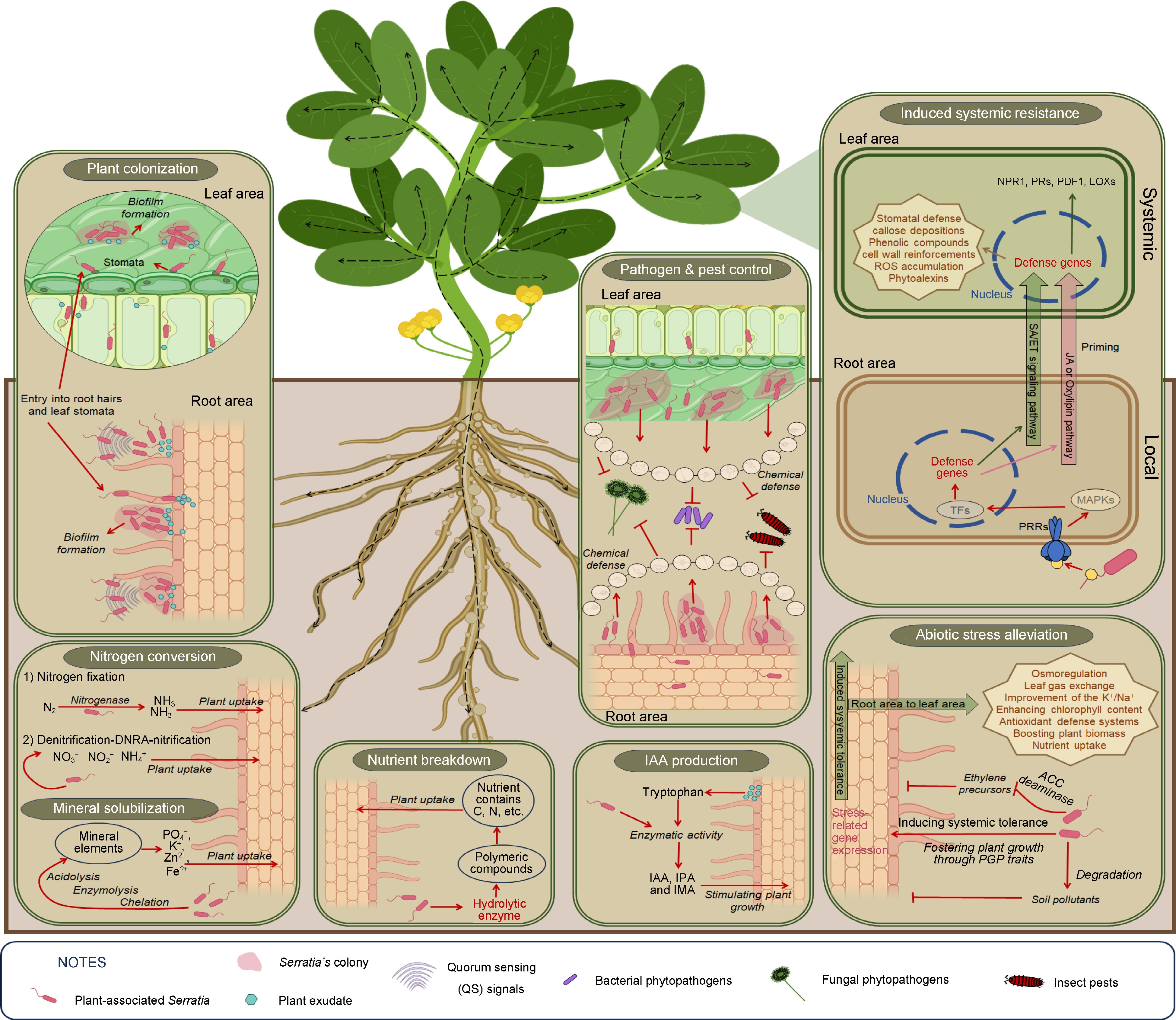
-
The genus Serratia is an intriguing member of the family Enterobacteriaceae. Presently, 23 species with validly published and correct names have been documented (referencing LPSN). Initially described in Italy as the producer of blood-like spots on polenta, Serratia marcescens was named by Bizio in 1823 and is considered the type species of this genus in recent studies[1]. The presence of red spots on food was often attributed to the activity of red-pigmented bacteria, such as Serratia marcescens, which were believed to be responsible for food spoilage as 'saprophytes'. The genus Serratia occupies a broad range of habitats, being isolated from soil, water, plants, insects, vertebrates, and hospitalized patients. Various studies have demonstrated that multiple species within the Serratia genus inhabit the interior or surface of plant tissues, classifying them as either endophytic or epiphytic bacteria[2−4]. Serratia associated with plants exhibit a plethora of traits that promote plant growth, including the production of phytohormones, the augmentation of nutrient availability, and protect plants from the deleterious effect of biotic and abiotic stressors[5,6]. The red pigment, known as prodigiosin, has been found to effectively combat various phytopathogens in crop plants[7−9]. Besides, various other secondary metabolites produced Serratia spp. have been demonstrated as antimicrobial, which have also been applied for controlling phytopathogens[10−12]. The increasing demand for sustainable agriculture has highlighted the importance of microbial pesticides and biostimulants in research, drawing significant attention to Serratia spp. However, the communication between plants and Serratia, along with their respective metabolites, remains ambiguously defined. On the other hand, several Serratia spp. are considered pathogens for plants and humans. This lack of clarity has spurred us to consolidate information on plant-associated Serratia, prompting a deeper exploration into the intricate interactions between plants and the Serratia genus. The strategy for safe exploitation of potential Serratia bacteria in agriculture is also the focus of this review.
-
The genus Serratia consists of Gram-negative, catalase-positive, and oxidase-negative rods found in diverse ecological environments[13]. Many species within this genus are distinguished by their ability to produce the red pigment prodigiosin, a pyrrole alkaloid, which makes them easily identifiable from other members of the family Enterobacteriaceae. However, it is important to note that not all Serratia strains produce prodigiosin; these non-pigmented strains are classified as non-pigment Serratia[1].
Regarding Serratia from plants, Serratia proteamaculans was first isolated in 1919 from a leaf spot disease of the tropical plant Protea cynaroides and was later shown to cause a hypersensitivity reaction in detached leaves of the plant. Many other Serratia spp. were isolated from plants. A comprehensive survey conducted by Grimont in 1981 revealed that bacteria from eight of the 10 known Serratia species of the time (S. marcescens, S. plymuthica, S. liquefaciens, S. proteamaculans, S. grimesii, S. rubidaea, S. odorifera, and S. ficaria) were isolated from various plants, including vegetables, mushrooms, mosses, decaying materials, grasses, small plants, and trees. S. liquefaciens and S. proteamaculans were particularly prevalent, followed by S. marcescens[14]. Pigmented S. marcescens biotypes were rarely isolated from plants. Several S. marcescens biogroups frequently encountered in nosocomial infections in humans were also isolated from plants. There is a hypothesis that vegetables used in salads might bring Serratia strains to hospitals and contaminate the patients' digestive tracts[1].
To date, 13 additional Serratia species have been isolated, including Serratia oryzae, an endophyte of rice stem; Serratia inhibens, a new antifungal species isolated from potato, and Serratia rhizosphaerae, isolated from rhizospheric soil and recognized as a novel plant resistance inducer against soft rot disease in tobacco[15−17].
Many species within the genus Serratia, including S. plymuthica, S. liquefaciens, S. proteamaculans, S. grimesii, S. nematodiphila, S. rubidaea, and S. marcescens, have been reported as plant growth-promoting bacteria. Additionally, some less common species, such as S. ficaria, S. fonticola, S. odorifera, S. entomophila, and S. quinivorans, have been recognized for their roles in stimulating plant growth[18].
These bacteria promote plant growth through various mechanisms, such as directly enhancing nutrient availability by breaking down high molecular weight organic compounds, converting nitrogen compounds, and solubilizing minerals etc. They also produce phytohormones and help alleviate salinity and drought stress in plants. Furthermore, the production of secondary metabolites such as prodigiosin, pyrrolnitrin, and biosurfactants enable Serratia spp. to combat phytopathogens and insect pests, thereby stimulating plant growth even under stressful conditions[19−21].
Serratia spp. possess mechanisms that enable them to adapt to diverse lifestyles, distinguishing them from other plant growth-promoting bacteria[22]. For instance, genome analysis of Serratia grimesii BXF1, isolated from pinewood nematode (Bursaphelenchus xylophilus), revealed the presence of several genes associated with antagonistic traits, plant growth regulation, and nematode development modulation. Moreover, most of the BXF1 genes are involved in environmental and genetic information processing, consistent with its ability to sense and colonize various niches. These findings enhance our understanding of the role and evolution of strain BXF1 as a mediator of interactions in complex disease systems and provide insights into the broader evolution of Serratia and Enterobacteriaceae towards multitrophic interactions[23]. Similarly, S. nematodiphila, initially reported as a symbiont of the entomopathogenic nematode Heterorhabditidoides chongmingensis[24], has been also found in the rhizosphere, exhibiting plant growth-promoting activities[25,26]. Unfortunately, eight species (Serratia liquefaciens, Serratia ficaria, Serratia fonticola, Serratia grimesii, Serratia odorifera, Serratia plymuthica, Serratia quinivorans, and Serratia rubidaea were reported to be associated with human infection, among which S. marcescens, S. liquefaciens, and S. odorifera are best known[27]. Of all Serratia species, S. marcescens is not only the most common nosocomial pathogen, but also considered the causal agent of plant diseases[22]. Some S. marcescens strains are known to cause leaf spot disease on industrial hemp (Cannabis sativa)[28]. Additionally, S. marcescens strain HFP01 has been identified as the causal agent of cucurbit yellow vine disease, a devastating disease for cucurbit production[22,29].
Recent advancements in whole genome sequencing and comparative genomic studies have significantly contributed to distinguishing clinical strains from other strains of Serratia spp., particularly through the identification of antibiotic resistance genes and the presence of plasmids. Genomic analysis has been pivotal in determining the virulence factors of Serratia spp.[30,31]. Comparative genomic studies of plant pathogenic and nonpathogenic Serratia marcescens strains have revealed genetic markers specific to phytopathogenic strains[32]. This approach has further elucidated the mechanisms by which plant-associated Serratia spp. colonize plant tissues, uncovered various genes responsible for plant growth-promoting traits, biosynthesis of quorum sensing (QS) signals, production of secondary metabolites effective against phytopathogens and insect pests, and elicitors of induced systemic resistance (ISR)[33,34].
-
Plant-associated bacteria are recognized as indigenous members of the plant microbiome, residing in high densities on the plant and influencing plant development[35]. Recent research has revealed that bacterial populations in the rhizosphere can range between 107 and 109 CFU/g of rhizosphere soil, while populations on the rhizoplane range from 105 to 107 CFU/g fresh weight (fw). After establishing themselves in the rhizosphere and rhizoplane, bacterial endophytes can enter plant roots and colonize with sub-populations ranging from 105 to 107 CFU/gfw. Once inside the roots, these endophytic bacteria can spread systemically to colonize above-ground tissues, establishing population densities in stems and leaves between 103 and 104 CFU/gfw under natural conditions[35]. In a study involving various rice varieties, endophytic bacteria were isolated from surface-sterilized roots and stems. The bacterial population in the roots ranged from approx. 105 to 106 CFU/gfw, whereas the bacterial population in the stems ranged from approx. 102 to 104 CFU/gfw. One strain, Serratia marcescens IRBG500, was identified as a diazotrophic bacterium capable of systemic spread. It colonized the roots (approx. 107 CFU/gfw on the surface and internal areas), stems (approx. 101 CFU/gfw on the surface and 105 CFU/gfw in the internal area), and leaves (approx. 102 CFU/gfw on both the surface and internal areas), positively influencing plant growth[2]. Several studies have indicated that biofilm formation is critical for the colonization process of plant-associated Serratia species, generally regulated by QS signal molecules[36,37]. Indeed, the biofilm-forming Serratia glossinae GS2 (a later synonym of Serratia fonticola), isolated from the rhizosphere of a sesame plant, as a producer of two quorum-sensing signal molecules: N-hexanoyl-L-homoserine lactone and N-octanoyl-L-homoserine lactone, boosts plant biomass and chlorophyll content[38]. An interesting assay revealed that the population density of root-associated bacterial communities varies on the roots of transgenic plants engineered with either AHL or acylhomoserine lactonase (AiiA) genes. The density of Gram-negative bacterial increased in AHL-expressing plants, whereas it significantly decreased in AiiA-expressing plants[39]. These findings suggest that plant colonization by Gram-negative bacteria like Serratia may rely on quorum sensing (QS) signal molecules, specifically AHLs, which also regulates other potential cell surface adhesins, including exopolysaccharide and outer membrane proteins[40]. Additionally, swarming motility, regulated by the flhD gene, enhances biofilm formation by promoting cell surface attachment. Major fimbriae subunits encoded by fimA and fimC are essential for adhesion and colonization. Genes bsmA and bsmB also regulate biofilm formation, with mutations impairing this process. The gene swrR acts as an AHL-dependent regulator. Downregulation of fimA, fimC, bsmA, bsmB, flhD, and swrR reduces attachment capacity, biofilm formation, and swarming motility[41,42].
Biofilm-forming bacteria aid plants cope with biotic and abiotic stressors by upregulating induced systemic resistance, balancing cytotoxicity, and inhibiting pathogens with substances like exopolysaccharide (EPS) matrix, antibiotics, and enzymes. They also trigger the production of defense hormones, antioxidants, and osmolytes, enhancing plant growth through mechanisms such as indole-3-acetic acid (IAA) production, mineral solubilization, nitrogen compound production, and ACC deaminase production[43]. Specifically, Serratia species form an EPS matrix that serves as a scaffold for biofilm structure, facilitating intercellular communication and surface adhesion. They also produce the red pigment prodigiosin and several extracellular virulence factors, including DNase, lipase, hemolysin, proteases, chitinase, and biosurfactants. These factors primarily contribute to biofilm formation through quorum sensing (QS) mechanisms[41].
In this intricate communication, plant exudates play a key role in attracting and stimulating the colonization and biofilm formation of beneficial Serratia to address the challenges that the plant faces[44,45]. One study demonstrated that seedlings treated with high red/far red (R/FR) light maintained high populations of S. plymuthica A21-4 in the roots, while populations of S. plymuthica A21-4 markedly reduced in seedlings treated with low R/FR light. The authors suggested that the composition of plant exudates changed with different light ratios. They also reported that plants secrete 10%–40% of their photosynthetically fixed carbon into the rhizosphere as root exudates, potentially affecting plant growth-promoting rhizobacteria (PGPR). In their experiment, high R/FR ratio exposure induced the secretion of certain tomato root exudates such as L-Leucine, L-Methionine, L-Glutamine, 6-Aminocaproic acid, and D-melezitose, which promoted the chemotaxis response, biofilm formation, and root colonization of S. plymuthica A21-4 on tomato seedlings[46]. Additionally, the growth of plants and the population of inhabitant Serratia influence each other. The authors of a previous study have indicated that the density of S. plymuthica HRO-C48 on plants influences both plant growth and its ability to compete with phytopathogens[47]. During the plant colonization process, S. plymuthica G3 and S. plymuthica HRO-C48 positively regulate antifungal activity and exoenzyme production but negatively regulate indole-3-acetic acid (IAA) production during plant-microbe interactions[36,48]. Plant-associated Serratia are known to produce phytohormones, particularly IAA, which promotes plant growth. When the bacterial population is low, higher amounts of IAA are released, supporting plant growth and supplying additional nutrients to the bacteria. Conversely, at high population densities, IAA production decreases to prevent damage and disease promotion[48]. In the presence of indigenous Serratia species as plant inhabitants, a plethora of plant growth-promoting traits and features for reducing plant pests are expressed, as depicted in Fig. 1. However, QS-deletion mutants in S. fonticola GS2 showed considerably fewer PGP activities compared to the wild type[49]. Interestingly, AHL-producing Serratia can inhabit both host and non-host plants, expressing various PGP traits and protecting plants from pathogens[50]. Therefore, plant colonization by Serratia focuses on chemotaxis response and biofilm formation, influenced by plant exudates under normal or environmentally changing conditions.
Intriguingly, the presence of an appropriate amount of plant cell wall-degrading enzymes, such as cellulase, xylanases, and peptinase activity establishes pathways for bacteria to access the root endosphere through the colonization process, enabling them to exhibit growth-promoting activities[51,52]. Notably, the cell wall-degrading enzymes of endophytic Serratia sp. S119 are influenced by root exudates from three plants: peanuts, soybeans, and maize[53]. These observations have led to the hypothesis that root exudates function as rewards, attracting beneficial microbes to collaborate in supporting plant growth.
-
In the intricate web of communication between plants and microbes, beneficial microorganisms, particularly bacteria, play a pivotal role. These bacteria, identified as plant growth-promoting bacteria (PGPBs), play a crucial role in fostering plant health. PGPBs exhibit traits that contribute significantly to plant well-being, including the production of substances vital for plant vitality and the breakdown of organic compounds in the soil into more accessible nutrients for enhanced plant uptake. Additionally, they serve as formidable defenders, shielding plants against both abiotic and biotic stressors[18,20]. A synthetic review further underscores the advantages of specific bacteria, such as Serratia, as elucidated in Table 1. This comprehensive overview highlights the numerous benefits these microorganisms provide in plant-microbe interactions, emphasizing their mechanisms in promoting plant growth.
Table 1. Plant growth promoting traits of Serratia species.
No. Plant growth-promoting traits Mechanisms Serratia species Ref. 1 Colonization Biofilm formation S. marcescens
S. plymuthica
S. fonticola
S. oryzae
Serratia spp.[2,36−39,44,46−50,53] Chemotaxis Entry into root hairs and leaf stomata 2 IAA production Trp-dependent pathway S. marcescens
S. liquefaciens,
S. plymuthica
S. Rubidaea S. glossinae
S. nematodiphila
Serratia spp.[5,8,26,38,48,57−59,90] 3 Nitrogen conversion Nitrogen fixation S. marcescens
S. nematodiphila
Serratia spp.[2,26,58,63,64] Denitrification-DNRA-nitrification 4 Minerial solubilization Organic acid production S. marcescens
S. nematodiphila
S. plymuthica
Serratia spp.[26,44,58,65,67−69,71,74,123] H+ extrusion Exopolysaccharide production Siderophores producion Solubilizing enzyme production 5 Nutrient breakdown Extracellular hydrolytic enzyme
productionS. marcescens
Serratia spp.[53,64] 6 Abiotic stress alleviation ACC deaminase production S. marcescens
S. liquefaciens
S. proteamaculans
Serratia spp.[5,6,44,58] Induced systemic tolerance PGP traits stimulation Cadmium (Cd) detoxification Pyrene degradation 7 Induced systemic resistance Elicited by AHLs,
bacterial flagellin (flg22), translation elongation factor Tu (elf18), and
2,3-butanediolS. marcescens
S. liquefaciens
S. rhizosphaerae
Serratia spp.[17,39,82,83] IAA production
-
In plant-microbe interactions, bacterial auxin from rhizospheric bacteria is crucial for colonization[54]. Researchers have revealed that 80% of rhizosphere bacteria produce IAA, which enhances biofilm formation, aiding bacterial adherence and stress resilience[55]. Bacterial IAA biosynthesis can be tryptophan-dependent or independent. While the tryptophan-independent pathway is not yet understood, the tryptophan-dependent pathway involves several known genes and enzymes, with plants providing tryptophan through exudates as a precursor for bacterial IAA synthesis[55]. To illustrate, a study revealed that root-associated S. marcescens RSC-14 contains the trpEGDCBA gene cluster, encoding key enzymes in the tryptophan biosynthesis pathway, linked to various biological processes, including IAA biosynthesis[5]. The RSC-14 genome includes ipdC for indolepyruvate decarboxylase and dhaS for indole-3-acetaldehyde dehydrogenase, essential for converting tryptophan to IAA. The pathway involves tryptophan-2-monooxygenase (IaaM) and indoleacetamide hydrolase (IaaH), though RSC-14 only has a putative iaaH gene and lacks iaaM. Similarly, S. fonticola strain GS2 from the rhizosphere contains many of the same genes as S. marcescens RSC-14[56]. Additionally, Serratia species isolated from various plants, such as common wheat, soybean, turmeric, and nepenthes, have been identified as IAA producers[8,26,36,48,57,58]. Interestingly, after colonizing the rhizosphere, S. marcescens PLR synthesizes IAA using tryptophan provided by root exudates, exporting it back to the roots along with its precursor's indole-3-pyruvate (IPA) and indole-3-acetamide (IAM). This up-regulates the expression of multiple auxin biosynthesis genes and nutrient transporter genes in plants, accelerating root development[59]. Thus, plant-associated Serratia may produce IAA to colonize and stimulate plant growth through tryptophan provided by plant exudates in the plant-bacteria communication.
Nitrogen conversion
-
Nitrogen is essential for proteins, nucleic acids (DNA and RNA), ATP, and NAD in all living cells. It is a key component of chlorophyll, necessary for photosynthesis, thus critical for plant growth. Dinitrogen (N2) is the most abundant atmospheric gas, but plants can only assimilate reactive forms like oxidized (NOx) and reduced (NH3 and amines) nitrogen. N2 is converted to usable nitrogen (NH3) by biological nitrogen fixation by certain bacteria and archaea (diazotrophic prokaryotes)[60]. Additionally, root development through IAA produced by beneficial plant-microbe interaction can facilitate plant access to available nitrogen compounds fixed by microbes[61]. For instance, certain diazotrophic bacteria like Serratia, when present as endophytes in plants like rice and soybean, exhibit significant nitrogen fixation activity through the enzyme nitrogenase. These bacteria not only fix nitrogen efficiently but also colonize plant tissues effectively, contributing to sustained nitrogen availability. In particular, studies have shown that Serratia localize densely around plant root tips shortly after inoculation[2,58], a critical area where plants release plant exudates. Moreover, other bacteria like S. nematodiphila RGK have been found to produce ammonia, further supporting plant growth[26]. Nitrogenase is a bacterial enzyme that reduces dinitrogen (N2) to ammonia (NH3) using ATP. It consists of two proteins: the catalytic molybdenum-iron protein (MoFeP) and the iron protein (FeP). The enzyme's activity involves synchronized electron and proton transfers, driven by ATP-dependent interactions between FeP and MoFeP[62]. Interestingly, a study has revealed that Biological Nitrogen Fixation (BNF) fueled by As(III) oxidation is a novel biogeochemical process. Over 20 Serratia genomes from NCBI contained essential genes for As(III) oxidation and BNF (aioA and nifH), indicating that As-dependent BNF may be common in Serratia species[63]. Moreover, the isolated S. marcescens (OK482790) from the lupin rhizosphere, possesses genes involved in denitrification-DNRA-nitrification pathways. This strain plays a crucial role in converting nitrate to nitrite and other nitrogen compounds, thereby enhancing plant growth and nutrient availability[64]. It can be seen that Serratia may provide usable nitrogen to plants through several microbiological processes.
Mineral solubilization
-
Interestingly, under phosphorus deficiency, plants secrete exudates to attract phosphorus-solubilizing Serratia, which aids in nutrient acquisition[44]. Plant-associated Serratia produces organic acid, gluconic acid through its phosphate solubilization mechanisms[65]. Organic acids form soluble complexes with metal ions in insoluble phosphates, releasing the phosphate. Gram-negative bacteria like Serratia are more efficient at dissolving mineral phosphates than Gram-positive bacteria due to their secretion of organic acids from glucose metabolism. This metabolism can occur via phosphorylation to glucose-6-phosphate or direct oxidation to gluconate, leading to the Entner–Doudoroff pathway. Phosphate solubilization results from acidification of the periplasmic space by acids produced during glucose oxidation by quinoprotein glucose dehydrogenase (PQQGDH), which converts glucose to gluconic acid[66]. In Serratia, phosphorus (P)-limitation, and root exudates from P-limited peanut plants enhance pqqE gene expression and pqq promoter activity. Under P-limitation, both pqqE expression and promoter activity increase, varying with root exudate concentration and bacterial growth phase[67]. Further studies have shown that organic acids such as malic acid, lactic acid, and acetic acid are produced during the phosphate solubilization process in Serratia sp. PSB-37[68]. Additionally, High Performance Liquid Chromatography (HPLC) analysis of culture supernatants revealed that gluconic acid is secreted through the direct oxidation pathway of glucose in S. marcescens CTM 50650 when grown on National Botanical Research Institute's phosphate (NBRIP) medium containing glucose as the sole carbon source[69]. Various organic acids, particularly 2-ketogluconic acid, and gluconic acid, play key roles in the phosphate solubilization process via excretion by Phosphate Solubilizing Microorganisms (PSMs)[70]. Moreover, H+ extrusion aids in the mineral solubilization process of beneficial microorganisms. Ammonium (NH4+) in soil is assimilated by PSMs for amino acid synthesis and converted to ammonia (NH3), releasing excess H+ into the cytoplasm. This acidifies the surrounding medium, aiding in the dissolution of insoluble phosphates[70]. In addition to phosphate solubilization, Serratia species can also solubilize compounds containing zinc (Zn) and potassium (K), transforming them into minerals available for direct plant uptake[26,71]. The mechanisms for K- and Zn-solubilization similarly involve organic acid production and H+ release by beneficial bacteria[72,73].
In organic phosphate solubilization, enzymes such as acid and alkaline phosphatases and phytase, phosphonatases/carbon–phosphorus (C–P) lyases produced by microorganisms facilitate the dissolution of organic phosphates through mineralization[70]. One study has shown the effect of low phosphate concentration and acidic pH on the alkaline phosphatase (APase) activities of S. marcescens. Under low phosphate conditions, some strains of S. marcescens synthesize two different types of APases: constitutive (CAPase) and inducible (IAPase)[74]. Conversely, another study found that Serratia sp. PSB-37 significantly produces acid phosphatase under high pH conditions to decrease pH, enhancing soil fertility and contributing to sustainable agriculture[68].
Nutrient breakdown
-
Extracellular enzymes in soils degrade organic matter through hydrolytic reactions. These enzymes cleave specific bonds in complex organic materials like plant debris, requiring microbial cells to transform organic matter into smaller molecules for cellular uptake, exemplified by the decay of cellulose and lignin, and contributing to the carbon-cycling process. Hydrolytic extracellular enzymes produced by microbes can break down polymers into monomers derived from deceased organisms (including plants and other living entities), serving as valuable sources of carbon, nitrogen, and other nutrients for plant uptake and proliferation[75]. Numerous studies indicate that plant-associated Serratia species produce various degrading enzymes, including protease, lipase, cellulase, and chitinase effectively promoting plant growth[64,76].
-
Plant abiotic stresses can be alleviated by chemical substances synthesized by beneficial microbes. For example, under salt stress, ethylene production in plants increases, exacerbating stress. The rhizobacterial strain S. proteamaculans M35 produces 1-aminocyclopropane-1-carboxylate (ACC) deaminase, which reduces ethylene levels, alleviates stress, and enhances wheat productivity[77]. ACC deaminase production is a key trait for promoting plant growth in Serratia[44]. During abiotic stress alleviation, Serratia also fosters plant growth through phosphate solubilization, siderophore and IAA production, nitrogen fixation, and ammonia production[6,20]. Besides that, using Serratia can induce plant tolerance to abiotic stresses. For example, when plants face salinity stress, S. marcescens CDP-13 modulates osmoprotectants and enhances antioxidant enzyme activities in wheat under salinity stress, minimizing oxidative damage[6]. Plants primed with AHLs (quorum-sensing molecules in Serratia, as described above) or AHL-producing rhizobacteria can better cope with abiotic stresses through induced systemic tolerance. AHLs activate Mitogen-activated protein kinases (MAPKs) and upregulate transcription factors, glutathione-S-transferase GST6, and heat shock protein Hsp60 genes, priming plants via the salicylic acid (SA) and oxylipin pathways. This modulates osmoprotectants, improves gas exchange, reduces the Na+/K+ ratio, enhances antioxidant enzymes, and boosts plant biomass and chlorophyll content. Specifically, inoculation with S. liquefaciens KM4 significantly reduces oxidative stress markers (hydrogen peroxide (H2O2), malondialdehyde (MDA), and electrolyte leakage (EL)), enhancing leaf gas exchange, osmoregulation (proline, soluble sugars, soluble protein, total free amino acids, phenols, and flavonoids), antioxidant defense systems (antioxidant enzymes: ascorbate peroxidase (APX), superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD); and levels of non-enzymatic antioxidants: ascorbic acid (AA) and glutathione (GSH)), and nutrient uptake under salt stress. These benefits are accompanied by the upregulation of stress-related genes, antioxidant genes (APX, CAT, SOD), Rubisco and photosynthesis-encoding genes (RBCS and RBCL), and genes mediating ion balance (H+-PPase, HKT1, NHX1), and the downregulation of the key gene in abscisic acid (ABA) biosynthesis (NCED)[78]. In soils containing pollutants like heavy chemicals, Serratia can help plants cope with these stressors. For example, introducing Serratia CP-13 to maize cultivars grown in cadmium (Cd)-stressed soil enhances Cd detoxification by modulating phytohormones and gas exchange[79]. Similarly, Serratia sp. Wed4 degrades pyrene and promotes plant growth through its PGP traits[58].
The positive effects of Serratia in alleviating abiotic stress bring various benefits to plant growth and health. According to the researchers, the potential strain S. proteomaculans demonstrated promising performance under abiotic stress conditions, significantly increasing plant height, root length, grain yield, and straw yield by up to 60%[77]. Introducing Serratia under abiotic stress conditions leads to substantial increases in plant biomass, seed germination, photosynthetic pigments, plant physiology, and nutrient uptake[58,79]. Overall, Serratia spp. make a significant contribution to plant growth and development across various environmental conditions.
-
Beneficial bacteria within plant tissues can activate the plant immune system to defend against phytopathogens and insect pests through Induced Systemic Resistance (ISR)[80]. Microbe-Associated Molecular Patterns (MAMPs) activate Pattern Recognition Receptors (PRRs), leading to hormonal defenses against pathogens and various plant immune responses. Elicitors from beneficial bacteria such as flagella, lipopolysaccharides, siderophores and quorum-sensing molecules act as MAMPs, recognized by PRRs, priming plant immunity[80,81].
To induce plant systemic resistance, numerous reports indicate that AHLs produced by Serratia act as elicitors, perceived by plants, leading to the activation of SA/ET and oxylipin signaling pathways in the ISR process[82]. For instance, a study showed that AHL produced by S. liquefaciens MG44 induces systemic resistance in plants through SA- and ET-dependent pathways[83]. Additionally, AHLs prime plants for controlling phytopathogens via SA-dependent and oxylipin pathways[84]. Phyto-oxylipins, including JA and its metabolites like cis-OPDA, methyl jasmonate, and JA-Ile, are unsaturated fatty acids produced by lipoxygenases (LOX) that oxidize lipid chains. Specifically, root treatment with S. marcescens B2 elevates lipoxygenase levels after pathogen inoculation, leading to induced systemic resistance to rice blast disease[85]. Plant roots primed with oxo-C14-HSL lead to the accumulation of SA and cis-OPDA in plant leaves[84]. Additionally, oxo-C14-HSL-treated Arabidopsis plants exhibit increased resistance to the hemibiotrophic bacterial pathogen Pseudomonas syringae pv. tomato DC3000. Oxo-C14-HSL promotes stronger activation of mitogen-activated protein kinases (MAPKs) AtMPK3 and AtMPK6 when challenged with flg22, followed by higher expression of defense-related transcription factors (TFs) WRKY22 and WRKY29, and the PATHOGENESIS-RELATED1 (PR1) gene[86]. Elicitors like flg22 and elf18 peptides in S. rhizosphaerae strain KUDC3025 also induces plant systemic resistance via SA and ET pathways[17]. Research on plant ISR elicited by 2,3-butanediol produced by S. marcescens 90-166, regulated by their AHLs, suggests that this strain may trigger plant ISR through both JA- and SA-dependent pathways[39]. Similarly, another study has shown that the intermediate response of plants was produced by metabolic elicitors from S. rubidaea strain N 12.34, which induced a strong differential expression of marker genes in the SA pathway and a marker in the JA/ET pathway[87]. However, these elicitors remain unexplored. Thus, further exploration of Serratia-induced plant immunity is needed in future studies.
Following hormonal signaling pathways, numerous defense genes are expressed in response to plant disease. For instance, the activation of defense genes like PR1a, 26 kDa acidic chitinase, and 30 kDa basic chitinase occurs when plant roots are treated with AHL-producing S. liquefaciens in the rhizosphere[83]. The induction of the SA/cis-OPDA pathway enhances stomatal defense by activating MPK3 and MPK6, which induce the guard cell lipoxygenase 1 (LOX1), leading to the peroxidation of polyunsaturated fatty acids into oxylipins and the subsequent accumulation of SA. Similarly, rice primed with S. plymuthica IC1270 accumulates reactive oxygen species (ROS) and phenolic compounds[88]. A study has shown that seedlings inoculated with S. rubidaea strain N 12.34 exhibit a significant increase in the differential expression of gene coding for defense-related proteins, including Nonexpressor of pathogenesis-related protein 1 (NPR1) (149.74-fold), PR2 (57.09-fold), Plant defensin 1 (PDF1) (675.98-fold), PR3 (41.37-fold), and LOX2 (32.79-fold), compared to negative controls[87]. Various actions induced by ISR elicited by Serratia include cell wall reinforcement, phenolic compound deposition, and callose formation. Other pathogenesis-related (PR) proteins involved in these defenses include chitinase, glucanase, phenylalanine ammonia-lyase (PAL), polyphenol oxidase (PPO), and peroxidases (POX). Additionally, secondary metabolites like phytoalexins play a crucial role in these defense mechanisms[18].
-
In the interaction between plant growth-promoting bacteria (PGPB) and plants, many Serratia spp. effectively trigger the plant immune system through induced systemic resistance (ISR) as previously discussed. Additionally, Serratia employs direct antagonism to overcome ecological competitors. Serratia species are renowned producers of a diverse array of bioactive compounds, encompassing hydrolytic enzymes and secondary metabolites (Table 2). These compounds exhibit direct inhibitory effects on the growth of plant pathogens. When confronted with ecological competitors such as harmful fungi, plant bacteria employ a diverse array of cell wall-degrading enzymes, including protease, chitinase, and β-1,3 glucanase. These enzymes can break down peptidic and glycosidic linkages present in the protein and various polysaccharides of the cell wall of phytopathogens, thereby reducing the density of plant pathogenic cells. Although various studies have extensively explored genes encoding protease and chitinase[22,89], genes coding for β-1,3 glucanase remain less explored. However, numerous studies have indicated that various Serratia species can produce β-1,3 glucanase (Table 2). This suggests that future studies need to explore the genes coding for β-1,3 glucanase in Serratia more thoroughly. The hydrolytic enzymes synthesized by Serratia species are recognized as crucial antifungal compounds in various studies focused on reducing plant fungal diseases[47,48,64,76,90].
Table 2. Common compounds produced by Serratia for the control of plant diseases and insect pests.
No. Compounds produced by
Serratia spp.Analyzed biosynthetic genes Mechanisms of action Ref. 1 Protease yfgC Degrading the cell walls of most of fungi and the insect's midgut [8,22,47,48,76,89,90,111] 2 Chitinase chiA,B, C Degrading the cell walls of most of fungi
Degrading the insect exoskeletons3 β-1,3 glucanase Gene encoding β-1,3-glucanase
in glycoside hydrolase familyDegrading the cell walls of most of fungi 4 Lipase lipA,B,C,D Degrading the insect's mitgut [140] 5 Phospholipase phlA Degrading the peritrophic membrane lining the midgut [109] 6 Serralysin
(alkaline metalloprotease)prtA1-prtA4 Promoting hemolymph bleeding in insect pests [22,114−116] 7 Prodigiosin
Tripyrrole alkaloidpig gene cluster (pig A-N) Inhibiting spore germination and growth of hyphae
Inhibiting immune system enzymes protease phosphatase, acid phosphatase, and acetylcholine esterase
Reducing pH in the insect's midgut[7−9,57,91,117−121] 8 Pyrrolnitrin - tryptophan-derived secondary metabolite prnABCD Damaging the cell membrane in combination with phospholipids
Inhibiting fungal growth by inhibiting the respiratory electron transport system[8,37,48,90,93−95] 9 Oocydin A - chlorinated macrocyclic lactone ooc gene cluster Anti-oomycete activity [4,141] 10 Biosurfactants
Serrawettin W1, Serrawettin W2swrW, swrA Swarming activity of Serratia spp.
Biosurfactant activity disturbing the cell membrane of phytopathogenic cells[21,96,97] 11 Serratiochelin A, B, C, D
Bis catecholate siderophoressch gene cluster Iron competition with phytopathogenic cells [48,90,139,142] 12 Volatile organic compounds (VOCs)
Sodorifen
DMDS
acetic acid, methylthiolacetate
Terpene synthase gene (Q5A_011535)
mdeA
Methyltransferase gene (Q5A_011540)Inhibition of fungal growth [99−101] The most prominent secondary metabolite of several Serratia spp. is prodigiosin. This tripyrrol alkaloid, synthesized under the instruction of the pig gene cluster (pigA-N) has been suggested to be associated with surface adherence and enhancing bacterial dispersal in addition to its antibacterial, and antifungal activities. Moreover, the biosynthesis of prodigiosins might function as a 'metabolic sink' by consuming the overflow of NAD(P)H or proline from primary metabolism[91]. Multiple studies underscore the pivotal role of prodigiosin in conferring Serratia's proficiency in controlling plant diseases induced by deleterious bacteria or fungi[8,9,57]. The wide spectrum of activity against phytopathogenic bacteria or fungi has been substantiated across various plant diseases. Prodigiosin induces bactericidal activity indicative of programmed cell death (PCD), involving DNA fragmentation, ROS generation, and proteins with caspase-like activity. Prodigiosin localizes in the membrane and nuclear fractions of bacterial cells, facilitating its binding to bacterial DNA. Regarding its fungicidal activity, prodigiosin has been reported to inhibit spore germination and hyphal growth. These findings suggest that the red-pigmented S. marcescens holds promise as a biocontrol agent for combating diverse plant diseases in crop cultivation[7,92].
Another noteworthy compound is pyrrolnitrin, which exhibits targeted efficacy against a broad range of plant diseases. The pyrrolnitrin biosynthesis with the prn gene cluster by rhizobacteria presumably has a key role in their life strategies and in the biocontrol of plant diseases[93]. The biosynthetic operon that encodes the pathway that converts tryptophan to pyrrolnitrin is composed of four genes, prnA through D[93]. The impact of pyrrolnitrin, produced by Serratia species has been observed across various crop diseases such as oilseed rapes, wheat, and cucumbers[8,37,48,90]. Pyrrolnitrin's mechanism of action is believed to involve damaging the cell membrane in combination with phospholipids[94] and inhibiting fungal growth by suppressing the respiratory electron transport system[95].
To combat fungal diseases caused by oomycetes such as Pythium or Phytophthora, plant-associated Serratia strains produce oocydin A and other haterumalides, which are chlorinated macrocyclic lactones. These compounds hold potential as antimycotics in agricultural applications, particularly for crop protection[4,10].
Lipopeptide biosurfactants, such as serrawettin W1 and W2 produced by S. marcescens strains, are responsible for the swarming activity of Serratia spp. These biosurfactants are synthesized by nonribosomal peptide synthetase genes, swrW and swrA, respectively. Their amphiphilic structures enable them to act as chemical defenders by disrupting the cell membranes of phytopathogenic cells[96,97]. Additionally, a new hybrid non-ribosomal peptide–polyketide antibiotic (serratamid) produced by S. plymuthica C1 can control 15 bacterial phytopathogens. It displays strong antibacterial activity against Ralstonia solanacearum and four Xanthomonas spp. The mode of action of this compound is expected to inhibit the carboxyltransferase step of the bacterial acetyl-CoA carboxylase-catalyzed reactions[98].
Another strategy involves limiting ions like available Fe3+ in the environment to impede the growth of plant pathogens. Iron-chelating siderophores serratiochelin A, B, C, D produced by Serratia spp. competitively acquire ferric iron in plant environments from phytopathogenic cells[48,90].
Crucially, the antagonistic compounds produced by Serratia are regulated by quorum sensing (QS) signals, such as AHLs. The AHL-negative mutants in S. plymuthica HRO-C48 significantly reduced the production of antifungal compounds[48]. This phenomenon is believed to be influenced by mechanisms involved in microorganism–plant interactions[90], and the Serratia population increases in the presence of rich plant nutrients until it reaches a critical bacterial size, triggering the QS system[48]. Thus, the establishment of the plant-associated Serratia cell with the host plant is deemed a key process, influencing the expression of its capabilities in controlling plant diseases.
Recently, the study of microbial interspecies communication and the mode of action of various antagonistic interactions, particularly those mediated by volatile organic compounds (VOCs), has garnered significant research interest. Bacteria respond to fungal VOCs by altering gene and protein expression related to motility, signal transduction, energy metabolism, cell envelope biogenesis, and secondary metabolite production. For instance, in response to VOCs emitted by the phytopathogen Fusarium culmorum, S. plymuthica PRI-2C, isolated from the maize rhizosphere, produces sodorifen, an unusual antifungal terpene[99]. Similarly, S. ureilytica strain ILBB 145 emits significant amounts of dimethyl disulfide (DMDS) when challenged by fungal phytopathogens, which inhibit the growth of Pythium cryptoirregulare[100]. Exposure to the fungal volatilome shifts the metabolism of S. plymuthica HRO-C48 into a defense mode by downregulating genes involved in cellular functioning while upregulating defense mechanisms at the volatile level. The activation of acetic acid and methylthiolacetate production in response to volatiles from fungal pathogens Rhizoctonia solani, Leptosphaeria maculans, and Verticillium longisporum suggests a common defense mechanism in S. plymuthica HRO-C48 against volatile-mediated fungal attacks. Notably, three-quarters of the VOCs produced by S. plymuthica HRO-C48 are antimicrobial substances, underscoring the significant contribution of VOCs to the biocontrol efficacy of S. plymuthica[101].
In the struggle for ecological niches and nutrients, microorganisms employ several mechanisms simultaneously. For instance, among 18 strains of S. plymuthica, S. rubidae, and S. liquefaciens isolated from the rhizosphere of oilseed rape, many exhibited strong antifungal activities against three phytopathogens: Verticillium dahliae, Rhizoctonia solani, and Sclerotinia sclerotiorum. Their mechanism of action typically involves a combination of at least two mechanisms, including antibiosis (production of prodigiosin and/or pyrrolnitrin), iron capture, and the production of hydrolytic enzymes (chitinases and/or β-1,3-glucanases)[8]. The broad-spectrum antimicrobial activity observed for Serratia spp. is hypothesized to result from a synergistic effect[21]. Additionally, Serratia produces other potent antimicrobial compounds, such as stephensiolides, glucosamine derivatives[102,103], and zeamines[104,105], which are effective against a wide range of Gram-positive and Gram-negative bacteria. Antibiotics such as althiomycin and carbapenem, along with glycolipid biosurfactants such as rubiwettins and rhamnolipids, are also expected to control plant diseases[106]. These compounds have not been extensively studied for controlling plant diseases and pests, suggesting that future research should explore their potential to mitigate the negative impacts of phytopathogens and protect plant health.
Serratia is widely recognized for its entomopathogenic properties, with various studies revealing that plant-associated bacteria, including endophytes and epiphytes, also exhibit entomopathogenic activity against insect pests. For instance, the endophytic S. marcescens SMR, isolated from summer squash flowers displayed entomopathogenic activity against Helicoverpa armigera larvae[107]. Another study highlighted the entomopathogenic effect of rhizospheric S. marcescens NPKC3_2_21 on Spodoptera litura, providing valuable insights into the potential of S. marcescens for pest control in plants[108]. These bacteria can persist in insect bodies by inhabiting the gut and hemolymph, overcoming host defenses, and rapidly proliferating throughout the gut and hemocoel. This process results in the reddening of the insect body due to the production of a red pigment called prodigiosin, ultimately leading to the insect's demise[107]. Research also suggests that S. marcescens functions as an insect pathogenic disease, taking residence in the insect's gut and hemolymph. The virulence of Serratia disease is associated with various enzymatic activities such as protease, lipase, phospholipase, and chitinase, which have been implicated in the degradation of the insect exoskeletons and gut epithelial tissue[109]. Notably, motility, demonstrated through swimming and swarming activities, has been identified as a significant factor contributing to Serratia's pathogenicity[110]. A pangenome analysis of the 73 S. marcescens's genomes indicates a preference for insects as hosts, associated with diverse enzymes such as hydrolases, isochorismatase, and N-acetyltransferase, the latter possibly exerting a neurotoxic effect[13]. Infections are believed to result from the interaction between insects and plant-associated bacteria, where insects either directly feed on plant materials or Serratia infects them through enzymes that hydrolyze and destroy the surface and periplasmic structure of insects[111−113]. Additionally, serralysin an alkaline metalloprotease (synthesized with prtA1 to prtA4), produced by Serratia spp., acts as an insecticidal toxin for reducing insect pests, offering potential applications in insect control[114,115]. The pathogenesis of serralysin producer S. marcescens involves inhibiting wound healing, leading to a substantial loss of hemolymph from silkworm larvae[116]. Red-pigment prodigiosin has demonstrated insecticidal activity in many studies[117−119]. It induces a drop in pH in the insect's midgut, leading to reduced nutrient uptake and ultimately resulting in the insect's death[120]. Furthermore, prodigiosin has been reported to inhibit insect immune system enzymes such as protease phosphatase, acid phosphatase, and acetylcholine esterase when exposed to insects[121]. However, a separate study suggests that prodigiosin may not be an essential virulence factor in entomopathogenic Serratia[122]. Regarding compounds produced by Serratia spp. that induce the plant immune system to respond to insect pests, limited information is available. Therefore, further studies are necessary to gain insights into the beneficial role of Serratia in triggering induced systemic resistance in plants.
-
To date, several examples illustrate the beneficial effects of using Serratia on plant growth (Table 3). For instance, a study on Serratia marcescens demonstrated that rice plants inoculated with S. marcescens IRBG500 exhibited a significant increase in both root length and root dry weight compared to uninoculated control plants. Specifically, the root length and dry root weight of plants treated with IRBG500 were 1.32 and 1.39 times higher, respectively, than those of the control plants[2]. Furthermore, S. marcescens OK482790, when applied to soil, effectively promoted wheat growth. Soil inoculation significantly enhanced shoot and root lengths by 30.5% and 109.07%, respectively, over control plants. Additionally, soil inoculation led to the most significant increases in plant biomass, with fresh and dry shoot weights increasing by 295% and 172%, respectively, compared to controls[64]. In addition to its benefits under normal conditions, Serratia shows promise under abiotic stress. S. nematodiphila EU-PW75 enhanced barley growth under low-temperature conditions, effectively improving growth and physiological parameters[123]. Similarly, S. proteamaculans M35, as a wheat rhizospheric inhabitant, increased wheat height, root length, and grain yield under various salinity stress conditions compared to controls[77]. Furthermore, plants grown under low-nutrient conditions, including petunia, impatiens, and pansy, showed significant biomass increases of 24%, 41%, and 51%, respectively, when treated with S. plymuthica MBSA-MJ1[124]. Serratia species also demonstrated the ability to reduce soil contaminants and stimulate plant growth. For example, Serratia sp. Wed4 increased the dry weight of barley by 18.9%–33.9% and reduced pyrene residues by 46.4% in shoots, and 32.1% in roots[58]. Additionally, Serratia CP-13 showed potential in enhancing cadmium (Cd) detoxification in maize crops, promoting plant biomass and seed germination under soil Cd stress ranging from 0 to 30 µM, suggesting its promise as a biofertilizer for future soil Cd bioremediation efforts[79]. Overall, Serratia is a beneficial species for stimulating plant growth under both normal and abiotic stress conditions, promising a potential approach in agricultural applications.
Table 3. Application of Serratia species for enhancing plant growth and plant protection.
No. Strains Hosts Applications Ref. 1 S. marcescens IRBG500 Rice (Oryza sativa L.) Improving plant biomass [2] 2 S. marcescens OK482790 Wheat (Triticum aestivum L.) Improving plant biomass [64] 3 S. nematodiphila EU-PW75 Barley (Hordeum vulgare L.) Reducing plant abiotic stress
Improving plant biomass[123] 4 S. proteamaculans M35 Wheat (Triticum aestivum L.) Reducing plant abiotic stress
Improving plant biomass[77] 5 S. plymuthica MBSA-MJ1 Petunia (Petunia × hybrida), impatiens (Impatiens walleriana), and pansy
(Viola × wittrockiana)Reducing plant abiotic stress
Improving plant biomass under[124] 6 Serratia sp. Wed4 Barley (Hordeum vulgare L.) Reducing plant abiotic stress
Improving plant biomass[58] 7 Serratia CP-13 Maize (Zea mays L.) Reducing plant abiotic stress
Improving plant biomass[79] 8 S. nematodiphila (code SN01) Rice (Oryza sativa L.) Biocontrol of Rice blast caused by
Magnaporthe oryzae Improving grain yield[127] 9 S. nematodiphila RGK Turmeric rhizome (Curcuma longa L.) Biocontrol of Pythium aphanidermatum
Improving plant biomass and bioactive compounds[26] 10 Serratia liquefaciens, S. plymuthica and S. rubidaea Oilseed rape (Brassica napus L.) Biocontrol of Verticillillm dahliae , Rhizoctonia solani and Sclerntinia sclerotiorum [8] 11 S.liquefaciens CL80 Dutch white cabbage
(Brassica oleracea L.)Biocontrol of B. cinerea [125] 12 S. plymuthica HRO-C48 (Fragaria × ananassa Duch.) Biocontrol of Verticillium dahliae and
Phytophthora cactorum[47] 13 Cucumber (Cucumis sativus L.), bean (Phaeseolus vulgaris L.),and tomato (Lycopersicon esculentum L.) Biocontrol of Pythium apahnidermatum and
Botrytis cinerea[37] 14 Oilseed rape (Brassica napus L.) Biocontrol of Verticillium wilt [48] 15 S. nematodiphila CT-78 Rice (Oryza sativa L.) Biocontrol of leaf blight disease [25] 17 S. marcescens Dolichos bean (Lablab purpureus L.) Biocontrol of Anthracnose disease caused by Colletotrichum lindemuthianum [132] 18 Serratia sp. G3 Wheat (Triticum aestivum L.) Biocontrol of Botrytis cinerea, Cryphonectria parasitica, Rhizoctonia cerealis and Valsa sordida [90] 20 S. marcescens NPKC3_2_2 Rice (Oryza sativa L.) Reducing the damages from rice stem borer attacks (Scirpophaga innotata) in rice crops [108] 21 S. marcescens HJ-01 The red palm weevil (RPW), Rhynchophorus ferrugineus (Olivier) Reducing the red palm weevil (RPW), Rhynchophorus ferrugineus (Olivier) larvae [111] 22 S. marcescens B2 Rice (Oryza sativa L.) Eliciting plant ISR against rice blast caused by Pyricularia oryzae [85] 23 S. liquefaciens MG1 Tomato (Lycopersicon esculentum L.) Inducing systemic resistance against Alternaria alternata infection [83] 24 S. marcescens Tomato (Lycopersicon esculentum L.) Inducing systemic resistance against against root-knot nematode Meloidogyne incognita [126] 25 S. marcescens 90-166 Tobacco (Nicotiana tabacum L.) Inducing systemic resistance against Pectobacterium carotovorum subsp. carotovorum and Pseudomonas syringae pv. tabaci and cucumber mosaic virus [39] 26 S. rhizosphaerae KUDC3025 Tobacco (Nicotiana tabacum L.) Inducing systemic resistance against soft-rot disease caused by P. carotovorum subsp. carotovorum SCC1 [17] 27 S. marcescens GPS5 Groundnut (Arachis hypogaea L.) Inducing systemic resistance against late leaf spot disease [130] 28 S. marcescens N4-5 (extract) Cucumber (Cucumis sativus L.) Control of cucumber damping-off caused by
Pythium ultimum[128] 29 S. marcescens SE1 and SE2 (extracts) Peanut (Arachis hypogaea L.) Control of yellow mold disease caused by A. flavus [129] 30 S. plymuthica AED38
(extract)Avocado (Persea americana L.) Control of root rot caused by by Phytophthora cinnamomi [139] 31 Serratia sp. ARP5.1 (extract) Avocado (Persea americana L.) Control of fruit
body rot (Colletotrichum gloeosporioides)
and root rot (Phytophthora Cinnamomi)[138] To control biotic stress, Serratia species are used for effective management of plant diseases and pests, with multiple studies highlighting their potential. In the 1990s, various species of the Serratia genus, including S. liquefaciens, S. plymuthica, and S. rubidaea, were isolated from the rhizosphere of oilseed rape and screened as potent antifungal agents against Verticillium wilt (caused by V. dahliae var. longisporum), post-emergence damping-off, and brown girdling root rot (caused by R. solani), showing promise as potential plant biocontrol agents[8]. Additionally, S. liquefaciens CL80 effectively mitigates post-harvest rot of Dutch white cabbage, reducing fungal growth from 32%−82% to 15%−25% of leaf area covered[125]. Greenhouse trials demonstrate that bacterial treatment reduces Verticillium wilt by 18.5% and Phytophthora root rot by 33.4%. In three consecutive field trials on commercial strawberry farms in Germany, pre-planting dipping in S. plymuthica suspension decreased Verticillium wilt by 0% to 37.7% (average 24.2%) and boosted yield by 156% to 394% (average 296%). Phytophthora root rot was reduced by 1.3% to 17.9% (average 9.6%), with a yield increase of 60% compared to untreated controls[47]. Moreover, S. plymuthica HRO-C48 defends cucumbers against Pythium aphanidermatum, lowering disease incidence from 93.3% to 44.4% and inducing systemic resistance to B. cinerea, reducing lesion areas threefold in beans and tomatoes[37]. Additionally, the utilization of S. marcescens NPKC3_2_2 has significantly reduced damage from rice stem borer attacks (Scirpophaga innotata) in rice crops, decreasing incidence from 75% to below 25% under field conditions[108]. Another study demonstrated that S. marcescens HJ-01 exhibits remarkable pathogenicity against the red palm weevil (RPW), Rhynchophorus ferrugineus (Olivier) larvae. At a concentration of 1.0 × 108 CFU/mL, the mortality rate of RPW reached 82.22%, with a half-lethal time (LT50) of 4.72 d[111]. Various applications of ISR-eliciting Serratia have shown promising results for large-scale use. Root treatment with S. marcescens B2 enhances resistance against rice blast caused by Pyricularia oryzae, reducing disease incidence to 59% in rice plants[85]. Inoculating tomato plants with AHL-producing S. liquefaciens MG1 triggers ISR, reducing necrotic cell death from A. alternata infection by over 70%[83]. The root-knot nematode Meloidogyne incognita is controlled by S. marcescens, enhancing plant fitness by 2.25−2.29 times, and reducing egg masses by 3.92−4.17 times and eggs by 7.23–8.45 times[126]. Applying S. marcescens 90−166 to plant roots enhances ISR against bacterial pathogens such as Pectobacterium carotovorum subsp. carotovorum and Pseudomonas syringae pv. tabaci, as well as cucumber mosaic virus[39]. Additionally, a novel species, S. rhizosphaerae KUDC3025, has been reported to suppress P. carotovorum subsp. carotovorum SCC1, the causal pathogen of soft-rot disease in tobacco, by priming the plant defense system[17]. These findings suggest that utilizing ISR-eliciting Serratia strains could be a promising approach in sustainable agriculture.
The versatile applications of Serratia in enhancing plant growth and protecting against diseases underscore its potential in sustainable agriculture. This review outlines various approaches researchers have used to harness Serratia for diverse agricultural purposes. For example, the direct application of S. marcescens cells to rice soil increases fresh biomass by 1.21−1.43 times compared to seed coating with a high bacterial density[64]. To combat bacterial leaf blight in rice, researchers have tested S. nematodiphila CT-78 through seed coating, foliar spraying, and soil drenching. Notably, foliar spraying 14 d before inoculation and seed coating proved most effective, reducing infected tillers by up to 83%, infected leaves by up to 88%, and disease severity by 4.5-fold, while also improving grain yield and quality[25]. Additionally, seed coating with one strain of S. nematodiphila provided blast control and grain yield comparable to microbial products applied as transplant dipping and post-transplanting foliar spray, as well as fungicide control[127]. Furthermore, the ethanol extract of S. marcescens N4-5, containing prodigiosin and serratamolides, effectively controls cucumber damping-off caused by Pythium ultimum. Seeds treated with a ¼ dilution of the N4-5 extract showed significantly greater plant stand compared to non-treated and ethanol-only controls at pathogen inoculum levels of 30 and 50 sporangia/cm³[128]. Recent research has focused on enhancing the production of antifungal compounds in Serratia by supplementing fungal biomass or chitin in culture broths, aiming to control fungal diseases in plants. For instance, seed treatment with S. marcescens strains SE1 and SE2 induced by fungal biomass effectively controlled yellow mold disease caused by A. flavus in peanut plants, reducing disease severity index from approximately 70% to 40%[129]. Additionally, chitin-supplemented foliar application of S. marcescens GPS5 significantly enhanced control of late leaf spot disease in groundnut by inducing plant defense-related enzymes, reducing lesion frequency by 64% compared to chitin-treated controls and by 67% compared to phosphate buffer-treated controls[130]. Research also indicates that combining PGP Serratia with other plant-associated bacteria, such as Microbacterium arborescens and Enterobacter sp., significantly enhances wheat growth parameters in pot and field trials compared to individual strain applications[131]. To reduce Anthracnose disease caused by Colletotrichum lindemuthianum, researchers have combined S. marcescens cells with Allium sativum extracts in both seed treatment and foliar spray approaches, achieving superior results compared to commercial fungicide[132]. In another innovative study, alginate beads containing S. marcescens cells and chitinolytic enzymes demonstrated promising results, achieving a 60% reduction in bean disease compared to a 10% reduction with free S. marcescens in greenhouse experiments[133]. Commercially, potent species exhibiting robust bioactive activities can be formulated into conventional products suitable for application in bio-agriculture. For instance, liquid or granular formulations generated from S. entomophila, following specific procedures, can be effectively utilized in biological pest and disease control[134]. Overall, this diversity in approaches underscores the vast potential of Serratia and its metabolites in agricultural applications.
Despite various studies highlighting the agricultural benefits of Serratia, it also presents potential risks to human and plant health. Numerous studies have documented that several Serratia species can cause diseases in both humans and plants. In unfavorable conditions, microbial inoculants may behave as parasites or pathogens, escaping their intended use, invading other ecosystems, disrupting native microbiomes and posing threats to human and plant health[135]. In scenarios involving opportunistic pathogenic Serratia, the absence of adequate control measures could lead to adverse consequences in treated lands and surrounding areas. Therefore, research has emphasized the importance of implementing stringent biosafety testing to mitigate these risks. Before the routine field application of bioinoculants, it is essential to establish robust regulations and a comprehensive biosafety assessment framework for plant growth-promoting bacteria (PGPB) strains, encompassing considerations for humans, animals, plants, and the environment[136,137]. Furthermore, the use of metabolites from Serratia spp. is emerging as a rapid alternative approach to curb the proliferation of opportunistic pathogens. For instance, crude extracts from the cell-free supernatant of S. marcescens SE1 and S. marcescens SE2 have effectively alleviated symptoms of yellow mold disease caused by Aspergillus flavus in peanut plants[129]. Similarly, extracts from Serratia sp. ARP5.1 have demonstrated significant efficacy in combating both root rot infections caused by Phytophthora cinnamomi and postharvest body rot infections caused by Colletotrichum gloeosporioides[138]. Additionally, siderophore-containing extracts from Serratia plymuthica AED38 have proven to be a potent strategy against avocado root rot induced by Phytophthora cinnamomi[139].
Thanks to advancements in genome editing techniques, we can now create non-pathogenic Serratia strains with robust plant growth-promoting (PGP) traits and strong biocontrol activities, paving the way for their commercialization. While various studies have explored mutant Serratia strains for their antibiotic production capabilities, research on mutants targeting Serratia's virulence factors has been limited. We propose further research on the agricultural application of these mutant Serratia strains.
-
Serratia species are found in diverse environments, including soil, water, plants, and animals, and have developed mechanisms that enable them to adapt to various lifestyles. These bacteria exhibit numerous plant growth-promoting traits and produce bactericidal, fungicidal, or insecticidal compounds. Many Serratia strains demonstrate beneficial roles by inducing systemic resistance (ISR) in plants, sparking significant interest in their potential applications for biological control and plant growth stimulation in crop cultivation.
Despite this interest, specific aspects of the interactions between plants and Serratia, such as the particular compounds produced by Serratia that trigger plant immune responses against insect pests, remain relatively unexplored. Further studies are essential to delve deeper into these interactions and unravel the intricacies of plant-Serratia relationships. By investigating these relationships more comprehensively, we can better understand the potential benefits and implications for utilizing Serratia species in agriculture and beyond.
While this review primarily focuses on plant-associated Serratia, various species residing in different habitats, such as insects, nematodes, soils, waters, or mammals, may also exhibit plant growth-promoting traits and the ability to protect plants against phytopathogens by producing antimicrobial and insecticidal compounds or by eliciting ISR. Therefore, we recommend considering non-plant-associated Serratia for use in bio-agriculture, as their positive effectiveness has been demonstrated in plants.
The application of live Serratia cells is the best way to exploit all their plant-growth-promoting and protective activities. To achieve this, it is critical to exclude pathogenic strains that are harmful to plants and animals, including humans. Identifying the genetic markers of pathogenic strains is essential in this regard. Alternatively, cell-free supernatants or their crude solvent extracts could be researched to develop biostimulation and bioprotection agents for sustainable agriculture.
-
The authors confirm contribution to the paper as follows: study conception and design: Trinh LL, Nguyen HH; data collection: Trinh LL, Nguyen HH; analysis and interpretation of results: Trinh LL, Nguyen HH; draft manuscript preparation: Trinh LL, Nguyen HH. All authors reviewed the results and approved the final version of the manuscript.
-
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
-
The authors declare that they have no conflict of interest.
- Copyright: © 2024 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Trinh LL, Nguyen HH. 2024. Role of plant-associated microbes in plant health and development: the case of the Serratia genus. Technology in Agronomy 4: e028 doi: 10.48130/tia-0024-0025
Role of plant-associated microbes in plant health and development: the case of the Serratia genus
- Received: 02 February 2024
- Revised: 04 July 2024
- Accepted: 24 July 2024
- Published online: 08 October 2024
Abstract: The genus Serratia, a member of the family Enterobacteriaceae, is found in diverse ecological environments. Recently, Serratia has emerged as a multifaceted contributor to plant growth promotion and defense against plant diseases and insect pests. This review examines the mechanisms by which Serratia spp. induce plant growth and alleviate both abiotic and biotic stresses. Their seamless integration within the plant ecosystem allows Serratia spp. to produce quorum-sensing molecules, N-acyl homoserine lactones (AHLs), facilitating colonization of plant tissues and capitalizing on nutrition from plant exudates. This intricate web of communication enables Serratia to produce phytohormones and break down essential nutrients from the soil for plant uptake. When confronted with ecological competitors, many Serratia strains showcase remarkable adaptability by producing a diverse array of hydrolytic enzymes and antibacterial, antifungal, or insecticidal compounds, effectively controlling harmful bacteria, fungi, and insect pests. Furthermore, beneficial Serratia strains also use induced systemic resistance (ISR) and tolerance (IST) to alleviate biotic and abiotic stresses, respectively. Various agricultural applications of Serratia include the direct use of bacterial cells for seed coating, foliar spraying, and soil inoculation, or the application of their bioactive compounds alone or in combination with other materials on various plant parts. These efforts aim to bolster plant health, curb diseases, and manage pest populations. Despite promising applications, there have been reports of opportunistic pathogenicity in plants and animals. Therefore, several safety approaches and the use of virulence factor mutant strains should be considered. The trend toward the application of Serratia in agriculture is expected to continue.