings rely on LPOR to turn green. LPOR requires light-induced NADPH as a reducing agent to catalyze the Pchlide conversion. In regular plant development, LPOR-Pchlide-NADPH forms a ternary complex that accumulates during the dark morphogenesis of the etioplast; the NADPH hydride on the nicotinamide ring is transferred to the C17 position of Pchlide through light induction (Fig. 2). Then, the conserved Tyr residue transfers a proton to the C18 position of Pchlide. Thus, the C17 and C18 double bonds of the Pchlide-D ring are reduced, resulting in the creation of Chlide. Then, Chlide undergoes esterification and further modifications to form Chl a and Chl b[47,54].
Method of LPOR entering plastids
-
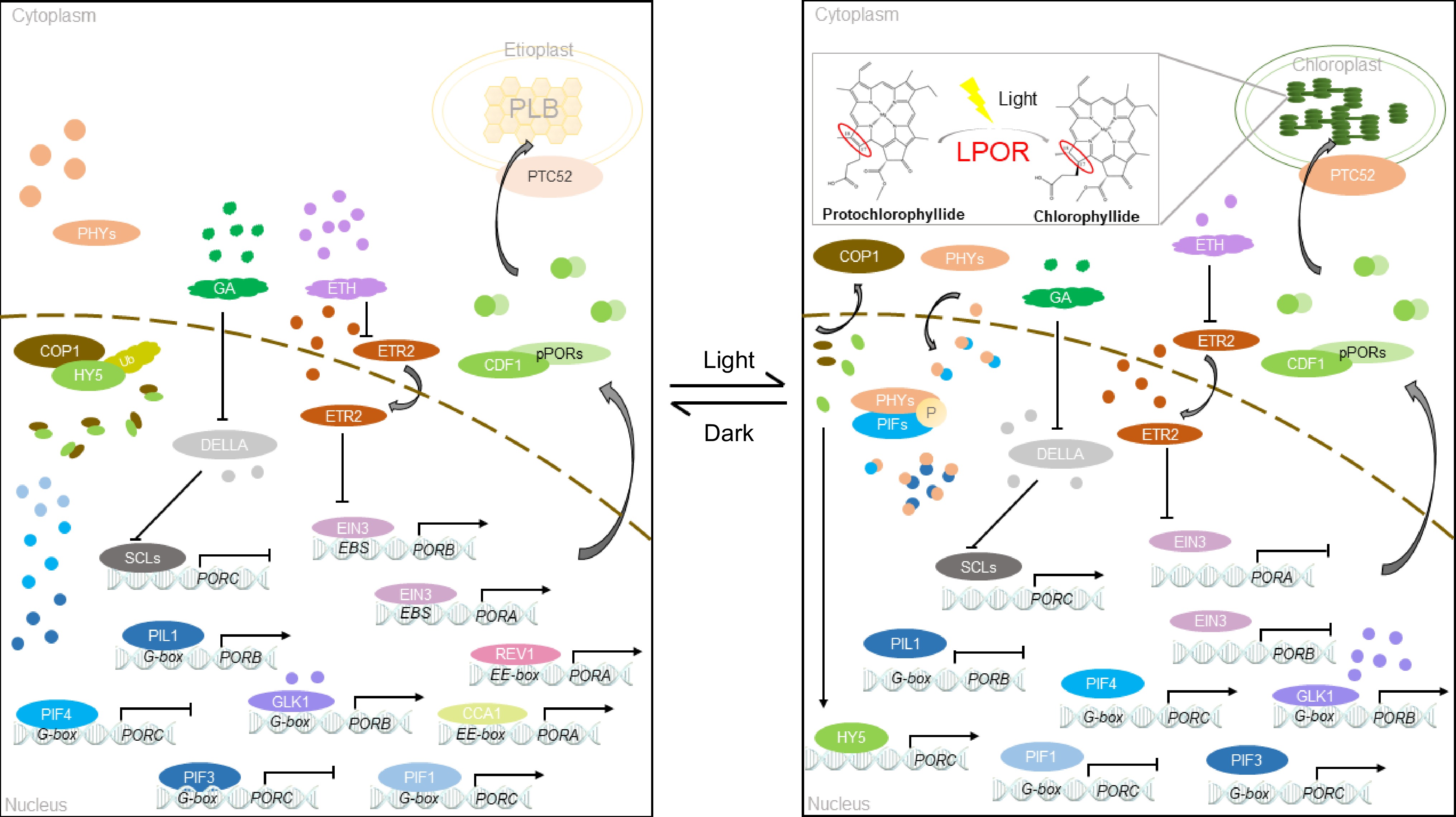
LPOR is a protein encoded in the nucleus, synthesized into large pPORs in the cytoplasm, and then modified to enter the plastid[55] (Fig. 3). The process of LPORs entering chloroplasts is distinct[56]. Prior studies verified that pPORA relies on substrates to enter chloroplasts, whereas pPORB does not require any substrate for entry[57]. TOC33 is an essential core component in the complex of PORA and PORB import channels in cotyledons and leaves[56,58]. PTC52 is a unique Pchlide a oxygenase complex located in the plastid envelope. It is responsible for associating the synthesis of Pchlide b with the import of pPORA[59]. In RNAi plants lacking PTC52 transcripts and proteins, pPORA cannot be imported to plastids normally. This phenomenon causes an excessive accumulation of Pchlide and leads to ROS accumulation and cell death during greening[59]. In addition, CDF1 present on thylakoids and capsules interacts with the LPOR subtype and plays a crucial role in the introduction and stability of LPOR[55]. Deletion of CDF1 leads to a decrease in LPOR protein accumulation, which normally hinders chlorophyll synthesis, damages PLB formation, affects chloroplast development under light, causes photobleaching of plants under light, and inhibits plant growth[55,60]. Various pathways of LPOR import into plastids guarantee normal chloroplast development and chlorophyll biosynthesis. This finding suggests that factors beyond the components of the core complex of the import channel participates in the transportation of nuclear-encoded plastid proteins. Plastid-localized membrane-bound factors, such as TTP1, play a role in LPOR-directed import into chloroplasts. TTP1 deficiency leads to the accumulation of glutamate receptors, enhances pentosamine ketoglutarate synthesis and reduces POR levels, which in turn leads to increased sensitivity to reactive oxygen species and slower greening of yellowing seedlings[61].
-
For the reduction of Pchlide to occur, LPOR catalysis requires photoactivation[23]. The catalytic efficiency of the reaction varies with different light qualities, and the quantum yield of the reaction is 3–7 times higher under red light (647 nm) than under blue light (407 nm)[62]. Moreover, LPOR's catalytic reaction efficiency varies when combined with different substrates. Several studies suggested that cabbage’s photobleaching is primarily caused by short-wavelength light (625–630 nm) of 7 μmol photons m−2∙s−1. Light with wavelengths longer than 630–640 nm causes bleaching and photoreduction, whereas that with wavelengths higher than 640 nm predominantly causes photoreduction[26]. Similar conclusions can be reached through various methods, such as behavioral spectroscopy, pigment content measurement, and kinetic analysis. These studies indicated that under low light intensity, the excitation energy of the short-wavelength-absorption-type Pchlide causes the photoreduction of long-wavelength-absorption-type Pchlide; in comparison, photobleaching occurs under high photochemical light[63].
The active site of LPOR plays a critical role in its catalytic activity. Tyr275 and Lys279, which are essential active sites of LPOR, along with four conserved Cys residues involved in substrate binding, have a very important function in the operation of LPOR. Tyr275 and Lys279 regulate the efficiency of LPOR photoactive state formation by participating in the coordination of NADPH and Pchlide at enzyme catalytic sites[64]. LPOR photoactivity is regulated by several Cys binding sites with different substrate affinities[58]. Any mutation at these sites can affect LPOR photoactivity[65].
Furthermore, proteins interacting with LPOR regulate their enzyme activity in various ways. The structural stability of the LHPP complex is crucial for plant greening. The presence of TLs has been demonstrated to influence the activation of LPORs in response to regulators[66]. Research indicated that galactosyl diacylglycerol impacts the catalytic reaction of LPOR by influencing the creation and breakdown of the Pchlide-LPOR-NADPH complex, which affects the biosynthesis of plant chlorophyll[67]. Feedback regulation is usual in chlorophyll biosynthesis. Fluorescent protein is one of the tetrapyrrole biosynthesis negative feedback regulators in higher plants. It directly interacts with LPOR and cooperates with LPOR negative feedback to regulate chlorophyll biosynthesis[68]. The protein LIL3 regulates plant greening through direct interaction with LPOR. The mutation in lil3 causes a considerable loss of LPOR protein, affecting its posttranslational modifications, which lead to abnormal chlorophyll synthesis[69]. Pchlide transformation catalyzed by LPOR involves protein phosphorylation. Previous research demonstrated that certain plastid ADP-dependent kinase impacts the membrane association of LPOR via reversible protein phosphorylation regulates PLB formation, promotes light-dependent diffusion, and ultimately facilitates chloroplast development[70].
-
Light is necessary for angiosperms to turn green, and the reaction involved is Pchlide reduction[56]. Several researchers examined prevalent transcription factors in light signaling, and they summarized and predicted the availability of multiple light response elements on the promoter of genes involved in chlorophyll biosynthesis, further highlighting the importance of light signaling in chlorophyll synthesis in plants[71]. Under dark conditions, PORC expression is extremely reduced or completely inhibited because of the suppression of PIF3, histone deacetylase 1, and SCL proteins. Research indicated that the phytochrome proteins PHYA and PHYB, which are photosensitive receptors found in plants, play a role in chlorophyll biosynthesis. When exposed to white light, PHYA and PHYB positively regulates LPOR, promoting chlorophyll biosynthesis[33]. Terminal flower 2 protein located downstream of the PHYA signal regulates the expression of PORA and promotes chlorophyll synthesis[72]. Under shading or low-intensity far-red light, the expression levels of PORB and PORC in the phyA mutant is substantially suppressed[73,74]. Far-red light exposure leads to the far-red-blocked greening phenomenon in plants. PHYA suppresses LPOR gene expression under far-red light, resulting in irreversible plastid damage that restrains proper greening ability of seedlings[75]. The sigma factor is a nuclear-encoded protein regulated by PHYA that participates in the regulation gene expression in chloroplasts and influences reverse signaling from plastids to nuclei to promote plant greening and plastid development[75].
In the dark, PORA activity is initiated by acetylation in the presence of HDAC, which regulates chlorophyll synthesis[76]. Meanwhile, phytochrome interacting factor 1 binds to the G-box DNA sequence element (CACGTG) of the PORC promoter and positively regulates PORC expression[77] (Fig. 3). After exposure to light, PIF3 is phosphorylated and inactivated, and histone H4 is acetylated[78]. Moreover, light facilitates the expression of miR171 and hinders gibberellic acid synthesis, thereby promoting the expression of DELLA protein. However, miR171 and DELLA inhibit the SCL transcription, resulting in a significant increase in the PORC expression under light[79,80] (Fig. 3). In addition, light positively regulates the expression of the transcription factor HY5, which binds directly to PORC to promote its gene expression[81]. HY5 interacts with PHYB in the dark through COP1/SPA1; after exposure to light, the transcription factor HY5 is released. HY5 cooperates with the biological clock of PIFs to regulate the transcription level of PORC[81,82] (Fig. 3).
The dark-to-light transition enables plants to transition from the skotomorphogenesis to the photomorphogenesis state. This process is often accompanied by changes in phytohormones, which regulate the expression of different PORs in different ways[83]. EIN 3 and EIN 3-like 1 positively regulate PORA and PORB[84]. Cytokinins significantly enhance the transcription of POR mRNA and accelerate plant greening, whereas abscisic acid has the opposite effect[85]. Auxin binds to the promoters of PORA and GUN5 through ARF2 and ARF7 to inhibit their expression directly, with the help of IAA14[86]. Additionally, growth hormones are known to inhibit chlorophyll biosynthesis. Inhibition of their expression directly inhibits chlorophyll biosynthesis[86]. In Arabidopsis, the structures of PORA and PORB are mostly the same, except for the initial transit peptide. However, they perform different functions and cannot replace each other. PORA solely performs during the initial stage of light exposure in yellowing seedlings, and light significantly inhibits PORA. Following light exposure, the expression of PORA declines rapidly, whereas PORB stays constantly expressed.[25].
The regulatory mechanisms of PORA, PORB, and PORC are interconnected and not completely independent. When ethylene is applied under light, EIN3 regulates the transfer of COP1 from the cytoplasm to the nucleus. Thus, the activity of extranuclear COP1 is blocked, and the expression of PORC is inhibited. When ethylene is absent under light, COP1 mainly exists in the cytoplasm, and HY5 initiates PORC transcription[87] (Fig. 3).
The expression of LPOR mRNA exhibits noteworthy cyclic variations[88]. Reveille 1 directly binds to the PORA promoter through the EE motif (AAAATATCT) and regulates the transcription of PORA[88] (Fig. 3). AtPORB expression is regulated by the biological clock, whereas AtPORC expression is independent of the biological clock. This finding corresponds to the regulation of OsPORB expression observed in rice grown under short-day conditions. The LPOR enzyme in cucumber is encoded by a single gene, with its expression under light being six times greater than that under dark treatment[31]. Upon exposure to light, the LPOR level decreases slightly, followed by a gradual increase in LPOR expression from 3 to 12 h[31].
Research on light signal regulation of LPOR primarily focuses on plants turning green during the transition from darkness to light. Further experiments must be conducted to determine whether changes occur in the expression, content, and enzyme activity of LPOR during shading (including changes in light intensity and quality) and whether it is an essential enzyme affecting chlorophyll biosynthesis during variations in the light environment.
Regulation of LPOR under abiotic stress
-
Changes in LPOR enzyme activity are one of the important factors affecting chlorophyll biosynthesis under abiotic stress[89,90] (Table 1). Shading environments are ubiquitous in nature. Increasing the content of photosynthetic pigments and reducing Chl a/b are important shade tolerance mechanisms for plants[91]. However, the molecular mechanism regulating chlorophyll synthesis under shade has not yet been studied. Previous proteomic studies indicated that compared with the level of LPOR protein in the soybean seedling leaves grown under normal light, that in the soybean seedling leaves grown under shade increases[92−94]. This increase may be a significant reason for the augmentation observed in plants’ chlorophyll content under shade conditions.
Table 1. Effects of different stresses on the LPOR activity, protein, and transcription levels.
Abiotic stress Species Response to stress (transcript and protein expression and enzyme activity) Ref. Water Rice LPOR content decreases. [90] Salt/drought Peanut The expression of AhPORA is downregulated during drought and upregulated during postdrought recovery through AhGLK. [95] Rice LPOR activity is downregulated by 60% in salt-treated seedlings. [89] Chill Rice and LPOR activity is downregulated. [41] Corydalis bungeana Turcz. LPOR's transcript and protein content slightly decline at 4 °C but dramatically decrease at −4 °C with time. [96] Wheat and cucumber LPOR level is not reduced in light-exposed chill-stressed seedlings. [97] Heat Wheat and cucumber LPOR content is greatly reduced in response to light in heat-stressed seedlings. [97] Shade Camellia sinensis L. and soybean LPOR is significantly upregulated after shading, but downregulated by low R/FR ratio [93,98,99] Rice OsPORA expression is repressed by light, and OsPORB expression is rapidly upregulated by high-light treatment. [100] When plants undergo the process of greening, low temperatures significantly hinder chlorophyll biosynthesis, leading to a decline in chlorophyll accumulation. However, the extent of this decline differs between various species[97]. One of the primary factors contributing to this phenomenon is the inhibition of the conversion of Pchlide to Chlide, which significantly diminishes LPOR activity and downregulates protein and transcriptional expression levels at low temperatures[41,97,101], This results in the obstruction of chlorophyll synthesis and the accumulation of ROS at low temperatures[41]. These ROS subsequently have the potential to cause oxidative damage during the greening process[41]. However, spraying exogenous carotenoids can improve the downregulation of LPOR transcriptional levels at low temperatures, thereby reducing their impact on chlorophyll biosynthesis[102]. Furthermore, the exogenous application of ALA and H2S was found to significantly enhance the content of chlorophyll and its upstream precursors[103]. Previous research showed that LPOR plays an important role in the cold resistance of plants[96,104,105,107]. CbPORB is resistant to cold in Chorispora bungeana[96]. CbPORB transcription and protein content decrease slightly at 4 °C but significantly decrease over time at −4 °C. Conversely, in A. thaliana[106] and wheat[105], low temperatures upregulate the HY5 expression at the transcriptional level, and HY5 regulates the transfer of COP1 from the nucleus to the cytoplasm, thereby promoting PORC expression (Fig. 3). A comparison of winter wheat XN1376 with its albino line XN1376B revealed that the expression of TaPOR2D in albino leaves with methylation of its promoter at low temperatures was an important factor influencing chlorophyll accumulation at low temperatures[108]. The restricted decrease in Pchlide is the primary cause of the impact on chlorophyll biosynthesis during periods of high-temperature stress[97,101,109,110]. Although the activity of the LPOR enzyme in green seedlings increases under high-temperature conditions[101], the LPOR protein content decreases significantly[97], resulting in a decrease in chlorophyll content. Developing seedlings could regulate the balance between ROS and Chl levels by regulating the production of LPOR enzymes[111]. PORB plays a significant role in the thermoregulation of chlorophyll biosynthesis in phototrophic seedlings and FCA (Flowering Control Locus A) induces the expression of PORA and PORB by promoting the DNA accessibility of RNA polymerase II to the gene promoters, thereby maintaining protein levels at a constant temperature[111]. Some studies suggest that melatonin can enhance plant stress resistance and improve the impact of heat stress on plant chlorophyll synthesis by upregulating the PORA expression[110]. A high-temperature stress-responsive protein, Ta2CP, was discovered in a heat-adapted wheat variety that is also involved in regulating chlorophyll biosynthesis under high-temperature stress[109]. The results indicated that Ta2CP positively regulates chlorophyll biosynthesis via interaction with TaPORB. Silencing Ta2CP expression downregulates TaPORB expression and decreases chlorophyll content, whereas Ta2CP overexpression upregulates TaPORB expression and increases chlorophyll content.
Under salt stress, the decrease in LPOR enzyme activity is a key contributor to the decline in chlorophyll levels[89]. The expression of genes related to chlorophyll synthesis and photosynthesis in peanuts (Arachis hypogaea) decreases significantly under drought stress. During recovery, the transcript and protein expression levels of AhPORA upregulate significantly, leading to the recovery of chlorophyll biosynthesis and photosynthesis[95]. This finding is consistent with the results observed in rice, where the accumulation of chlorophyll in seedlings developed under water stress is significantly reduced because of a decrease in the accumulation of intermediate precursors for chlorophyll synthesis. In particular, the decrease in the activity of LPOR enzymes, protein, and gene expression leads to damage to the Shibata shift, resulting in a decrease in Pchlide photoreduction[90]. Similarly, when cucumbers experience water stress, the LPOR enzyme content and the transcriptional content directly impact chlorophyll accumulation[112]. Arsenic significantly reduced the growth rate, chlorophyll content, and photosynthetic rate of melon plants. In contrast, iron oxide nanoparticles and selenium treatments up-regulated the expression of chlorophyll synthase and LPOR and increased the chlorophyll content of melon plants under arsenic stress[113]. In summary, LPOR plays a crucial role in regulating chlorophyll biosynthesis during adverse conditions. Plants improve their stress response capability by regulating their LPOR transcription, protein level expression, and enzyme activity. However, the regulatory mechanism is not completely clear. Thus, further exploration is needed.
-
In angiosperms, chlorophyll synthesis is dependent on light-induced activity of LPOR, which reduces Pchlide (Fig. 2) and promotes chloroplast development. Different types of LPOR were identified in multiple species, with varying expression patterns. The different expression patterns and the dependence of LPOR activity on the type of substrate can optimize preparations in the dark to ensure efficient chlorophyll synthesis with minimal impact on photosynthesis. The current research on light has focused on the greening process during the transition from darkness to light. A large number of studies have gradually revealed the regulatory mechanisms of light signaling factors. However, the complexity of light variations in natural environments, such as shaded or densely planted areas in forests, as well as cultivation techniques including strip cropping, result in varying degrees of light intensity and light quality. The precise regulation of LPOR in a variable environment and the efficient synthesis of the optimal amount of chlorophyll to ensure the utilization of light energy by plants remain unclear. Additionally, numerous studies have demonstrated that LPOR plays a pivotal role in abiotic stress response by regulating chlorophyll synthesis at the transcriptional and protein levels (Fig. 3, Table 1). However, the regulatory mechanisms at the posttranslational level have not been extensively investigated. Consequently, further investigation will provide a theoretical foundation for the breeding and development of plant germplasm resources with stress tolerance.
-
The authors confirm contribution to the paper as follows: draft manuscript preparation: Wang Q; manuscript revision: Gao J, Chen J, Tan X, Liu C, Yu L, Yang F, Yang W; conceptualization, funding acquision: Yang F, Yang W. All authors reviewed the results and approved the final version of the manuscript.
-
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
This work was supported by the National Natural Science Foundation of China (32071963).
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Wang Q, Gao J, Chen J, Tan X, Liu C, et al. 2024. Regulatory mechanism of a light-dependent protochlorophyllide oxidoreductase in chlorophyll biosynthesis and environmental adaptation. Technology in Agronomy 4: e023 doi: 10.48130/tia-0024-0019
Regulatory mechanism of a light-dependent protochlorophyllide oxidoreductase in chlorophyll biosynthesis and environmental adaptation
- Received: 29 January 2024
- Revised: 19 May 2024
- Accepted: 22 May 2024
- Published online: 19 August 2024
Abstract: Chlorophyll is a vital component of photosynthesis and must be produced throughout the plant life cycle. Light-dependent protochlorophyllide oxidoreductase (LPOR) is a pivotal enzyme in the chlorophyll biosynthesis pathway, catalyzing the conversion of Pchlide to Chlide. The presence of different types of LPOR ensures the efficient synthesis of chlorophyll in photosynthetic organisms during the dark-light transition. In addition to the transcriptional, translational, and post-translational regulation of LPOR function under different abiotic stresses, the nature of the substrate also influences LPOR function. Here, a perspective on chlorophyll synthesis and the development of chloroplasts is offered, the importance of LPOR in safeguarding plant light energy utilization is summarized, the gene expression pattern and structural-functional features of LPOR are outlined, as well as the role of LPOR in abiotic stress tolerance response, the catalytic mechanism of LPOR as well as the modulation of LPOR by light signals and other environmental factors are discussed. The aim is to provide references for the cultivation and innovation of plant germplasm resources with stress tolerance.
-
Key words:
- Chlorophyll biosynthesis /
- POR /
- Light signaling /
- Protochlorophyllide